Abstract
Aims—To investigate the disease causing event in patients with familial hypercholesterolaemia, carrying two mutations each, E256K in exon 6 and I402T in exon 9, of the gene encoding the low density lipoprotein (LDL) receptor. It was not known whether the mutations were positioned in cis or trans, or if they were each pathogenic separately or only when present together.
Methods—Polymerase chain reaction, denaturing gradient gel electrophoresis and sequencing were used to characterise the LDL receptor locus of the patients and family members. The different LDL receptor mutants, constructed in vitro by oligonucleotide directed mutagenesis, were expressed in LDL receptor deficient Chinese hamster ovary (CHOldlA7) cells, to determine the effects of the mutations on LDL receptor function.
Results—The two mutations were located on the same allele of the LDL receptor gene. All mutant constructs resulted in the production of a detectable protein in CHO cells. The cells expressing only the I402T mutation, or the combination of I402T and E256K mutations, were seriously affected in mediating uptake and degradation of LDL. Contrary to initial predictions, the cells expressing only the E256K mutation showed essentially the same binding, uptake, and degradation of 125I labelled LDL as cells transfected with normal LDL receptor cDNA. These results suggest that the pathogenic mutation in the patients heterozygous for the E256K/I402T allele is the I402T mutation, and that E256K alone is a rare sequence variation, which does not affect LDL receptor protein function. E256K was not detected either in DNA from a healthy population or in DNA from other hypercholesterolaemic patients studied.
Conclusions—Despite the information available on the structure–function relations between the LDL receptor and LDL receptor like proteins, predictions about the disease causing potential of a mutation are not reliable. These results suggest that the I402T mutation is pathogenic and that the substitution of E256K alone is a rare sequence variation, without a detectable phenotype modulating effect.
Keywords: low density lipoprotein receptor, expression, mutation, familial hypercholesterolaemia
Familial hypercholesterolaemia is one of the most prevalent monogenic traits, with a prevalence of about one in 500. It is inherited in a codominant fashion.1 The disease was described for the first time in the 1930s by Müller2 and by Thannhauser and Magendantz,3 who identified its clinical features—hypercholesterolaemia, atherosclerosis, xanthomas, and premature coronary artery disease. In 1976, Brown and Goldstein explained the raised low density lipoprotein (LDL) cholesterol levels in familial hypercholesterolaemia by a decreased function of the LDL receptor,4 a cell surface protein that mediates the specific uptake and degradation of LDL.
Since the characterisation of the LDL receptor gene in 19845 more than 300 gene defects have been detected.6–8 Many of these gene aberrations can be predicted a prima facie to be disease causing—for example, when a large deletion or insertion (causing a frameshift) or when a premature stop codon has been detected. Furthermore, by investigations of large families it is, in many cases, possible to demonstrate co-segregation of the disease and the gene defect. However, for a considerable number of the mutations found, it is an open question whether they are pathogenic or not. Such information is important not only for the individual patient but also for possible diagnostic and preventative action among family members. In an earlier study, a patient with a phenotype of heterozygous familial hypercholesterolaemia was found to carry two mutations, a G → A transition in exon 6, leading to the amino acid substitution E256K, and a T → C transition in exon 9, resulting in I402T.9 It could not be clarified whether the mutations were positioned in cis or trans or if they were pathogenic, either separately or when present together. In our study we have investigated additional members of the family and have expressed the LDL receptor mutants in Chinese hamster ovary (CHO) cells to determine the effects of the mutations on LDL receptor function.
Materials and methods
SUBJECTS
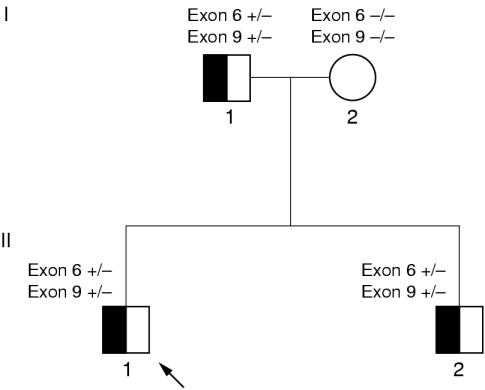
We describe a family of Swedish origin (fig 1 ▶) in which the father of the family had undergone coronary bypass surgery and suffered from a serious myocardial infarction at the age of 36 (table 1 ▶). His diagnosis of heterozygous familial hypercholesterolaemia was established clinically. His brother was also treated for hyperlipidaemia. The grandfather of the family died of myocardial infarction, at the age of 52, but it was not possible to establish retrospectively whether he had hyperlipidaemia or not. The index case and his brother both had heterozygous familial hypercholesterolaemia, as shown by raised cholesterol levels when they were 10–12 years old. Their mother does not show any signs of hyperlipidaemia. In a previous study,9 the father was screened for mutations in the gene encoding the LDL receptor and in the part of the apolipoprotein B gene encoding the receptor binding domain. The screening revealed two different mutations in the LDL receptor gene: E256K (G → A transition) in exon 6 and I402T (T → C transition) in exon 9. Southern blotting analysis of DNA digested with EcoRI and BglII indicated that no large deletions or other large rearrangements in the LDL receptor gene were present in the DNA from the father.
Figure 1.
Pedigree for the family with two low density lipoprotein receptor mutations on the same allele. Exon 6, heterozygous for the missense mutation E256K; exon 9, heterozygous for the missense mutation I402T; open symbol, patient with a normal phenotype; half filled symbols, patients were examined and had a heterozygous familial hypercholesterolaemia phenotype; diagonal arrow, the index case.
Table 1.
Lipid profile and clinical characteristics of the members of the family available for examination
| Patient | Sex | Birth (year) | Sample (year) | TC (mM) | LDLC (mM) | HDLC (mM) | TG (mM) | Notes |
| I:1 | Male | 1946 | 1991 | 6.5 | 4.6 | 0.79 | 2.4 | |
| II:1 | Male | 1974 | 1991 | 7.0 | 5.4 | 1.00 | 1.3 | |
| 1995* | 7.6 | 5.4 | 1.39 | 1.6 | Fat reduced diet | |||
| II:2 | Male | 1977 | 1988 | 6.6 | 1.10 | 0.74 | No treatment | |
| 1993* | 6.4 | 4.9 | 1.10 | 0.95 | No treatment | |||
| 1996* | 8.6 | 7.0 | 1.11 | 1.19 | Non-fasting, no treatment |
*Serum analyses.
HDLC, high density lipoprotein cholesterol; LDLC, low density lipoprotein cholesterol; TC, total cholesterol; TG, triglycerides.
DNA specimens from a healthy population (n = 343) and other patients with hypercholesterolaemia (n = 245 patients with serum cholesterol > 8 mmol/litre) were screened for the E256K mutation. The latter group was also screened for the I402T mutation.
BLOOD SAMPLES AND LIPID ANALYSES
Whole blood samples were collected into EDTA containing tubes. The samples were centrifuged, followed by DNA preparation from the leucocyte fraction.10 Enzymatic methods were used to determine total cholesterol (TC), high density lipoprotein cholesterol (HDLC) and triglyceride (TG).11 Low density lipoprotein cholesterol (LDLC) was calculated by means of Friedewald's formula.12
DNA ANALYSES
DNA samples from the index case, his brother, and parents were examined by denaturing gradient gel electrophoresis (DGGE), as described previously.9 In short, DNA fragments corresponding to exons 6 and 9 of the LDL receptor gene were amplified by the polymerase chain reaction (PCR).13 The amplified GC clamped fragments were analysed with DGGE on dedicated equipment (CBS Scientific, Del Mar, California, USA). The denaturating gradients used were 50–75% denaturant for exon 6 and 60–80% for exon 9 (100% denaturant: 7 M urea, 40% (vol/vol) deionised formamide). Electrophoresis was carried out for 22 hours at approximately 100 V (5 V/cm). The gel was stained in ethidium bromide (3 mg/ml) and the band pattern was then visualised in UV light.
EXPRESSION OF MUTANT LDL RECEPTOR GENES IN VITRO
Individual point mutations were introduced into the human LDL receptor cDNA by oligonucleotide directed mutagenesis of an EcoRI-SacI fragment subcloned into pSELECT, as described previously.14 The oligonucleotides were as follows (with the base changes in bold and underlined): for the mutant E256K, 5′-TGA CACTCTGCAAGGGACCCAAC and for the mutant I402T, 5′-ACCAGCCTCACCCCC AACCTG. The double mutant E256K/I402T was constructed by digestion of the pSELECT plasmids carrying each single mutation with StuI and EcoRI, followed by re-ligation of a mixture containing approximately equal proportions of each digest. Transformants that contained both mutations were identified by PCR and restriction enzyme digestion of appropriate fragments with MnlI and HphI. The EcoRI-SacI fragments were then subcloned into pLDLR4 digested with EcoRI, and SacI to produce the full length mutant LDL receptor cDNAs; the presence of the desired mutation in an otherwise normal sequence was verified by nucleotide sequencing of the entire coding region for the LDL receptor in each plasmid. From these, an XbaI-EcoICR I (Promega, Madison, Wisconsin, USA) fragment, including the coding region, was then subcloned into the mammalian expression vector pcDNA3.1(−) (Invitrogen, Carsbad, California, USA) digested with XbaI and EcoRV.
The normal and mutant plasmids were transfected into LDL receptor deficient CHOldlA7 cells15 by electroporation, and stable geneticin resistant transformants were selected and cultured as described by Patel and colleagues.16 Semi-quantitative immunoblotting of cell extracts with specific antibodies to the LDL receptor protein was used to determine the relative amounts of the LDL receptor protein in each cell line, as described previously.14,17 To confirm that the cells contained the expected LDL receptor cDNA construct, genomic DNA was isolated from the stably transformed cells and analysed by PCR and restriction enzyme digestion.
Measurement of LDL catabolism by the cells was essentially as described elsewhere.18 Cells were plated in 35 mm diameter dishes (1 × 105 cells/dish) and incubated in medium containing fetal calf serum for approximately 30 hours, then for 16–18 hours in medium containing lipoprotein deficient serum (10% vol/vol) and sterols (30 μg/ml of cholesterol and 6 μg/ml of 25(OH)-cholesterol). Saturable binding, uptake, and degradation of LDL were determined in triplicate four hour incubations as the difference in cell associated radioactivity (binding plus uptake) or soluble non-acid precipitable iodide radioactivity in the medium (degradation) between cells incubated with 125I labelled LDL in the presence or absence of an excess (1 mg/ml) of unlabelled LDL. The results are representative of two separate experiments.
Results
LIPID CONCENTRATIONS
Patients I:1, II:1, and II:2 showed moderate hypercholesterolaemia (table 1 ▶). Patient II:2 was not on a fat reduced diet or having medical treatment.
DNA ANALYSES
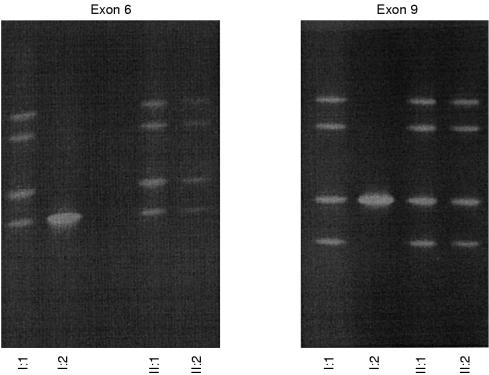
The DGGE patterns indicating mutations in exon 6 and in exon 9 in the DNA from the father were also found in DNA from both of his children (figs 1 and 2 ▶ ▶). The DGGE analyses of DNA from the mother showed no indications of sequence alterations in exon 6 or in exon 9, indicating a normal genotype regarding both exons of the LDL receptor gene. Thus, the two mutations were located on the same allele of the LDL receptor gene in the father and his two sons.
Figure 2.
Denaturing gradient gel electrophoresis (DGGE) patterns for exon 6 and exon 9 of the low density lipoprotein receptor gene from four of the family members. I:1, II:1, and II:2 show a band pattern indicating mutations in exon 6 and exon 9. The mother of the family (I:2) has normal DGGE patterns for the two exons.
PRODUCTION OF THE MUTANT LDL RECEPTOR PROTEINS IN VITRO
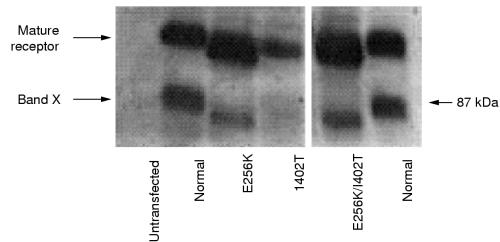
The E256K and I402T mutations were introduced individually into a full length cDNA for the human LDL receptor, and a cDNA was also constructed that contained both the E256K and I402T mutations. These LDL receptor cDNA constructs were expressed as stable transformants in CHOldlA7 cells that lack endogenous LDL receptor activity, and the proteins produced were analysed by immunoblotting with specific antibodies to the LDL receptor (fig 3 ▶). All of the cells expressing the mutant cDNA variants produced a receptor protein of the size of the normal mature protein as well as a band “X”, presumed to be a degradation product of the mature receptor, which has been seen previously in cells producing high amounts of the LDL receptor.14
Figure 3.
Immunoblotting with specific antibodies to the low density lipoprotein (LDL) receptor was carried out to assess heterologous LDL receptor protein production in a system with stably transformed, LDL receptor deficient CHOldlA7 cells. Under non-reducing conditions extracts of the cells (50 μg/lane) were fractionated by sodium dodecyl sulphate polyacrylamide gel electrophoresis and then transferred to nitrocellulose. The blots were sequentially incubated with the anti-LDL receptor antibody MAb10A2 and antimouse IgG conjugated with horseradish peroxidase. Bound antibodies were detected by chemiluminescence; the blots were exposured to x ray film for one minute. Lane 1, untransfected CHO cells; lane 2, normal LDL receptor; lane 3, E256K; lane 4, I402T; lane 5, I402T/E256K; lane 6, normal LDL receptor. The position of marker protein (BioRad, Hemel Hampstead, UK) is indicated by an arrow and its corresponding weight (kDa) is indicated to the right. Band “X” is presumed to be a degradation product of the mature receptor, which has been seen previously in cells synthesising large amounts of the LDL receptor.
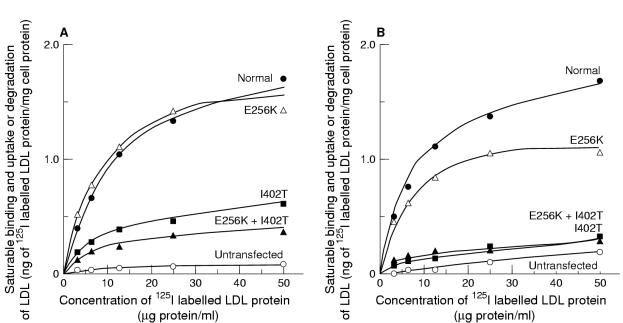
To compare the activities of the different mutant proteins, the relative amounts of mature receptor protein in each cell line were assessed by densitometric scanning of immunoblots similar to that shown in fig 3 ▶, but at lower exposures; different amounts of each cell extract were loaded on to the blots to ensure the linearity of the response (data not shown). LDL receptor function in cells expressing the mutant genes was assessed by measurement of the uptake and degradation of labelled LDL at 37°C by intact cells (fig 4 ▶). Although all the cells produced a detectable protein, cells expressing the I402T and I402T/E256K mutation were seriously affected in their ability to mediate uptake and degradation of LDL. On the other hand, cells expressing only the E256K mutation showed essentially the same binding and uptake of labelled LDL as cells transfected with the normal LDL receptor cDNA, although the relative amount of LDL degraded seemed slightly reduced. This probably has a technical rather than biological explanation, related to the fact that that the cell line obtained for the E256K mutation expressed the gene encoding the LDL receptor at a very high level. Under such conditions, all the receptors bind and internalise ligands; however, the degradation system becomes saturated, and thus, less degradation in relation to binding and uptake is seen.
Figure 4.
(A) Binding and uptake, and (B) degradation of 125I labelled low density lipoprotein (LDL) by cells stably transfected with the normal and mutant LDL receptor cDNAs. The cells were pre-incubated for 18 hours with medium containing lipoprotein deficient serum and sterols, then with 125I labelled LDL for four hours. The results shown are the mean of triplicate incubations and represent saturable cell associated or degraded LDL, determined as the difference between cells incubated with 125I labelled LDL in the presence or absence of an excess of unlabelled LDL. Non-specific cell associated or degraded LDL was always less than 5% of the total in cells expressing the normal LDL receptor cDNA. Similar results were obtained in two separate experiments with different preparations of LDL. Results are expressed as LDL protein/mg of cell protein, corrected for the amount of mature LDL receptor protein detected for each mg of cell protein by immunoblotting (relative to that in cells expressing the normal LDL receptor cDNA).
Even if the mutation does not have any detectable effect on LDL receptor function in vitro we cannot be sure that it does not have a subtle effect in vivo, which might interact with other genetic or environmental factors (such as apolipoprotein E genotype). However, taken together, these results suggest that the causal mutation in the patients heterozygous for the E256K/I402T allele is the I402T mutation, and that E256K alone is the result of a rare sequence variation that does not affect LDL receptor protein function.
Discussion
More than 300 gene defects have been detected in the gene encoding the LDL receptor.6–8 The gene is divided into 18 exons that encode five domains: the ligand binding domain, the epidermal growth factor (EGF) precursor homology domain, the domain with O-linked carbohydrates, the membrane spanning domain, and the cytoplasmatic part of the receptor.1 The ligand binding domain consists of seven repeats, each containing six cysteine residues, which form disulphide bonds within each repeat.1 Negatively charged amino acids are clustered at the C-terminal end of each repeat.
Both brothers investigated in our study (II:1 and II:2) had two different mutations, E256K in exon 6 and I402T in exon 9. Two or more sequence alterations occurring in one subject have been described previously.19–21 In our study, both mutations came from the father and thus reside on the same allele. E256K was not detected in DNA from a healthy population or in DNA from other patients with hypercholesterolaemia that we studied. The latter group was screened for the I402T mutation, with a negative result. At first glance, both mutations might be considered as pathogenic.
Substitution of lysine for the glutamate residue in position 256 (E256K) is the amino acid change that might be expected to lead to the strongest impact on protein function because it results in a change from a positively to a negatively charged residue in the ligand binding region. Furthermore, E256 is situated next to a cysteine residue involved in a disulphide bond, which is presumably crucial for the conformation of the seventh repeat of the first domain.22 Moreover, the E256K mutation was detected previously by Pereira et al in a small family of Cuban origin.23 The entire coding sequence and the promoter were sequenced, and the mutation co-segregated with hypercholesterolaemia in the family consisting of three siblings and their parents; all three individuals carrying the mutation suffered from hypercholesterolaemia and the two family members lacking the mutation had normal cholesterol values. The co-segregation in this small family suggests that the E256K mutation is pathogenic. It was somewhat surprising, therefore, that in our experiments the cells expressing the E256K mutation showed essentially the same binding, uptake, and degradation of labelled LDL as cells transfected with normal LDL receptor cDNA. Our results show that E256K alone has no impact on LDL receptor protein function, and should be considered as a rare sequence variation. In support of this, the glutamic acid at position 256 is not a strongly conserved residue.24 Although the LDL receptor 1 gene of Xenopus laevis and the rabbit LDL receptor gene encode glutamic acid as residue 256, the equivalent residue is an aspartic acid in hamster, house mouse, and rat LDL receptor genes, a leucine in the human very low density lipoprotein (VLDL) receptor gene, and a glutamine in the X laevis LDL receptor 2 gene.
The other mutation found in this family results in the substitution I402T at a residue located in the EGF precursor homology domain. In contrast to the E256K exchange, the isoleucine to threonine substitution would not be expected to alter protein function drastically. However, isoleucine is conserved at this residue in many species.24 Hamster, rabbit, rat, and X laevis all have isoleucine, and the house mouse has the structurally similar residue, leucine. Some of the closest relatives in the LDL receptor gene family, such as the human apolipoprotein E receptor 2 and the human VLDL receptor, have an isoleucine and the similar valine residue, respectively, in this position. Our functional results of the protein variants found in LDL receptor deficient CHOldlA7 cells showed clearly that the I402T variant alone, or together with the E256K substitution, were seriously defective at uptake and degradation of 125I labelled LDL.
Our study shows that, in spite of all the information available about structure–function relations between the LDL receptor and LDL receptor like proteins, one should be cautious in predicting the effect of mutations on function. Loux and colleagues25 described a mutation in exon 15, T705I, designated FH-Paris 9, which was initially observed in France, in a patient suspected to be compound heterozygous for familial hypercholesterolaemia. The other mutation could not be detected. The T705I mutation could not be detected in a control population and was therefore suspected to be pathogenic. Fibroblasts from the patient showed a low receptor activity compared with normal fibroblasts,6 but the mutation was not expressed in a cell system. Later Lombardi et al described a family with familial hypercholesterolaemia in which the T705I mutation did not co-segregate with hypercholesterolaemia.26 The T705I mutation was also found in a control normocholesterolaemic population, suggesting that it should be regarded as a sequence variation without functional importance.
Gudnason et al reported the A370T substitution,27 corresponding to the StuI polymorphism in exon 8 of the LDL receptor. No differences could be seen in the uptake or degradation of 125I labelled LDL between cells stably transfected with genes encoding the A370 and T370 LDL receptor variants. However, in the general Icelandic population, it was shown that men with the T370 allele had significantly higher TC, LDLC, and apolipoprotein B values than those with the more common A370 variant. Thus, some mutations might have a very subtle impact on the individual phenotype. Such a marginal functional impairment might be difficult to detect by conventional investigations, but might nevertheless influence plasma lipid concentrations at the population level.
Therefore, a recurrent problem is the interpretation of the clinical importance of sequence alterations. The most obvious criteria for pathogenicity are the following: (1) the mutation implies an early truncation, deleterious for the protein; (2) the gene alteration is shown to segregate with familial hypercholesterolaemia in large families; and (3) in vitro expression shows a defective function. Modern technology has made it easy to detect mutations in various genes. The future challenge lies in the issue of pathogenicity; that is, to decide when information on a certain genetic aberration is sufficient to be applied in clinical decision making.
Acknowledgments
G Nilsson provided excellent technical assistance. The authors are grateful to Dr D Russell for providing plasmid pLDLR4, containing a full length cDNA for the human low density lipoprotein receptor, and to Dr M Krieger for LDL receptor deficient CHOldlA7 cells. Many thanks to Dr H Hobbs and Dr D Russell for sharing valuable LDL receptor sequences, to Dr H Nissen, Odense, and Dr H Jensen, Aarhus, Denmark, for generous exchange of primer sequences, and to Dr S Humphries and Dr G Eggertsson who provided control DNA for the apolipoprotein B-3500 assay. We acknowledge Dr LS Lerman for providing the computer programs MELT 07/87. This work was supported by grants from the Physiographic Society of Lund; the medical faculty, University of Lund; the Lund Medical Society; Påhlsson's Foundation; and the Swedish Medical Research Council (project number 04966).
References
- 1.Goldstein JL, Brown MS. Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, et al, eds. The metabolic basis of inherited disease, 6th ed. New York: McGraw-Hill, 1989:1215–50.
- 2.Müller C. Xanthoma, hypercholesterolemia, angina pectoris. Acta Med Scand Suppl 1938;89:75–84. [Google Scholar]
- 3.Thannhauser SJ, Magendantz H. The different clinical groups of xanthomatous diseases; a clinical physiological study of 22 cases. Ann Intern Med 1938;11:1662–746. [Google Scholar]
- 4.Brown MS, Goldstein JL. Receptor-mediated control of cholesterol metabolism. Science 1976;191:150–4. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto T, Davis CG, Brown MS, et al. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 1984;39:27–38. [DOI] [PubMed] [Google Scholar]
- 6.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat 1992;1:445–66. [DOI] [PubMed] [Google Scholar]
- 7.Varret M, Rabés J-P, Collod-Béroud G, et al. Software and database for the analysis of mutations in the human LDL receptor gene. Nucleic Acids Res 1997;25:172–80 (website: http://www.umd.necker.fr:2004/4DMETHOD/Sortirhtml/%23%23977944469.18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The low density lipoprotein receptor (LDLR) gene in familial hypercholesterolemia. Database. Collation made by Heath K, Whittall R, Wilson D, et al in the Division of Cardiovascular Genetics, Department of Medicine, UCL Medical School, The Rayne Institute, London, UK (website: http://www.ucl.ac.uk/fh/maptext.html).
- 9.Ekström U, Abrahamson M, Sveger T, et al. An efficient screening procedure detecting six novel mutations in the LDL receptor gene in Swedish children with hypercholesterolemia. Hum Genet 1995;96:147–50. [DOI] [PubMed] [Google Scholar]
- 10.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sveger T, Fex G, Borgfors N. Hyperlipidemia in school children with family histories of premature coronary heart disease. Acta Paediatr Scand 1987;76:311–15. [DOI] [PubMed] [Google Scholar]
- 12.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma without the use of the preparative ultracentrifuge. Clin Chem 1972;18:499–502. [PubMed] [Google Scholar]
- 13.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via polymerase catalyzed chain reaction. Methods Enzymol 1987;155:335–50. [DOI] [PubMed] [Google Scholar]
- 14.Sun XM, Patel DD, Webb JC, et al. Familial hypercholesterolemia in China. Identification of mutations in the LDL-receptor gene that result in a receptor-negative phenotype. Arterioscler Thromb Vasc Biol 1994;14:85–94. [DOI] [PubMed] [Google Scholar]
- 15.Sege RD, Kozarsky KF, Krieger M. Characterization of a family of gamma-ray-induced CHO mutants demonstrates that the ldlA locus is diploid and encodes the low-density lipoprotein receptor. Mol Cell Biol 1986;6:3268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel DD, Forder RA, Soutar AK, et al. Synthesis and properties of the very-low-density-lipoprotein receptor and a comparison with the low-density-lipoprotein receptor. Biochem J 1997;324:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun XM, Patel DD, Knight BL, et al. Comparison of the genetic defect with LDL-receptor activity in cultured cells from patients with a clinical diagnosis of heterozygous familial hypercholesterolaemia. The familial hypercholesterolaemia regression study group. Arterioscler Thromb Vasc Biol 1997;17:3092–101. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein JL, Brown MS. Binding and degradation of low density lipoproteins by cultured human fibroblasts. Comparison of cells from a normal subject and from a patient with homozygous familial hypercholesterolemia. J Biol Chem 1974;249:5153–62. [PubMed] [Google Scholar]
- 19.Lombardi P, Kamerling SW, Defesche JC, et al. Identification of a double mutation in the low-density lipoprotein receptor gene causing familial hypercholesterolemia. Clin Genet 1996;50:525–6. [DOI] [PubMed] [Google Scholar]
- 20.Jensen HK, Jensen TG, Faergeman O, et al. Two mutations in the same low-density lipoprotein receptor allele act in synergy to reduce receptor function in heterozygous familial hypercholesterolemia. Hum Mutat 1997;9:437–44. [DOI] [PubMed] [Google Scholar]
- 21.Kotze MJ, de Villiers JN, Loubser O, et al. A double mutant LDL receptor allele in a Cypriot family with heterozygous familial hypercholesterolemia. Hum Genet 1997;100:101–3. [DOI] [PubMed] [Google Scholar]
- 22.Sudhof TC, Goldstein JL, Brown MS, et al. The LDL receptor gene: a mosaic of exons shared with different proteins. Science 1985;228:815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira E, Ferreira R, Hermelin B, et al. Recurrent and novel LDL receptor gene mutations causing heterozygous familial hypercholesterolemia in La Habana. Hum Genet 1995;96:319–22. [DOI] [PubMed] [Google Scholar]
- 24.National Center for Biotechnology Information, US National Library of Medicine, National Institute of Health, Bethesda, MD, USA. Database (website: http://www2.ncbi.nlm.nih.gov/genbank).
- 25.Loux N, Saint-Jore B, Collod G, et al. Screening for new mutations in the LDL receptor gene in seven French familial hypercholesterolemia families by the single strand conformation polymorphism method. Hum Mutat 1992;1:325–32. [DOI] [PubMed] [Google Scholar]
- 26.Lombardi P, Sijbrands EJ, Kamerling S, et al. The T705I mutation of the low density lipoprotein receptor gene (FH Paris-9) does not cause familial hypercholesterolemia. Hum Genet 1997; 99:106–7. [DOI] [PubMed] [Google Scholar]
- 27.Gudnason V, Patel D, Sun XM, et al. Effect of the StuI polymorphism in the LDL receptor gene (Ala 370 to Thr) on lipid levels in healthy individuals. Clin Genet 1995;47:68–74. [DOI] [PubMed] [Google Scholar]