Abstract
Aims—To establish a robust method of extracting DNA from paraffin wax embedded bone marrow trephine (PBMT) biopsies for the amplification of relatively long polymerase chain reaction (PCR) products.
Method—Xylene and ethanol were used to remove paraffin wax from eight formalin fixed, EDTA decalcified PBMT biopsies and DNA extraction was performed using a Qiagen QIAamp tissue kit. The DNA samples were amplified using nine different PCR primers sets, including those used to detect chromosomal translocations t(11;14) and t(14;18), and clonal B cell populations. A t(11;14) PCR product of approximately 600 base pairs (bp) was sequenced using dye terminator cycle sequencing.
Results—All eight DNA samples extracted from PBMT biopsies were amplified successfully to generate DNA fragments up to 643 bp in length. Chromosomal translocations and immunoglobulin gene rearrangements were detected by PCR in some of the samples. Sequencing of the t(11;14) PCR product demonstrated the presence of chimaeric sequences, which included both bcl-1 and immunoglobulin heavy chain (IgH) gene sequences, consistent with the presence of this translocation.
Conclusions—This method enables PCR analyses of PBMT biopsies that were not previously possible, offering the prospect of improved accuracy of diagnosis and the monitoring of patients with bone marrow disease.
Keywords: bone marrow trephine, DNA extraction, polymerase chain reaction
The examination of histological and cytological preparations of bone marrow is an important part of the diagnosis and staging of lymphoid malignancy. Although bone marrow aspirates are easy to obtain and can demonstrate the presence of abnormal lymphoid cell populations, they provide little information on the micro-anatomical relations that contribute to the diagnosis of non-Hodgkin's lymphoma. In addition, the cell populations sampled might not be wholly representative of the bone marrow, because lymphoma cell aggregates might be strongly adherent to stromal cells and reactive populations. These limitations necessitate the examination of bone marrow trephine (BMT) biopsies.
Although histological examination of BMT biopsies might confirm the presence of non-Hodgkin's lymphoma, distinguishing morphologically between the various subcategories of low grade non-Hodgkin's lymphoma can be problematical. In particular, the distinction between mantle cell, lymphocytic, and follicle centre lymphomas can be difficult, especially if the peripheral blood is not involved or there are no accessible lymph nodes for examination.
New molecular genetic techniques have been established as diagnostic tools for haematological malignancies. The highly sensitive polymerase chain reaction (PCR) can be used to detect clonal B cell populations,1,2 clonal T cell populations,3–5 and chromosomal translocations including t(14;18),6 t(11;14),7 and t(9;22).8 The ability to detect clonal B and T cell populations and translocations is useful for the diagnosis, classification, prognosis, and monitoring of minimal residual disease in non-Hodgkin's lymphoma.
Paraffin wax embedded bone marrow trephine (PBMT) biopsies provide a potentially important source of archival DNA for the retrospective molecular analysis of bone marrow. A recent study compared PCR detection of immunoglobulin gene rearrangements with morphological analysis and found the level of detection of immunoglobulin gene rearrangements in bone marrow aspirates to be much lower than expected.9 This was thought to be the result of dilution of the bone marrow cells with peripheral blood, sampling errors, or both. Therefore, the authors suggested that PBMT biopsies might provide a better source of DNA than bone marrow aspirates for the detection of immunoglobulin gene rearrangements.
Successful extraction of high molecular weight DNA from frozen bone marrow trephines has been described.10,11 However, most BMT biopsies taken for diagnosis are fixed, decalcified, and embedded in paraffin wax. The only previously published method of DNA extraction from PBMT biopsies is complex and time consuming.12 Only six of the 10 DNA samples extracted by these authors could be amplified to generate a PCR product of 294 base pairs (bp). The inability to amplify a relatively short PCR product from the remaining four DNA samples suggested that the extracted DNA was degraded, or that the samples contained PCR inhibitors.
The aim of our study was to establish a simpler method of extraction, suitable for routine use in a diagnostic molecular genetics laboratory. Ideally, this should yield DNA of a consistent quality that can be amplified to produce longer PCR products, including the 600 bp t(11;14) MTCA PCR product.
Materials and methods
BMT PROCESSING
BMT biopsies were placed in 10% neutral buffered formalin overnight for fixation. The fixative was then replaced by approximately 20 ml of a saturated EDTA solution to decalcify the biopsy for 48 hours with agitation. Samples requiring urgent processing were placed in a 37°C incubator. After decalcification, BMT biopsies were removed from EDTA and processed using a Tissue-Tek VIP tissue processor (Miles Incorporated, Elkart, Indiana, USA). On completion of processing, BMT biopsies were embedded in paraffin wax.
PATIENTS
PBMT blocks were obtained from five patients: four patients with lymphoma and one with anaemia and no known haematological malignancy (table 1 ▶). DNA extraction from blocks 2, 4, and 5 was performed in duplicate.
Table 1.
Paraffin wax embedded bone marrow trephine blocks used for the extraction of DNA
| PCR results using PBMT DNA samples | ||||||||
| IgH clonality | ||||||||
| Patient | Block | DNA | Histology | t(11;14) | t(14;18) | FR2 | FR3 | PCR results using DNA extracted from other tissues |
| 1 | 1 | 1 | N | Negative | Negative | Polyclonal | Polyclonal | No other tissues tested |
| 2 | 2 | 2A,2B | FCL | Negative | Negative | Polyclonal | Polyclonal | t(14;18) not detected in blood or bone marrow aspirate |
| Polyclonal B cell profile generated from blood sample | ||||||||
| 3 | 3 | 3 | FCL | Negative | Positive | Oligoclonal (4 dominant bands) | Polyclonal | t(14;18) detected in biopsy from small intestine |
| 4 | 4 | 4A,4B | MCL | Positive | Negative | Clonal | Clonal | t(11;14) detected in lymph node and peripheral blood |
| 5 | 5 | 5A,5B | MCL | Positive | Negative | Clonal | Clonal | t(11;14) and clonal B cell population detected in lymph node |
FCL, follicle centre lymphoma; MCL, mantle cell lymphoma; N, normal; PBMT, paraffin wax embedded bone marrow trephine.
REMOVAL OF PARAFFIN WAX FROM BMT SECTIONS
Ten PBMT sections of 10 μm were cut from each block and incubated in 1 ml of xylene at 55°C for 15 minutes. The samples were centrifuged at 20 000 ×g and the supernatant discarded. The residual pellet was washed twice using 1 ml of 1/1 xylene/ethanol and twice using 1 ml of absolute ethanol. Finally, the pellet was dried at 55°C for 10 minutes.
DNA EXTRACTION
DNA extraction was performed using a Qiagen QIAamp tissue kit (catalogue number 29304, Qiagen, Crawley, West Sussex, UK) according to the manufacturer's instructions. Briefly, 360 μl of ATL lysis buffer and 40 μl of proteinase K were added to each pellet, mixed thoroughly using a vortex mixer, and incubated at 55°C for between 72 and 96 hours, with intermittent vortex mixing. Any remaining tissue debris was pelleted by centrifugation and the supernatant used for the extraction of DNA using a Qiagen DNA extraction column. The DNA was eluted in a final elution volume of 400 μl. The yield was determined using a Pharmacia Biotech GeneQuant DNA spectrophotometer (Pharmacia, St Albans, Herts., UK) and the size of the extracted DNA was determined by agarose gel electrophoresis.
POLYMERASE CHAIN REACTIONS
The extracted DNA samples were first tested for their suitability as templates for PCR in three separate reactions. These included amplification of part of the gene encoding factor V13 and the 3′ and 5′ regions of exon 11 of the BRCA1 gene14 to yield products of 147, 482, and 643 bp, respectively. PBMT biopsy DNA samples were tested using either 2 μl of neat DNA or 2 μl of DNA diluted 1/10 with water in a reaction volume of 20 μl. The products were analysed on a 2% agarose gel and the concentration that gave the greatest yield was selected for all subsequent PCR amplifications.
PCR for detection of the t(11;14) (MTCA and MTCB) and t(14;18) (mbr and mcr) translocations was performed as described previously.1,6,7 These yield amplified products of approximately 600 bp t(11;14) MTCA, 400 bp t(14;18) mcr, and 200 bp t(11;14) MTCB and t(14;18) mbr.
B cell clonality PCR was performed using oligonucleotide primers to amplify two regions of the immunoglobulin heavy chain (IgH) gene sequence generating two sets of PCR products.1,2 One set, FR2A, ranges from 200 to 300 bp whereas the other, FR3A, measures 75 to 100 bp. Fluorescent R110-labelled dCTPs (Perkin Elmer, Warrington, Cheshire, UK; part number 402795) were used for internal labelling of B cell clonality PCR products, which were subsequently analysed by electrophoresis on a 4% polyacrylamide denaturing gel using an ABI 377 DNA sequencer. Positive (clonal patient) and negative (water blank) controls were included in all PCR amplifications.
SEQUENCING
The MTCA PCR products were purified using QIAquick PCR purification columns and both strands were sequenced using MTCA (forward) and VLJH (reverse) primers and an additional internal forward primer (5′-CGGTTAGACTGTGATTAGC-3′). The sequencing reactions were performed using a Big Dye Terminator cycle sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, Cheshire, UK) according to the manufacturer's recommendations and analysed on an ABI PrismTM 377 DNA sequencer.
Results
The DNA yields from the PBMT biopsies ranged from 1.4 to 7.2 μg (mean, 4.3; SD, 2.0), which is sufficient for at least 35 amplification reactions when using 40 ng of DNA for each reaction. The purified DNA ranged from 5 to 21 kilobases in length (results not shown) and was stable for at least one year when stored at −30°C.
The 147 bp factor V PCR product was amplified from all eight DNA samples. It was necessary to dilute one of the samples (DNA 2A) 1/10 in water to obtain this product, suggesting the presence of PCR inhibitors in this sample. The other seven samples generated a product both when used neat and at a dilution of 1/10. The intensity of the bands produced from DNA used neat or diluted 1/10 in water varied only slightly, and subsequent PCR amplifications were carried out using the concentration that appeared to give the greatest yield.
The amplification of BRCA1 exon 11A is the longest PCR (643 bp) performed routinely in our laboratory. This product was amplified from seven of the eight DNA samples (fig 1 ▶). Sample 2A failed to amplify, but when this sample was diluted 1/20 in water a product was obtained (data not shown). All of the samples could be amplified to produce a 482 bp exon 11B BRCA1 product (fig 1 ▶).
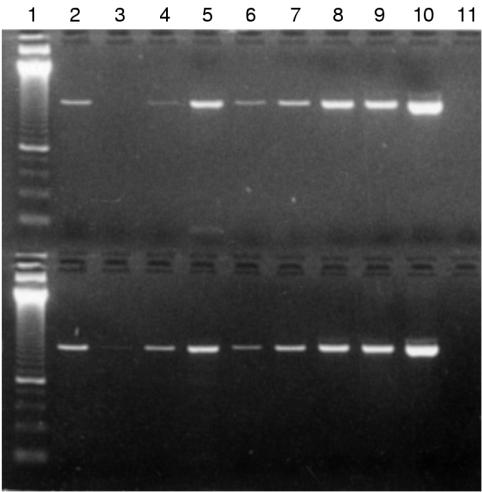
Figure 1.
Amplification of 643 bp BRCA1 exon 11A PCR product from DNA extracted from paraffin wax embedded bone marrow trephine samples. Top: BRCA1 exon 11A 643 bp PCR product; bottom: BRCA1 exon 11B 482 bp PCR product. Lane 1, 50 bp size standard; lane 2, DNA 1 1/10; lane 3, DNA 2A 1/10; lane 4, DNA 2B neat; lane 5, DNA 3 neat; lane 6, DNA 4A 1/10; lane 7, DNA 4B neat; lane 8, DNA 5A neat; lane 9, DNA 5B neat.
Having established that PCR products in excess of 600 bp could be amplified from the PBMT biopsy DNA samples, the samples were tested for the presence of chromosomal translocations and clonal B cell populations.
Patient 1 was anaemic but not known to have any haematological malignancy. Histological analysis of block 1 showed this BMT biopsy to be of normal cellularity with no lymphoid aggregates. No translocations or clonal B cell populations were detected using the DNA extracted from this block (DNA sample 1).
DNA samples from the BMT biopsy taken from patient 2 (who had been diagnosed as having follicle centre lymphoma) were not shown to contain any bcl-1 or bcl-2 chromosomal translocations or a B cell clonal population. The histology report on this BMT sample stated that the marrow was of normal cellularity, with one small lymphoid aggregate without neoplastic features, and concluded that there was no evidence of bone marrow involvement with low grade non-Hodgkin's lymphoma.
A biopsy from the small intestine of patient 3 had previously been found to yield an mbr t(14;18) PCR product. An identical mbr product was detected using DNA extracted from the PBMT biopsy (DNA sample 3; fig 2 ▶). This mbr product was not detected in DNA extracted from a bone marrow aspirate from this patient. Immunoglobulin heavy chain gene analysis with the FR2 primer set showed the presence of four dominant bands, whereas the FR3 reaction generated a polyclonal profile. Histologically, this BMT biopsy was found to be of normal cellularity, with some suspicious areas that appeared to be mononuclear, but no definite involvement by lymphoma could be seen.
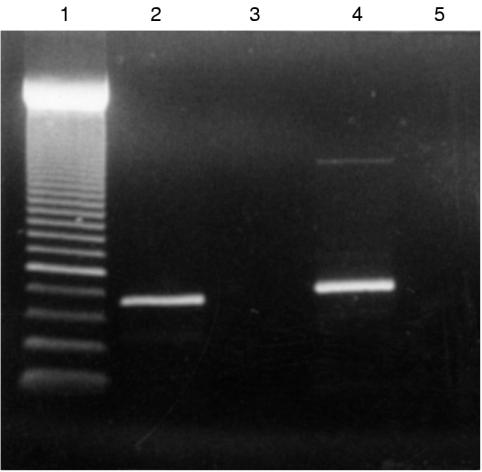
Figure 2.
Detection of t(14;18) mbr translocation using PCR. Lane 1, 50 bp size standard; lane 2, sample from a positive patient; lane 3, sample from a negative patient; lane 4, positive control; lane 5, negative control.
A t(11;14) translocation was present in a peripheral blood sample from patient 4 and in lymph node biopsies from patients 4 and 5. The 200 bp MTCB t(11;14) PCR product was detected in all four PBMT biopsy DNA samples from these two patients (DNA samples 4A, 4B, 5A, and 5B), although it was only possible to amplify the 600 bp MTCA t(11;14) PCR product from two of these samples (DNA samples 4B and 5B). Sequencing of the MTCA PCR products amplified from DNA sample 4B and the MTCA positive control demonstrated chimaeric sequences, which included both bcl-1 and IgH gene sequences, consistent with the presence of the t(11;14) MTCA translocation (fig 3 ▶). The bcl-1 sequence amplified from the PBMT biopsy was 34 bases shorter than that of the positive control, and different bases were seen at three positions within the shared part of the sequence. In addition, the random nucleotide and IgH gene sequences were different.
Figure 3.
Sequencing of the t(11;14) MTCA PCR product amplified from a paraffin wax embedded bone marrow trephine (PBMT) biopsy DNA sample 4. The t(11;14) MTCA sequences generated from the positive control and PBMT biopsy DNA sample 4B are shown. The bcl-1, random nucleotide, IgH, and primer sequences are displayed in normal text, underlined, italic and bold, respectively. x, difference between sample 4B and the positive control in the bcl-1 sequence.
Clonal B cell profiles were generated from both DNA samples extracted from blocks 4 and 5 (DNA samples 4A, 4B, 5A, and 5B) and from the lymph node of patient 4 (fig 4A ▶).
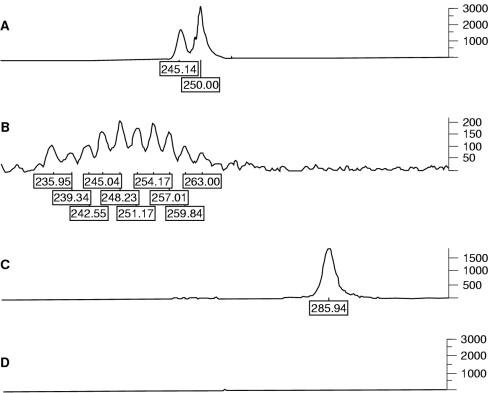
Figure 4.
Detection of B cell clonality (FR2A) using fluorescent PCR in conjunction with an ABI 377 DNA sequencer. (A) Paraffin wax embedded bone marrow trephine (PBMT) biopsy DNA 5A showing a clonal B cell profile; (B) polyclonal control; (C) clonal control; (D) negative control.
Histological examination showed that the BMT biopsy from patient 4 had increased cellularity, with both a focal and diffuse infiltration of small lymphoid cells, increased reticulin, and slightly reduced haematopoiesis. It was concluded that low grade non-Hodgkin's lymphoma was present in this sample. The BMT biopsy from patient 5 was hypercellular and heavily infiltrated by lymphoma, with moderate haematopoietic reserve.
Discussion
We report a simple technique for the extraction of DNA from PBMT biopsies that yields DNA suitable for the detection of clonal B cell populations and cells containing a variety of chromosomal translocations.
To our knowledge this is the first report of the successful amplification of PCR products in excess of 600 bp using DNA extracted from PBMT biopsies. A previous study in 1992 reported amplification of a shorter PCR product of 294 bp from six of 10 PBMT biopsy DNA samples.12 All eight DNA samples extracted from the five PBMT biopsies in our study were amplified successfully to generate DNA fragments up to 643 bp in length. Although the numbers of samples in these two trials do not permit statistical comparison, the ability to amplify a 643 bp product from eight PBMT biopsy DNA samples is a considerable achievement. The ability to amplify long products will allow PCR analyses of PBMT biopsies that would previously not have been possible, including the detection of cells bearing the t(11;14) translocation MTCA breakpoint product, which is approximately 600 bp in length.
The results of the translocation and clonality analyses were consistent with the patients' diagnoses and previous test results on other tissues. The patient with follicle centre lymphoma whose PBMT biopsy did not contain the t(14;18) translocation showed no evidence by histological analysis of bone marrow involvement and had tested negative for the translocation in previous blood and bone marrow aspirate samples. A t(14;18) PCR product identified in DNA extracted from the small intestine of patient 3 was also found in the PBMT biopsy DNA but not in the bone marrow aspirate DNA, highlighting the importance of using PBMT samples rather than bone marrow aspirates for the detection of molecular abnormalities in the bone marrow of patients with solid cell disease. Definite bone marrow involvement in this patient could only be confirmed using PCR analysis of the PBMT biopsy.
The t(11;14) translocation was detected in all four PBMT biopsy DNA samples from the two patients with mantle cell lymphoma. The 200 bp MTCB product was amplified from all four samples. It was possible to amplify the 600 bp MTCA product from both patients, although amplification was only seen in one of the two samples from each patient. The BRCA1 PCR results suggest that this method of DNA extraction is suitable for the amplification of long DNA templates, but there may be occasions when the number of cells present in the PBMT biopsy bearing the translocation is low, and therefore it might be difficult to extract sufficient intact, translocation bearing DNA to amplify long PCR products.
Sequence analysis of the MTCA PCR products from patient 4 (sample 4B) and the positive control identified a number of differences in the sequences of the two products. Therefore, it is unlikely that the positive result seen in this patient was the result of carryover contamination. This provides further evidence for the successful amplification of long PCR products (600 bp) from DNA extracted from PBMT biopsies using our method.
Clonal B cell populations were detected in five of the seven PBMT biopsy DNA samples from the four lymphoma patients. The two samples in which a clonal population could not be detected were both from patient 2, in whom there was no evidence of bone marrow involvement.
We attribute the success of the technique to the fact that the BMT biopsies were decalcified using EDTA, which (unlike formic acid) does not degrade DNA, and to the efficiency with which DNA is extracted using the Qiagen QIAamp tissue kit. However, other methods of DNA extraction were not investigated once the efficiency of this kit was established and hence the DNA extraction methodology might not be the crucial determinant. Formic acid was used to decalcify the BMT samples investigated in 1992,12 which might explain the inability of these investigators to amplify four of the 10 DNA samples. We have not attempted to extract DNA from PBMT biopsies that have been prepared using other decalcification techniques, and without a comparative study can only speculate that the use of EDTA has contributed to the success of this technique. EDTA decalcification is used routinely in this department and, despite the minor delay involved in tissue processing, has consistently facilitated both high quality immunohistochemical and tinctoral staining in PBMT biopsies.
There are occasions when bone marrow is the only accessible tissue for the diagnosis of non-Hodgkin's lymphoma. In this context, PBMT biopsies potentially provide a more informative source of bone marrow for examination than aspirate samples. The latter often contain a large number of peripheral blood cells9 and might not contain representative cells from adherent lymphomatous infiltrates. The distinction between subtypes of low grade B cell lymphoma can often be problematical when light microscopic examination alone is used.15
The ability to extract DNA suitable for molecular studies from PBMT biopsies provides an additional aid to the diagnosis and monitoring of patients with lymphomatous bone marrow involvement.
Acknowledgments
The authors thank the following members of staff at the Royal Devon and Exeter NHS Healthcare Trust for their invaluable assistance with this work: Dr R Lee, Dr M Pocock, the histology department, and all members of the molecular genetics laboratory. We are particularly grateful to Dr Lynas, Derriford combined laboratory, Plymouth NHS Healthcare Trust for her advice. This work was funded jointly by the Exeter Leukaemia Fund and Amgen UK Ltd. The support of the Royal Devon and Exeter NHS Healthcare Trust is appreciated.
References
- 1.Brisco MJ, Tan LW, Orsborn AW, et al. Development of a highly sensitive assay, based on the polymerase chain reaction, for rare B-lymphocyte clones in a polyclonal population. Br J Haematol 1990;75:163–7. [DOI] [PubMed] [Google Scholar]
- 2.Ramasamy I, Brisco M, Morley A. Improved PCR method for detecting monoclonal immunoglobulin heavy chain rearrangement in B cell neoplasms. J Clin Pathol 1992;45:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynas C, Howe D. Simple and reliable detection of T cell clones by PCR-LIS-SSCP analysis of TCRγ rearrangement. Mol Cell Probes 1998;12:41–8. [DOI] [PubMed] [Google Scholar]
- 4.Lynas C, Howe D. Additional TCRVB primers and minor method modifications improve detection of clonal T cell populations by RT–PCR. J Clin Pathol 1997;50:53–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynas C, Howe D, Copplestone JA, et al. A rapid and reliable PCR method for detecting clonal T cell populations. J Clin Pathol: Mol Pathol 1995;48:M101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gribben JG, Feedman AS, Woo SD, et al. All advanced stage non-Hodgkin's lymphomas with a polymerase chain reaction amplifiable breakpoint of bcl-2 have residual cells containing the bcl-2 rearrangement at evaluation and after treatment. Blood 1991;78:3275–80. [PubMed] [Google Scholar]
- 7.Williams ME, Swerdlow SH, Meeker TC. Chromosome t(11;14)(q13, q32) breakpoints in centrocytic lymphoma are highly localized at the bcl-1 major translocation cluster. Leukaemia 1993;7:1437–40. [PubMed] [Google Scholar]
- 8.Cross NCP, Junia VM, Feng L, et al. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR–ABL fusion mRNAs in haematological disorders. Leukemia 1994;8:186–9. [PubMed] [Google Scholar]
- 9.Coad JE, Olson DJ, Christensen DR, et al. Correlation of PCR detected clonal gene rearrangements with bone marrow morphology in patients with B-lineage lymphomas. Am J Surg Pathol 1997;12:1047–56. [DOI] [PubMed] [Google Scholar]
- 10.Fey MF, Theilkas L, Tobler A. Bone marrow trephine biopsies as a source of nucleic acids for molecular diagnosis of haematological neoplasms. Br J Haematol 1990;74:229–31. [DOI] [PubMed] [Google Scholar]
- 11.Hodges E, Stacey G, White D, et al. Histologic, immunotypic and genotypic analyses of bone marrow trephines from patients with non-Hodgkin's lymphoma. Leuk Res 1991;15:1117–24. [DOI] [PubMed] [Google Scholar]
- 12.Provan AB, Hodges E, Smith AG, et al. Use of paraffin wax embedded bone marrow trephine biopsy specimens as a source of archival DNA. J Clin Pathol 1992;45:763–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beauchamp NJ, Daly ME, Hampton KK, et al. High prevalence of a mutation in the factor V gene within the UK population: relationship to activated protein C resistance and familial thrombosis. Br J Haematol 1994;88:219–22. [DOI] [PubMed] [Google Scholar]
- 14.Gayther SA, Harrington P, Russell P, et al. Rapid detection of regionally clustered germ-line BRCA1 mutations by multiplex hetroduplex analysis. Am J Hum Genet 1996;58:451–6. [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid C, Isaacson PG. Bone marrow trephine biopsy in lymphoproliferative disease. J Clin Pathol 1992;45:745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]