Abstract
Kv1.1 and Kv1.4 potassium channels have different pore region determinants that were found to affect their cell-surface levels positively and negatively [Zhu, Watanabe, Gomez and Thornhill (2001) J. Biol. Chem. 276, 39419–39427; Zhu, Watanabe, Gomez and Thornhill (2003) J. Biol. Chem. 278, 25558–25567; Zhu, Watanabe, Gomez and Thornhill (2003) Biochem. J. 375, 761–768]. In the present study, we focused on the deep pore region of Kv1 members to test whether a cell-surface trafficking code was dictated by two amino acids. Kv1 channels with a threonine/lysine amino acid pair in a non-contiguous pore region promoted high surface levels, whereas a serine/tyrosine amino acid pair inhibited high surface expression by inducing a high level of partial endoplasmic reticulum retention. Our work suggests that a possible positive trafficking amino acid pair coding here for the Kv1 subfamily is Thr/Lys>Thr/Val>Thr/Tyr>Thr/Arg∼Thr/His>Ser/Val>Ser/Tyr>Ser/Lys. The Kv1 trafficking code was not transferable to a Kv2 family member and thus it appears that it only governs surface levels in the context of its Kv1 native pore loop region and/or its S5 and S6 regions. All members of a given Kv2, Kv3 or Kv4 potassium channel subfamily have identical amino acids at similar positions in their deep pore regions (Thr/Tyr or Thr/Val), which suggests that any difference in surface levels among members is not dictated by these amino acids. Thus a major determinant for cell-surface trafficking of Kv1 potassium channels is an amino acid pair in their deep pore regions, whereas the cell-surface levels of a given Kv2, Kv3 or Kv4 subfamily member are probably not affected by these amino acids.
Keywords: amino acid pair, cell-surface expression, deep pore regions, endoplasmic reticulum, potassium channel, trafficking
Abbreviations: CHO, Chinese-hamster ovary; endo H, endoglycosidase H; ER, endoplasmic reticulum
INTRODUCTION
Potassium channels are expressed by essentially all cells and play critical roles in cell and animal physiology, such as repolarizing and shaping action potential waveforms in excitable tissue [1]. Voltage-gated potassium channels (Kv) comprise a large gene family and mutagenesis work has identified various amino acid determinants involved in channel function [2,3]. Most members of a Kv potassium channel subfamily can be expressed in cell systems as tetrahomomers or tetraheteromers and, in native tissue, they may be associated with beta subunits [2,3]. Ion channels that are not trafficked to the cell surface in appropriate numbers may cause human disorders [4].
Kv1 potassium channels are components of the brain dendrotoxin-binding protein [5] and individual members share high amino acid identity [6] but show differences in expression/function [2,3,7–12]. Previous work presented evidence that Kv1.1 and Kv1.4 pore loops between the S5 and S6 transmembrane domains have non-contiguous pore regions, one in the outer pore and two in the deep pore, that were involved in trans-Golgi glycosylation and cell-surface expression [10–12]. A pertinent amino acid substitution at these positions altered trans-Golgi glycosylation and surface expression by approx. 5–20-fold. In addition to pore region determinants, Kv1.4 has a cytoplasmic C-terminus determinant (VXXSL) that promoted high surface levels [13], but this determinant appeared to exert its effect only in the presence of a deep pore region determinant [14]. In contrast with those Kv1 subfamily members that have variable amino acid sequences in their outer pore regions and their deep pore regions, other Kv subfamilies (e.g. Kv2, Kv3 and Kv4) have different amino acids in their outer pore regions but identical amino acids in their deep pore regions (Figure 1). Thus amino acids in deep pore regions of Kv1 subfamily members may be major determinants for channel processing and cell-surface expression.
Figure 1. The pore-loop region amino acid sequences of some members of the Kv family.
(A) Kv1 members. The deep pore region is underlined and bracketed. Dashes indicate amino acid identity to Kv1.1. X1 and X2 denote amino acids that are different in the deep pore region in Kv1 members. (B) Kv2 members. The deep pore region is underlined and X1 and X2 amino acids are identical in the subfamily. Dashes indicate amino acid identity to Kv2.1. (C) Kv3 members. The deep pore region is underlined and X1 and X2 amino acids are identical in the subfamily. Dashes indicate amino acid identity to Kv3.1. (D) Kv4 members. The deep pore region is underlined and X1 and X2 amino acids are identical in the subfamily. Dashes indicate amino acid identity to Kv4.1.
Given the above observations, we sought to examine, in the present study, (i) whether a trafficking code is associated with the deep pore regions of Kv1 subfamily members and what are the possible mechanism(s) involved and (ii) whether any identified Kv1 trafficking code was transferable to other Kv potassium channel subfamily members, such as Kv2.1.
EXPERIMENTAL
Cell lines and transfections
CHO (Chinese-hamster ovary) pro5 cells and COS7 cells, obtained from A.T.C.C. (Rockville, Maryland), were maintained in Dulbecco's modified Eagle's medium with 0.35 mM proline or α-minimum essential medium with 10% (v/v) fetal bovine serum at 37 °C under 5% CO2. Rat brain Kv1.4, Kv1.2, Kv1.1 and Kv2.1 cDNAs were used for standard mutagenesis by PCR or replication, to produce different cDNAs in pcDNA3 [10]. cDNA integrity was confirmed by DNA sequencing. Cells were transiently transfected with 0.5 μg of Kv-containing plasmid per 35 mm dish and incubated for 24 h as described previously [10,11,14].
Cell-surface Kv protein estimation by surface biotinylation and immunoblotting
Cell-surface biotinylation on intact cells used the hydrazide-LC-biotin (Pierce, Tattenhall, Cheshire, U.K.) reagent that is specific for carbohydrates or for Kv2.1, which is not glycosylated, or Kv N-glycosylation mutants. The sulphosuccinimidyl-2-(biotinamide) ethyl-1,3-dithiopropionate (Pierce) reagent was used as we have described in [10,11,15]. An aliquot was taken from the solubilized cells, before streptavidin-bead precipitation, to estimate the total amount of cell Kv proteins by immunoblotting. The eluted biotinylated surface proteins from streptavidin beads were run on 9% (w/v) SDS gels and processed for immunoblotting with Kv1.4 (1:2000 monoclonal; Upstate Biotechnology, Lake Placid, NY, U.S.A.), Kv1.1 (1:500 polyclonal), Kv1.2 (1:2000; Upstate Biotechnology) or Kv2.1 (1:1000; Upstate Biotechnology) antibodies. After washing horseradish peroxidase-linked anti-specific secondary antibodies were added and the bound antibodies were visualized with enhanced chemiluminescence (ECL® detection kit; Amersham Biosciences) and a preflashed X-ray film (AR5; Kodak, Rochester, NY, U.S.A.). All visualized bands in a lane were used for quantification. Signals on immunoblots were quantified as described below. Non-transfected CHO or COS cells did not express endogenous Kv1.4, Kv1.2, Kv1.1 or Kv2.1 proteins by immunoblotting (results not shown). Immunoblots of whole cell lysate were probed with anti-actin antibodies (clone AC-40; Sigma) as a control for any difference in cell density between wells. A Microtex 8700 scanmaker (dynamic range of 0–3.5) was used to scan a film image and densitometry analysis was performed with the NIH Image 1.6 software with an internal and external standard [10,11]. Cell-surface protein levels of test Kv channels were expressed as values normalized to a control, which was taken as 100.0±S.E.M. Total cell protein levels of test Kv channels were also expressed as values normalized to a control, which was taken as 100.0±S.E.M. Relative surface expression levels were calculated by dividing the normalized surface protein by the normalized total protein to compare with a control value of 1.0. The ECL® detection system was linear over an approx. 1–20-fold range by an immunoblot assay on serially diluted total cell membranes from transfected cells (results not shown).
Whole cell Kv protein localization pattern by immunofluorescence microscopy
Transiently transfected COS cells on glass coverslips were incubated for 20 h after transfection and fixed/permeabilized to visualize the whole cell Kv1 protein localization patterns as we have described in detail previously [11,14]. COS7 cells were used because they have consistent ER (endoplasmic reticulum) and Golgi morphology from cell to cell [10]. Kv1.4, Kv1.1 or Kv1.2 antibodies were used at 1:1000 for an overnight incubation [10]. After washing with PBS plus 1% (v/v) Triton X-100, the cells were incubated in a secondary antibody conjugated with a fluorophore (Alexa dyes; Molecular Probes) for 1 h and then washed and mounted on glass coverslips. An Olympus BX50 microscope with a BX-FLA fluorescence attachment and an automatic camera setting were used for photographs. An important parameter in micrographs is the whole cell localization pattern and not the signal intensity. Non-transfected cells exhibited no fluorescence signal on incubation in primary and/or secondary antibodies (results not shown). We also used antibodies to resident proteins in the ER (BiP/Grp78 and calnexin from StressGen Biotechnologies, Victoria, BC, Canada) and the Golgi (GM130 and p230 from Transduction Laboratories, Lexington, KY, U.S.A.) to help determine the predominant localization of a Kv protein (results not shown).
RESULTS AND DISCUSSION
A possible Kv1 potassium channel surface trafficking code: two amino acids in the deep pore region dictate whether these proteins are expressed on the cell surface at high or low levels
Kv1.1 and Kv1.4 homomers are trafficked to the cell surface very differently. Kv1.1 showed high partial ER retention and low cell-surface levels, whereas Kv1.4 showed low ER retention and high cell-surface levels in three different cell lines [CHO, COS7 and neuronal-like CAD (Cath-a-differentiated) cells] ([10,11], see also [12]). Kv1.1 and Kv1.4 cell-surface levels were affected by their pore regions, which have only nine amino acids that are different in three non-contiguous regions (Figure 1A), because pore swapping or pertinent individual amino acid exchange affected the cell-surface expression of these channels [10–12]. In the deep pore region, there are only two amino acids that are different between Kv1.1 and Kv1.4 (Figure 1A). Kv1.1 has a serine residue in the X1 position and a tyrosine residue in the X2 position, whereas Kv1.4 has a threonine and a lysine residue in these positions respectively. We now address the following question: do the amino acids at these two positions predict whether a K+ channel in the Kv1 subfamily exhibits high or low cell-surface protein levels, and is there a surface-trafficking code in this subfamily?
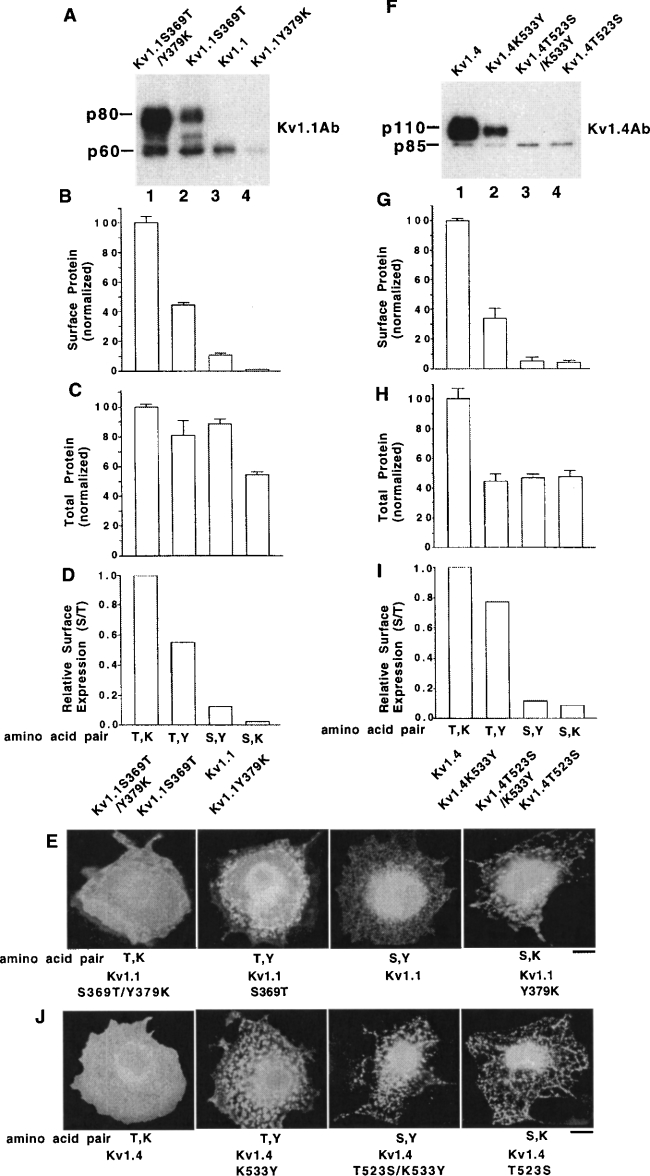
Kv1.1 was engineered with the above two amino acids exchanged as well as only a single amino acid exchanged and assayed for cell-surface expression levels in CHO cells by surface biotinylation and immunoblotting and by immunoblotting to estimate the total cell-protein levels. Surface protein levels (Figures 2A and 2B) and total protein levels (Figure 2C) for Kv1.1S369T/Y379K, Kv1.1S369T, Kv1.1 wild-type and Kv1.1Y379K were determined and normalized to Kv1.1S369T/Y379K control. Dividing the surface protein levels (Figure 2B) by total protein levels (Figure 2C) gave the relative surface expression levels (Figure 2D) versus Kv1.1S369T/Y379K. These relative surface expression levels can be considered as indicative of a trafficking code to the cell surface of other constructs versus Kv1.1S369T/Y379K. For relative surface expression levels, we found that Kv1.1S369T/Y379K (1.00)>Kv1.1S369T (0.58)>Kv1.1 wild-type (0.12)>Kv1.1Y379K (0.03) and that the amino acid pair order that promoted high relative surface expression, and thus the positive trafficking code order, was Thr/Lys>Thr/Tyr>Ser/Tyr wild-type>Ser/Lys (Figure 2D). These results suggest that these other Kv1.1 constructs may exhibit higher partial intracellular retention versus Kv1.1S369T/Y379K. Indeed, immunofluorescence localization patterns were also consistent with the above positive trafficking code order for amino acid pairs. Kv1.1S369T/Y379K had a pattern suggestive of high surface expression and low intracellular retention, and the other construct patterns were suggestive of differential lower surface expression and higher partial intracellular retention (Figure 2E) (see [11,14]). Thus Kv1.1S369T/Y379K had the highest relative surface expression level because it promoted high ER export (Figures 2D and 2E) and there was little effect on total protein levels for the other constructs, except Kv1.1Y379K which showed only approx. 50% total protein level compared with control (Figure 2C). As shown previously [10–12], wild-type Kv1.1 on the cell surface of transiently transfected CHO cells was mostly a high-mannose-type glycoprotein (p60, Figure 2A, lane 3), sensitive to endo H (endoglycosidase H) treatment (results not shown), and wild-type Kv1.4 on the cell surface was mostly a mature-type glycoprotein (p110, Figure 2F, lane 1) that was insensitive to endo H treatment (results not shown). The difference in the extent of mature glycosylation on Kv1.1 and Kv1.4 was governed by a pore determinant [10].
Figure 2. Kv1.1 and Kv1.4 cell-surface levels are governed by two amino acids in their deep pore regions: a Thr/Lys pair in Kv1.4 promoted high surface expression, whereas a Ser/Tyr pair of Kv1.1 inhibited high surface expression by inducing high partial ER retention.
Transiently transfected CHO cells were analysed for their cell-surface protein levels by surface biotinylation/immunoblotting and their total protein levels by immunoblotting. The Kv1 protein's whole cell immunofluorescence localization patterns in transiently transfected permeabilized COS7 cells are also shown. (A) Cell-surface protein profile of Kv1.1 constructs with different amino acid pairs in the deep pore by biotinylation/immunoblotting. (B) Group data for cell-surface expression. Normalized cell-surface protein levels by biotinylation/immunoblotting. The Kv1.1S369T/Y379/K construct value was taken as 100.0±S.E.M. (n=3) and the other construct values were normalized to it. (C) Group data for total protein expression of cells (immunoblots are not shown). The Kv1.1S369T/Y379/K construct value was taken as 100.0±S.E.M. (n=3) and the other construct values were normalized to it. (D) Relative surface expression levels, calculated by dividing the surface values in (B) by the total values in (C). (E) Immunofluorescence localization pattern of Kv1.1 constructs in COS7 cells. Scale bar, 10 μm. (F) Cell-surface protein profile of Kv1.4 constructs with different amino acid pairs in the deep pore by biotinylation/immunoblotting. (G) Group data for cell-surface protein expression. Normalized cell-surface protein levels by biotinylation/immunoblotting. The Kv1.4 construct value was taken as 100.0±S.E.M. (n=3) and the other construct values were normalized to it. (H) Group data for total protein levels of cells (immunoblots are not shown). The Kv1.4 construct value was taken as 100.0±S.E.M. (n=3) and the other construct values were normalized to it. (I) Relative surface expression levels, calculated by dividing the surface values in (G) by the total values in (H). (J) Immunofluorescence localization pattern of Kv1.4 constructs from COS7 cells. Scale bar, 10 μm. Note that, in immunofluorescence micrographs, the important parameter is the localization pattern and not the signal intensity.
Similar mutants for Kv1.4 were constructed and surface protein levels (Figures 2F and 2G) and total protein levels (Figure 2H) in CHO cells were estimated and normalized to Kv1.4 control. Dividing surface protein levels (Figure 2G) by total protein levels (Figure 2H) gave the relative surface expression levels (Figure 2I) versus Kv1.4 control. For relative surface expression levels, we found that Kv1.4 wild-type (1.00)>Kv1.4K533Y (0.77)>Kv1.4T523S/K533Y (0.10)>Kv1.4T523S (0.07) (Figure 2I). Thus the amino acid pair order that promoted the highest relative surface levels here, and thus a positive trafficking code order, was Thr/Lys wild-type>Thr/Tyr>Ser/Tyr>Ser/Lys and this was consistent with the order for Kv1.1. These results suggest that the other Kv1.4 constructs exhibited higher partial intracellular retention versus Kv1.4 and immunofluorescence localization patterns were also consistent with the above positive trafficking code order for amino acid pairs. Kv1.4 showed a immunofluorescence localization pattern that is characteristic of high cell-surface expression and low-ER retention, whereas the other Kv1.4 constructs exhibited a differential pattern more consistent with a higher partial ER retention and lower surface expression versus control (Figure 2J). Thus Kv1.4 control had the highest relative surface expression level (Figure 2I) when compared with other constructs because it promoted higher ER export, whereas the other constructs showed a higher intracellular retention. In addition, other Kv1.4 constructs had a decreased total protein level (∼40% of control level; Figure 2H), which is indicative of a decreased protein stability induced by the mutations.
The above results suggest that two amino acids in the X1 and X2 pore positions of Kv1.1 and Kv1. 4 affected trafficking and may be indicative of a trafficking code in the Kv1 subfamily. That is, a Thr/Lys pair (threonine residue in the X1 position and a positively charged amino acid, such as a lysine residue, in the X2 position) promoted the highest relative surface expression levels of the channel, whereas a Ser/Tyr or Ser/Lys pair here resulted in the lowest relative surface expression levels. It also appeared that a serine residue in the X1 region inhibited high relative surface expression regardless of the amino acid in the X2 region. The amino acids at the X1 and X2 positions in the deep pore regions of some Kv1 subfamily members (Figure 1A) suggest that, for relative surface homomeric protein levels, Kv1.4 (Thr/Lys)> Kv1.6 (Thr/Tyr)>Kv1.2 (Ser/Val)>Kv1.1 (Ser/Tyr).
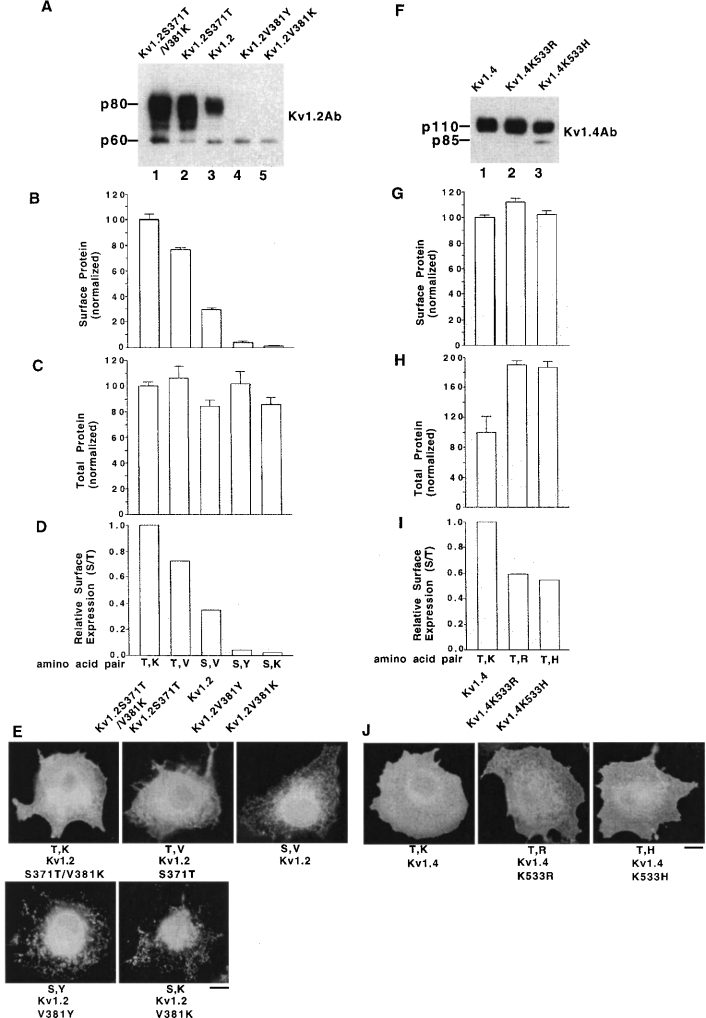
Kv1.2 was used next to test the above hypothesis further. Kv1.2 mutants were generated and analysed in CHO cells as above. Homomeric Kv1.2 was expressed on the cell surface at somewhat higher levels than Kv1.1 but at lower levels than Kv1.4 (results not shown), which is in agreement with the above Kv1 subfamily cell-surface level sequence. Surface protein levels (Figures 3A and 3B) and total protein levels (Figure 3C) in CHO cells were normalized to Kv1.2S371T/V381K control. Dividing surface protein levels (Figure 3B) by total protein levels (Figure 3C) gave relative surface expression levels (Figure 3D) versus Kv1.2S371T/V381K. For relative surface expression levels, we found that Kv1.2S371T/V381K (1.00)>Kv1.2S371T (0.75)> Kv1.2 wild-type (0.35)>Kv1.2V381Y (0.05)>Kv1.2V381K (0.03), and thus the amino acid pair order that promoted the highest relative surface protein levels, and thus the positive trafficking code order, was Thr/Lys>Thr/Val>Ser/Val wild-type>Ser/Tyr>Ser/Lys (Figure 3D), which was consistent with our hypothesis. These results suggest that these other Kv1.2 constructs may exhibit partial high intracellular retention versus Kv1.2S371T/V381K and the immunofluorescence localization patterns were also consistent with the above positive trafficking code order for amino acid pairs. Immunofluorescence localization pattern of Kv1.2 constructs in transfected cells showed that the high relative surface expression of Kv1.2S371T/V381K and Kv1.2S371T (Figure 3D) was mostly due to the release of high partial ER retention in wild-type Kv1.2 (Figure 3E). Thus Kv1.2S371T/V381K had the highest relative surface expression levels because it promoted high ER export (Figures 3D and 3E). There was only a small or no effect on total protein stability among the different constructs (Figure 3C).
Figure 3. Cell-surface levels of other Kv1 members are governed by similar amino acid pairs in their deep pore regions.
Transiently transfected CHO cells were analysed for their cell-surface protein levels and their total protein levels by biotinylation/immunoblotting and immunoblotting respectively. The Kv1 protein's whole cell immunofluorescence localization patterns in transiently transfected permeabilized COS7 cells are also shown. (A) Cell-surface protein profile of Kv1.2 constructs with different amino acid pairs in the deep pore by biotinylation/immunoblotting. (B) Group data for cell-surface protein levels. Normalized cell-surface protein levels by biotinylation and immunoblotting. The Kv1.2S371T/V381K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (C) Cell total protein levels by immunoblotting (immunoblots are not shown). The Kv1.2S371T/V381K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (D) Relative surface expression levels obtained by dividing the surface values in (B) by the total values in (C). (E) Immunofluorescence localization pattern of Kv1.2 constructs in COS7 cells. Scale bar, 10 μm. (F) Cell-surface protein profile of Kv1.4 constructs with different amino acid pairs in the deep pore by biotinylation/immunoblotting. Kv1.4K533R and Kv1.4K533H constructs have the same amino acid pair as Kv1.5 (Thr/Arg) and Kv1.3 (Thr/His) respectively (see Figure 1). (G) Group data for cell-surface protein levels from (F). The Kv1.4 construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (H) Group data for cell total protein levels (immunoblots are not shown). The Kv1.4 construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (I) Relative surface expression levels obtained by dividing the surface values in (G) by the total values in (H). (J) Immunofluorescence localization pattern of Kv1.4 constructs in COS7 cells. Scale bar, 10 μm. Note that, in immunofluorescence micrographs, the important parameter is the localization pattern and not the signal intensity.
Next, we mutated Kv1.4 K533 to one of the two amino acids arginine and histidine, which are found at equivalent positions in Kv1.5 and Kv1.3 respectively (Figure 1A). Both Kv1.5 and Kv1.3 have a threonine residue in their X1 position as does Kv1.4. Surface protein levels (Figures 3F and 3G) and total protein levels (Figure 3H) in CHO cells were normalized to Kv1.4 control. Dividing surface protein levels (Figure 3G) by total protein levels (Figure 3H) gave relative surface expression levels (Figure 3I) versus Kv1.4. Somewhat unexpectedly, for relative surface expression levels, we found that Kv1.4 (1.00)> Kv1.4K533R (0.58)∼Kv1.4K533H (0.54) and thus the pair order was Thr/Lys wild-type>Thr/Arg∼Thr/His (Figure 3I). These lower relative surface expression levels were indicative of higher partial intracellular retention for these two constructs but they had a somewhat similar immunofluorescence localization pattern as wild-type Kv1.4 (Figure 3J) and thus we could not readily detect an increased intracellular retention for them versus Kv1.4 by this assay. In addition, the two mutations to Kv1.4 increased total protein levels (Figure 3H), which indicated an increased protein stability. To test further the hypothesis, we also used Kv1.4K533V and Kv1.4T523S/K533V and found that, for relative surface expression levels, Kv1.4 (1.00)>Kv1.4K533V (0.83)>Kv1.4T523S/K533V (0.04) (immunoblots not shown) and thus the pair order here is Thr/Lys wild-type>Thr/Val>Ser/Val.
To summarize, our results for relative surface expression levels for Kv1.4, Kv1.2 and Kv1.1 with different mutations at the X1 and X2 pore regions suggest the following:
(i) Kv1.4: Thr/Lys wild-type>Thr/Val>Thr/Tyr>Thr/Arg∼Thr/His>Ser/Tyr>Ser/Lys>Ser/Val,
(ii) Kv1.2: Thr/Lys>Thr/Val>Ser/Val wild-type>Ser/Tyr>Ser/Lys and
(iii) Kv1.1: Thr/Lys>Thr/Tyr>Ser/Thr wild-type>Ser/Lys.
Although we have not tested every possible amino acid replacement in all constructs, we suggest that a possible positive trafficking code in the deep pore region for relative surface expression levels in this Kv1 subfamily is as follows: Thr/Lys>Thr/Val>Thr/Tyr>Thr/Arg∼Thr/His>Ser/Val>Ser/Tyr>Ser/Lys. The discrepancy found at the lower end of the rank order in a series (e.g. Ser/Tyr>Ser/Val in the Kv1.4 series, whereas Ser/Val>Ser/Tyr in the Kv1.2 series) may be due to effects of the other amino acids in the outer pore regions in each Kv1 channel. Thus, for the Kv1 subfamily, it is predicted that, for relative surface expression levels, Kv1.4 (Thr/Lys)>Kv1.6 (Thr/Tyr)>Kv1.5 (Thr/Arg)∼Kv1.3 (Thr/His)>Kv1.2 (Ser/Val)>Kv1.1 (Ser/Tyr).
The Kv1 potassium channel surface trafficking code was not transferable to a Kv2 subfamily member
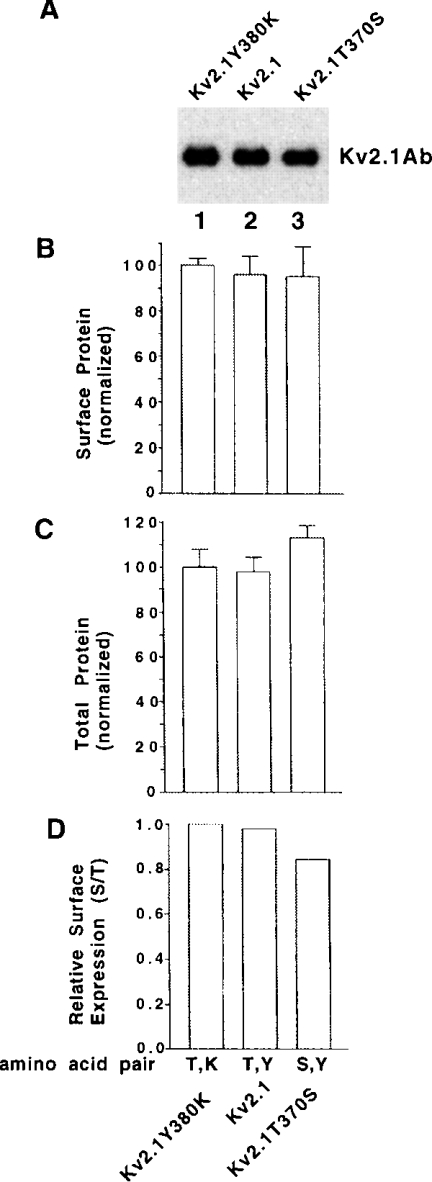
We next tested whether the Kv1 trafficking code was transferable to a Kv2 subfamily member. Kv2.1, which is not N-glycosylated [16], has a threonine residue in the X1 and a tyrosine in the X2 pore positions and Kv2.2 also has these amino acids (Figure 1B). Surface protein levels (Figures 4A and 4B) and total protein levels (Figure 4C) in CHO cells were normalized to Kv2.1Y380K control. Dividing surface protein levels (Figure 4B) by total protein levels (Figure 4C) gave relative surface expression levels (Figure 4D) versus Kv2.1Y380K. The relative surface expression levels, as well as the surface protein levels and total protein levels, were similar for all the constructs. Results here for Kv2.1 mutants suggest that the Kv1 trafficking code was not transferable to Kv2.1.
Figure 4. The proposed Kv1 trafficking code is not transferable to the Kv2.1 potassium channel.
Transiently transfected CHO cells were assayed for cell-surface protein levels as described in the legend to Figure 2. (A) Cell-surface protein profile of Kv2.1 constructs with different amino acid pairs in the deep pore by biotinylation/immunoblotting. (B) Group data for cell-surface expression. Normalized cell-surface protein levels by biotinylation/immunoblotting. The Kv2.1Y380K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (C) Total protein levels of cells by immunoblotting (immunoblots not shown). The Kv2.1Y380K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (D) Relative surface expression levels obtained by dividing surface values in (B) by total values in (C).
Kv1 members used in the present study were all N-glycosylated on their S1-S2 linkers in the ER. It is still not clear whether the effect of the amino acid pair on trafficking in the Kv1 family depends on the presence of a carbohydrate tree on the protein. Kv1.1N207Q and Kv1.4N354Q mutants that are not N-glycosylated but are expressed on the cell surface [15] were used as templates to construct other mutants to address the uncertainty above. For relative surface expression levels, Kv1.1N207Q/S367T/Y379K (1.00)>Kv1.1N207Q (0.23) and Kv1.4N354Q (1.00)>Kv1.4N354Q/T523S/K533Y (0.45) (immunoblots not shown). The relative surface protein expression levels for Kv1.1N207Q and Kv1.4N354Q were still affected by the amino acid pair tested, that is, Thr/Lys>Ser/Tyr was recorded for both conditions in the absence of glycosylation. Thus the trafficking code still exerted its effect in the absence of N-glycosylation.
The Kv1 channel surface trafficking code may not be transferable to other Kv channel subfamilies because of additional amino acid differences in their S5-pore loop-S6 regions
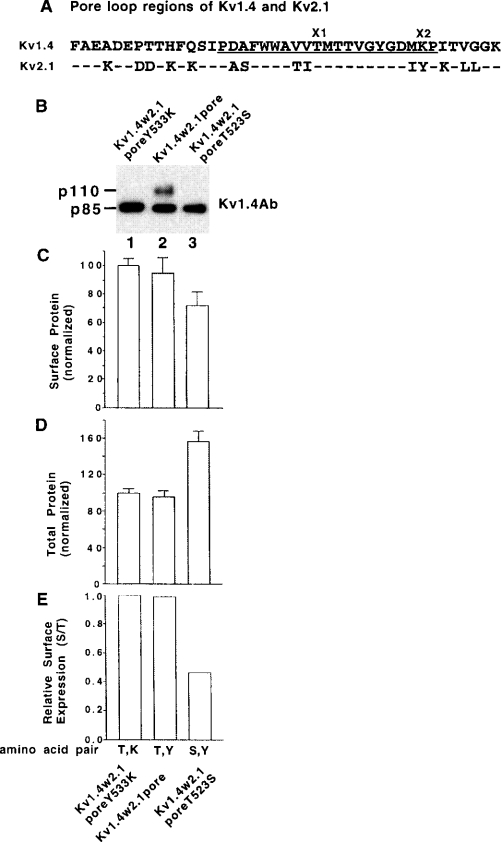
In KcsA and KvAP potassium channel crystal structures, the pore loop is surrounded by the S6 (inner helix) and the S5 (outer helix) [17,18]. Does the Kv1 trafficking code exert its effect only in the context of a complete Kv1 pore loop region or are S5 and S6 regions of Kv1 required? Kv1.4 and Kv2.1 pore loops have the same number of amino acids but there are 14 residues that are different (Figure 5A). To investigate the above question, a Kv1.4w2.1pore chimaera, which is Kv1.4 with the pore loop of Kv2.1 but without Kv2.1's S5 or S6 (Figure 5A), was generated and a number of mutations were introduced. Surface protein levels (Figures 5B and 5C) and total protein levels (Figure 5D) in CHO cells were normalized to Kv1.4w2.1poreY533K control. Dividing surface protein levels (Figure 5C) by total protein levels (Figure 5D) gave relative surface expression levels (Figure 5E) versus Kv1.4w2.1poreY533K. For relative surface expression levels, we found that Kv1.4w2.1poreY533K (1.00)∼Kv1.4w2.1pore (0.98)>Kv1.4w2.1poreT523S (0.43) and the amino acid pair order in the present study was Thr/Lys chimaera∼Thr/Tyr chimaera>Ser/Tyr chimaera (Figure 5E). The decreased relative surface expression level for Kv1.4w2.1poreT523S (Figure 5E), versus the other constructs, was consistent with a higher partial intracellular retention. In addition, the T523S (Thr523→Ser) mutation increased total protein levels (Figure 5D) and presumably protein stability also. Thus Kv1.4w2.1poreT523S did show a decreased relative surface expression, which is consistent with our Kv1 trafficking code. This finding suggested that an amino acid at X1 position in the chimaera now had an effect on trafficking, because it was in the environment of the Kv1 S5 and/or S6 region, which was required for at least a partial readout of the code in the chimaera. In contrast, Kv1.4w2.1poreY533K did not exhibit an increase in relative surface expression levels versus Kv1.4w2.1pore. Thus it appears that the X2 trafficking code position may require other Kv1 pore loop amino acids to exert an effect on this chimaera.
Figure 5. A complete readout of the proposed Kv1 trafficking code also requires other Kv1 S5, S6 and/or pore loop regions.
Transiently transfected CHO cells were assayed for cell-surface protein levels as described in the legend to Figure 2. (A) The amino acid sequence of the pore loop regions of Kv1.4 and Kv2.1. For Kv2.1, the dashes represent identical amino acids to Kv1.4. A Kv1.4w2.1pore chimaera, with the complete pore loop region of Kv2.1, was used to generate Y533K and T523S mutants. (B) Cell-surface protein profile of Kv1.4 chimaeras by biotinylation/immunoblotting. (C) Group data for cell-surface protein. Normalized cell-surface protein levels by biotinylation/immunoblotting. The Kv1.4w2.1poreY533K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (D) Cell total protein levels by immunoblotting (immunoblots not shown). The Kv1.4w2.1poreY533K construct value was taken as 100.0±S.E.M. and the other construct values were normalized to it (n=3). (E) Relative surface expression levels obtained by dividing the surface values in (C) by the total values in (D).
Members of the Kv2, Kv3 and Kv4 subfamilies have a threonine residue in the X1 position, whereas they have a different amino acid in the X2 position, but this amino acid is the same within a subfamily (Figures 1B–1D). A number of other Kv subfamilies (e.g. Kv5, Kv6.2, Kv6.3, Kv10.1 and Kv11.1) have a threonine residue or a serine residue in the X1 position of the deep pore, but most of these K+ channels do not express well, or at all, as homomers [3] and we will not consider them further in the present study. Our results suggest that the Kv1 trafficking code may not exert its effect on the Kv3 or Kv4 subfamily because of other amino acid differences in their pore loop regions and/or their S5 and S6 regions. Thus the Kv1 subfamily may have a unique trafficking code (see also [12]), but we suggest that the code would exert its effect on another Kv2, Kv3 or Kv4 subfamily member if a Kv1 pore-loop region and/or a Kv1 S5 and S6 region was also transferred on to it.
Conclusion and mechanisms of regulating cell-surface protein levels of potassium channels
We have shown that relative surface expression levels of homomeric Kv1 potassium channels were dictated by specific amino acids in their X1 and X2 pore loop positions and that an amino acid pair Thr/Lys here promoted high relative surface levels, whereas a Ser/Tyr or a Ser/Lys pair here promoted low relative surface levels due to high partial ER retention. We conclude that transferring the Kv1 trafficking code at positions X1 and X2 to Kv2.1 did not affect its relative surface expression because other Kv1 pore loop or S5 and S6 region amino acids were necessary for a complete readout of the code in this context.
Possible mechanisms involved in the readout of the Kv1 trafficking code include the following. We speculate that an unknown endogenous trafficking molecule(s) is associated with the ER in different cell lines (CHO, COS7 and CAD) and in brain cells and plays a role in the different trafficking programmes of Kv1 members. We further speculate that this ER-associated trafficking molecule(s) directly interacts with the X1 or/and X2 amino acid(s) and this interaction is (i) from the luminal side of the ER or (ii) from the cytoplasmic side of the ER (if the threonine or serine residue in the X1 position in the internal vestibule is only accessible from this side, then the cytoplasmic trafficking molecule would be an open-channel binder). An additional speculation is that there may be two trafficking molecules, one that interacts with the channel from the luminal side of the ER with position X2 and one that interacts from the cytoplasmic side with position X1. In any event, interaction of a trafficking molecule(s) either promotes partial high ER retention (a negative trafficking molecule) and/or promotes high ER export (a positive trafficking molecule). Other mechanisms are also possible if amino acids at X1 and X2 positions affect the conformation of other channel regions. It appears that this hypotheitcal trafficking molecule(s) may not play a role in the trafficking of Kv2, Kv3 or Kv4 subfamily members.
The electrical properties and signalling characteristics of excitable tissues are governed by the complement of ion channels they express on the cell surface [1] and thus mechanisms that regulate ion-channel surface levels are of considerable interest. Surface levels of ion channels can be controlled transcriptionally, post-transcriptionally and post-translationally. In post-translational control, the amino acid sequence or modifications to individual amino acids of ion channels may regulate their localization and numbers on the cell surface and thus modify the physiology of the cell. Understanding the amino acid determinants involved in traffic control in ion-channel biology would give insights into the cellular mechanism regulating the movement of these membrane proteins to the cell surface.
Acknowledgments
This work was supported by NIH grant no. NS29633 (to W.B.T.).
References
- 1.Hille B. 3rd edn. Sunderland, MA: Sinauer; 2001. Ionic Channels of Excitable Membranes. [Google Scholar]
- 2.Jan L. Y., Jan Y. N. Cloned potassium channels from eukaryotes and prokaryotes. Annu. Rev. Neurosci. 1997;20:91–123. doi: 10.1146/annurev.neuro.20.1.91. [DOI] [PubMed] [Google Scholar]
- 3.Coetzee W. A., Amarillo Y., Chiu J., Chow A., Lau D., McCormack T., Moreno H., Nadal M. S., Ozaita A., Pountney D., et al. Molecular diversity of K channels. Ann. N.Y. Acad. Sci. 1999;868:233–285. doi: 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 4.Ashcroft F. M. New York: Academic Press; 2000. Ion Channels and Disease. [Google Scholar]
- 5.Dolly J. O., Parcej D. N. Molecular properties of voltage-gated K channels. J. Bioenerg. Biomembr. 1996;28:231–253. doi: 10.1007/BF02110698. [DOI] [PubMed] [Google Scholar]
- 6.Stuhmer W., Ruppersberg J. P., Schruer K. H., Sakmann B., Stocker M., Giese K. P., Perschke A., Baumann A., Pongs O. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J. 1989;8:3235–3244. doi: 10.1002/j.1460-2075.1989.tb08483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trimmer J. S., Rhodes K. J. Localization of voltage-gated ion channels in mammalian brain. Annu. Rev. Physiol. 2004;66:477–519. doi: 10.1146/annurev.physiol.66.032102.113328. [DOI] [PubMed] [Google Scholar]
- 8.Papazian D. M. Potassium channels: some assembly required. Neuron. 1999;23:7–10. doi: 10.1016/s0896-6273(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 9.Deutsch C. Potassium channel ontogeny. Annu. Rev. Physiol. 2002;64:19–46. doi: 10.1146/annurev.physiol.64.081501.155934. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J., Watanabe I., Gomez B., Thornhill W. B. Determinants involved in Kv1 potassium channel folding in the endoplasmic reticulum, glycosylation in the Golgi, and cell surface expression. J. Biol. Chem. 2001;276:39419–39427. doi: 10.1074/jbc.M107399200. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J., Watanabe I., Gomez B., Thornhill W. B. Heteromeric Kv1 potassium channel expression: amino acid determinants involved in processing and trafficking to the cell surface. J. Biol. Chem. 2003;278:25558–25567. doi: 10.1074/jbc.M207984200. [DOI] [PubMed] [Google Scholar]
- 12.Manganas L. N., Wang Q., Scannevin R. H., Antonucci D. E., Rhodes K. J., Trimmer J. S. Identification of a trafficking determinant localized to the Kv1 potassium channel pore. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14055–14059. doi: 10.1073/pnas.241403898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D., Takimoto K., Levitan E. S. Surface expression of Kv1 channels is governed by a C-terminal motif. J. Biol. Chem. 2000;275:11597–11602. doi: 10.1074/jbc.275.16.11597. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J., Watanabe I., Gomez B., Thornhill W. B. Trafficking of Kv1.4 potassium channels: interdependence of a pore region determinant and a cytoplasmic C-terminal VXXSL determinant in regulating cell surface trafficking. Biochem. J. 2003;375:761–768. doi: 10.1042/BJ20030885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe I., Zhu J., Recio-Pinto E., Thornhill W. B. Glycosylation affects the protein stability and trafficking of Kv1.4 but not Kv1.1 potassium channels: a pore region determinant dictates the effect of glycosylation on trafficking. J. Biol. Chem. 2004;279:8879–8885. doi: 10.1074/jbc.M309802200. [DOI] [PubMed] [Google Scholar]
- 16.Shi G., Trimmer J. S. Differential asparagine-linked glycosylation of voltage-gated K+ channels in mammalian brain and in transfected cells. J. Membr. Biol. 1999;168:265–273. doi: 10.1007/s002329900515. [DOI] [PubMed] [Google Scholar]
- 17.Doyle D. A., Cabral J. M., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. The structure of the potassium channel: molecular basis of K conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. X-ray structure of a voltage-dependent K channel. Nature (London) 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]