Abstract
Background—Many lymph node abnormalities have been described in AIDS. These include opportunistic infections that sometimes result in spindle cell pseudotumours, Kaposi's sarcoma (KS), malignant lymphoma (Hodgkin's and nonHodgkin's), and florid reactive hyperplasia. Among these, reactive hyperplasia is the most common manifestation of AIDS related lymphadenopathy.
Aim—To examine whether human herpesvirus 8 (HHV-8), the aetiological agent of KS, can be localised in AIDS related lymphadenopathy and whether its appearance in such nodes is predictive of Kaposi's sarcoma development.
Methods—A series of human immunodeficiency virus (HIV) positive men (n = 21) with AIDS related lymphadenopathy who at the time of presentation had KS or subsequently developed KS (n = 5) were examined. The prevalence of HHV-8 was assessed in these patients using solution phase polymerase chain reaction (PCR), real time TaqMan quantitative PCR, and in cell amplification techniques (PCR in situ hybridisation (PCR-ISH) and labelled primer driven in cell amplification).
Results—Using standard solution phase PCR in a nested format, only two of the 21 patients with AIDS related lymphadenopathy were positive for HHV-8. The lymph node of one of these patients contained KS lesions. Three HHV-8 positive patients were identified using TaqMan PCR (the original two positive patients and one additional patient). All of the positive patients either subsequently developed KS (n = 2) or had KS at the time of diagnosis (n = 1). Two additional patients subsequently developed KS, but were negative for HHV-8 by solution phase PCR and TaqMan PCR. Using PCR-ISH, HHV-8 amplicons were identified in some lymphoid cells (in one patient) and in spindle cells of the KS lesion in another. The positive lymphoid cells were predominantly concentrated in B cell areas of the affected lymph nodes, confirming the B cell tropism exhibited by HHV-8.
Conclusions—The presence of HHV-8 in AIDS related lymphadenopathy is predictive of KS development and probably represents seeding of HHV-8 infected B cells from the peripheral blood. These findings support a role for HHV-8 in the pathobiology of KS.
Keywords: human herpesvirus 8, Kaposi's syndrome, AIDS related lymphadenopathy, TaqMan polymerase chain reaction
Many lymph node abnormalities have been described in AIDS. These include opportunistic infections that sometimes result in spindle cell pseudotumours, Kaposi's sarcoma (KS), malignant lymphoma (Hodgkin's and non-Hodgkin's), and florid reactive hyperplasia.1–3
Among these, reactive hyperplasia is the most common manifestation of AIDS related lymphadenopathy. It usually consists of collections of monocytoid B cells within the sinuses, scattered neutrophils, and features of dermatopathic lymphadenopathy. Several germinal centres show features of “follicle lysis”, which is characterised by the invagination of mantle zone lymphocytes into germinal centres. This results in the disruption of the germinal centres and gives rise to a “motheaten” appearance.4, 5 Within germinal centres, there is a predominance of follicular dendritic cells, which are assumed to be infected by the human immunodeficiency virus (HIV-1).6, 7
Occasionally, lymph nodes in HIV positive patients show advanced lymphocytic depletion with regressively transformed germinal centres.4, 6
A prominent vascular proliferation is sometimes seen in interfollicular areas, with the resulting picture acquiring a slight resemblance to Castleman's disease. In such cases, it is important to investigate and exclude the development of early KS.8, 9
Clinically, the term “persistent generalised lymphadenopathy” is given to the condition where there is enlargement of lymph nodes for at least three months duration, at two or more extra-inguinal sites, in an individual at risk of HIV.9
Here, we examine whether human herpesvirus 8 (HHV-8), the aetiological agent of KS, can be localised in AIDS related lymphadenopathy and whether its appearance in such nodes is predictive of KS development.
Materials and methods
A series of HIV positive men (n = 21) with AIDS related lymphadenopathy who at the time of presentation had KS or subsequently developed KS (n = 5) were examined. We have attempted to assess the prevalence of HHV-8 in this cohort and to identify which cell types are infected by HHV-8 in lymph nodes from these patients with AIDS related lymphadenopathy. All of the lymph nodes included in our study showed the changes described above for AIDS related lymphadenopathy. One lymph node contained intranodal KS lesions.
CULTURE OF THE BCP-1 CELL LINE FOR USE AS CONTROL MATERIAL
The BCP-1 cell line was derived from an HIV seronegative patient with body cavity based lymphoma. This is similar to a previously described HHV-8 infected, Epstein-Barr virus (EBV) negative cell line.10
The BCP-1 cell line was established in vitro from the malignant effusion of an index case,10 by culturing the lymphoma cells in RPMI 1640 supplemented with 20% autologous ascites, 50 μg/ml gentamycin, 0.05 mM 2-mercaptoethanol, 1 mM sodium pyruvate, and 2 mM L-glutamine at 37°C in a 5% CO2 incubator. On establishment and growth under these conditions, cells were gradually adapted to a medium containing 10% fetal calf serum by stepwise reduction of the amount of autologous ascitic fluid. Aliquots of BCP-1 were then taken and spotted on to Perkin Elmer in situ polymerase chain reaction (PCR) glass slides, for PCR in situ hybridisation (PCR-ISH) identification of HHV-8.
DETECTION OF HHV-8 BY SOLUTION PHASE PCR
Formalin fixed paraffin wax embedded tissue samples from patients with AIDS related lymphadenopathy were cut into sterile Eppendorf tubes, dewaxed, and suspended in 200 μl proteinase K digestion buffer (0.1 mg/ml proteinase K in 100 mM NaCl, 10 mM Tris, 25 mM EDTA, and 0.5% sodium dodecyl sulphate, pH 8.4) for three to five days at 37°C, followed by phenol/chloroform extraction and sodium acetate/ethanol precipitation. Samples were centrifuged, the supernatant was removed, and they were resuspended in HPLC (high performance liquid chromatography) water. The quality of extracted DNA was confirmed by β-globin gene amplification.
To detect HHV-8 by PCR, we used primers KS4: 5′-AGCACTCGCAGGGCAGTACG-3′ and KS5: 5′-GACTCTTCGCTGATGAAC- TGG-3′, derived from the putative minor capsid protein (homologue of open reading frame 26 (ORF-26) of Herpesvirus saimiri) and amplified for 25 cycles. To increase sensitivity and verify the specific nature of the amplicon obtained with these primers, a 2 μl aliquot of amplified product was reamplified (25 cycles) using internal primers KS1: 5′-AGCCGAAAGGATTCCACCAT-3′ and KS2: 5′-TCCGTGTTGTCTACGTCCAC-3′.
All positive cases were confirmed by amplification with non-overlapping primers derived from the major capsid protein of HHV-8 (ORF-25); outer primers: 5′-AGGCAACG TCAGATGTGAC-3′ and 5′-GAAATTACC CACGAGATCGC-3′; and inner primers: 5′-CATGGGAGTACATTGTCAGGACCTC-3′ and 5′-GGAATTATCTCGCAGGTTGCC-3′.
The detection sensitivity of single round HHV-8 PCR was 30 genome copies in 106 DNA molecules, and that of nested HHV-8 PCR was approximately five genomes in 106 DNA molecules.
PCR IN SITU HYBRIDISATION FOR DETECTION OF HHV-8 IN AIDS RELATED LYMPHADENOPATHY
Sections (5 μm thick) were cut on to silane coated in situ PCR glass slides (Perkin Elmer, Warrington, Cheshire, UK). Tissue sections were dewaxed with xylene for 15 minutes, and 100% ethanol for 10 minutes, followed by incubation in 0.02 N HCl for 10 minutes and digestion with 0.4 mg/ml proteinase K at 37°C for 15 minutes. Proteinase K was removed by washing the sections in cold phosphate buffered saline (PBS). Endogenous peroxidase activity was blocked with a 3% hydrogen peroxide solution in 0.1% sodium azide. PCR amplification was carried out using the GeneAmp in situ PCR 1000 system (Perkin Elmer). The following reaction conditions were used: 1 μM of each primer KS1 and KS2 for HHV-8, 200 μM dNTPs, 4–5 mM MgCl2, 1× PCR buffer II (Perkin Elmer), and 10 U Taq IS (Perkin Elmer) in a total reaction volume of 50 μl. Slides were assembled on the in situ PCR 1000 slide assembly unit (Perkin Elmer), which was held at 70°C to perform “hot start” PCR. An aliquot of 50 μl of the PCR mix was placed directly on the tissue section and the reaction mix covered with AmpliCover discs and clips (Perkin Elmer). The slides were transferred to the heating block and the following cycles applied: 94°C for 55 seconds (cycle 1), followed by 94°C for 50 seconds and 55°C for 90 seconds for a total of 30 cycles. Control sections from each sample were used for PCR-ISH, without Taq polymerase and/or without primers. After amplification, AmpliCover discs and clips were removed and slides were fixed in 100% ethanol for five minutes. For HHV-8 detection, a 5′-end biotin labelled 30 mer oligoprobe (5′-TGTTGGTGTACCACATC- TACTCCAAAATAT-3′; Oswell, Edinburgh, UK), which hybridises internally to the PCR amplicon, was applied at a concentration of 5–10 pmol/100 μl hybridisation buffer (5% dextran sulphate, 2× saline sodium citrate (SSC), and 10% formamide) to the centre of each section, which was then covered with glass coverslips.
Slides were denatured at 94°C for 15 minutes and then hybridised for 12 hours at 42°C. The coverslips were removed and the slides washed in SSC at different stringencies (4× SSC at 22°C for 15 minutes, 2× SSC at 30°C for 15 minutes, 2× SSC at 55°C for 15 minutes, or 1× SSC at 55°C for 15 minutes), followed by a final wash in TBT (100 mM NaCl, 40 mM Tris (pH 7.2), 3% bovine serum albumin (BSA), and 0.05% Triton X-100) for 10 minutes. Target specific signals were seen with washing stringencies of 2× SSC at 30°C and 2× SSC at 55°C. Only occasionally were signals seen at 1× SSC at 55°C, and fewer signals were seen at 0.1–0.2× SSC at this temperature. These conditions are appropriate for 30 mer oligoprobes. A further control included amplification with HHV-8 primers and the use of a human papilloma virus type 16 (HPV-16) 30 mer probe (5′-GACTCCTGAG GAGAAGTCTGCCGTTACTGC-3′).
For detection, the slides were incubated with avidin–horseradish peroxidase (1/100 in TBT with 5% skimmed dried milk). Unbound conjugate was removed by washing twice in Tris buffered saline (TBS) for five minutes each. The slides were then incubated with AEC development reagent (Histostain-SP kit; Zymed Lab Inc, San Francisco, California, USA) for 10–20 minutes, which yielded a red signal.
Parallel immunocytochemistry for B cell markers (CD20; Dako, Glostrup, Denmark) was carried out on all sections to confirm nodal and cellular localisation of the in cell PCR product. Combined immunocytochemistry and in cell PCR was not successful, as a result of shearing of cells and tissue sections during thermocyling.
The detection sensitivity of PCR in situ hybridisation for HHV-8 is one viral genome copy/cell.
LABELLED PRIMER DRIVEN IN SITU AMPLIFICATION OF HHV-8 IN BCP-1 CELLS
Aliquots of 103 cells were spotted on to silane coated in situ PCR glass slides (Perkin Elmer). Proteolysis was achieved using 0.01 mg/ml proteinase K at 37°C for 10 minutes. Proteinase K was removed by washing the sections in cold PBS. Endogenous peroxidase activity was blocked with a 3% hydrogen peroxide solution in 0.1% sodium azide.
Labelled primer driven amplification was carried out using the GeneAmp in situ PCR system 1000 (Perkin Elmer). The following reaction conditions were used: 1 μM of each primer KS1 (5′-biotin-AGCCGAAAGGATT CCACCAT-3′) and KS2 (5′-TCCGTGTT GTCTACGTCCAC-3′), 200 μM dNTPs, 4–5 mM MgCl2, 1× PCR buffer II, (Perkin Elmer), and 10 U Taq IS (Perkin Elmer) in a total reaction volume of 50 μl. Slides were assembled on the in situ PCR 1000 slide assembly unit (Perkin Elmer), which was held at 70°C to perform hot start PCR. An aliquot of 50 μl of PCR mix was placed directly on the tissue section and the reaction mix covered with AmpliCover discs and clips. The slides were transferred to the heating block and the following cycles applied: 94°C for 55 seconds (cycle 1), followed by 94°C for 50 seconds and 55°C for 90 seconds for a total of 30 cycles. Slides were maintained at 55°C after amplification.
AmpliCover discs and clips were removed and slides were washed in 2× SSC at 55°C for five minutes. Slides were then immersed in TBT for 10 minutes.
For detection of amplicon, the slides were incubated with avidin–horseradish peroxidase (1/100 in TBT with 5% skimmed dried milk). Unbound conjugate was removed by washing twice in TBS for five minutes each. The slides were then incubated with AEC development reagent (Histostain-SP kit; Zymed Lab Inc) for 10–20 minutes, which yielded a red signal.
Taqman pcr identification of hhv-8
All TaqMan PCRs were optimised first, as described previously.11 Duplicates were set up for each patient and a triplicate negative control “no DNA template” was included with each assay. Using primers KS1 and KS2 at 300 nM and including an internal TaqMan probe (FAM-CGCTATTCTGCAGCAGCT GTTGGTGTACCA-TAMRA) at 200 nM, 3−5 mM MgCl2, 0.023 U/ μl Taq DNA polymerase, and AmpErase UNG (0.01 U/μl), amplification was achieved using the following thermocycling protocol: 50°C for two minutes, 95°C for 10 minutes, followed by 94°C for one minute, 55°C for one minute, and 72°C for one minute for 40 cycles.
Product was detected by means of the LS 50B luminescence spectrophotometer (Perkin Elmer). In addition, five samples were subjected to quantitative TaqMan PCR or real time TaqMan PCR using the 7700 DNA sequence detector (Perkin Elmer).
The detection sensitivity of TaqMan PCR using a real time format (7700 DNA sequencing technology) is one viral genome copy in 107–108 DNA genomes; this is between 100 and 1000 times more sensitive than conventional solution phase PCR.
Results
Using standard solution phase PCR in a nested format, only two of the 21 samples from patients with AIDS related lymphadenopathy were positive for HHV-8. Interestingly, the lymph nodes of one of these patients contained KS lesions. Three HHV-8 positive patients were identified using TaqMan PCR (the original two positive patients and one additional patient). All of the positive patients either subsequently developed KS (n = 2) or had KS at the time of diagnosis (n = l). Two additional patients subsequently developed KS, but were negative for HHV-8 by solution phase PCR and TaqMan PCR.
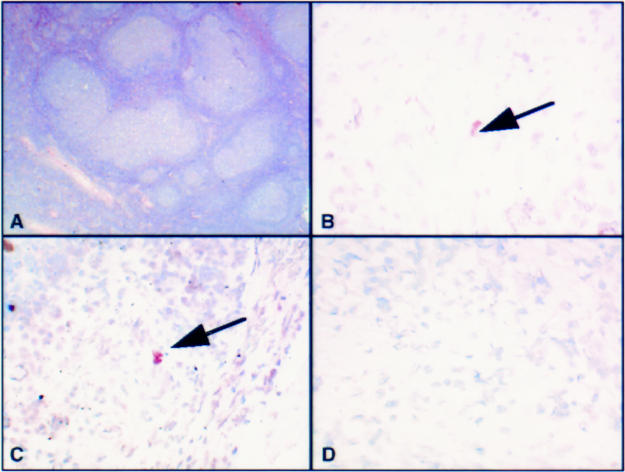
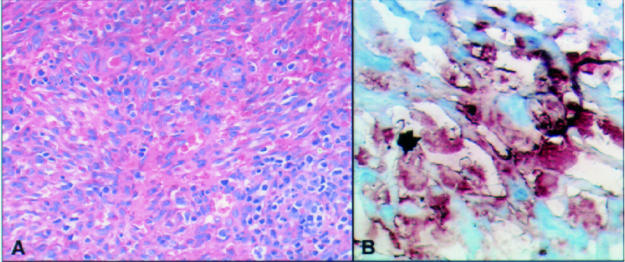
Using PCR-ISH, HHV-8 amplicons were identified in some lymphoid cells (in one patient) and in spindle cells of the KS lesion in another (figs 1 and 2 ▶ ▶). The positive lymphoid cells were mainly concentrated in B cell areas of the affected lymph nodes (as confirmed by examination of parallel sections by immunocytochemistry), confirming the B cell tropism exhibited by HHV-8.
Figure 1.
(A) Haematoxylin and eosin stained section of a lymph node from a patient with AIDS related lymphadenopathy, showing expansion of the germinal centres. (B) A lymphocyte containing human herpesvirus 8 (HHV-8) amplicons after polymerase chain reaction in situ hybridisation (PCR-ISH). (C) Another area of the same node showing numerous positive lymphocytes, containing HHV-8 amplicons. (D) Negative control with no amplicons visible. This result was obtained by omitting Taq DNA polymerase from the amplifying mixture or using an irrelevant primer pair.
Figure 2.
(A) Haematoxylin and eosin stained section of intranodal Karposi's sarcoma (KS). (B) Human herpesvirus 8 (HHV-8) amplicons in spindle cells and flattened endothelial cells in the KS specimen after PCR in situ hybridisation.
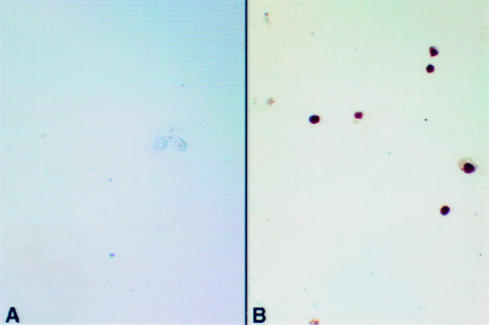
Figure 3 ▶ shows HHV-8 amplicons within the nuclei of cultured BCP-1 cells. All cells contain HHV-8 target. BCP-1 cells are estimated to contain approximately 50 copies of HHV-8, as determined by Southern analysis (C Boshoff, 1998, personal communication). The localisation of HHV-8 amplicons to the nucleus is appropriate for a herpes DNA virus, particularly a rhadinovirus. The results also show the versatility of labelled primer driven in situ amplification for the detection of viral targets in mammalian cells.
Figure 3.
Human herpesvirus 8 (HHV-8) labelled primer driven in situ amplification on the BCP-1 cell line. (A) DNase digestion abolishes the signal. (B) Intranuclear signals are seen in all cells.
The copy number of HHV-8 as assessed by TaqMan PCR was 100–500/100 ng of extracted DNA template, representing the DNA from 2000 cells. In a second set of experiments, microdissected individual cells positive by PCR-ISH contained approximately 10–30 copies of the HHV-8 genome in each cell.
Discussion
Recently, HHV-8 sequences have been identified in 27 of 43 cases of multicentric Castleman's disease and in three of 15 angioimmunoblastic lymphadenopathies,12–15 implicating HHV-8 in the pathobiology of these conditions.
The AIDS related lymphadenopathy cohort examined in our study comprises a unique group of HIV positive men, five of whom developed KS during the evolution of their disease. Of the five patients who developed KS at or subsequent to the time of lymph node biopsy, only three were shown to harbour HHV-8 DNA sequences in their lymph nodes. Importantly, one of the positive samples contained intranodal KS lesions at the time of diagnosis.
This is largely in keeping with the previous finding that approximately half of patients with AIDS and KS had detectable HHV-8 in their peripheral blood mononuclear cells,16 and the previous identification of HHV-8 in AIDS related lymphadenopathy.17, 18 However, our study is different in several aspects. First, the method of detection is 100–1000 times more sensitive than previously reported studies. Second, single copy gene detection is achieved at the single cell level.
The close association of HHV 8 with established or developing KS is again highlighted by our findings in AIDS related lymphadenopathy. The two patients who subsequently developed KS, but who were HHV-8 negative, may at the time of lymph node biopsy have been truly HHV-8 negative, or may have had a subpopulation of HHV-8 infected B cells in their circulation.
We are confident that our study had the highest level of sensitivity because we used TaqMan PCR, which has the ability to detect five copies of a viral target in 107–108 contaminating mammalian sequences (JJ O'Leary et al, 1999 unpublished data) and which our group has shown to be more sensitive than standard solution phase PCR and subsequent Southern blot analysis. In addition, PCR-ISH, with a detection sensitivity of one viral copy in formalin fixed paraffin wax embedded tissues, also failed to reveal any positive lymphoid cells. Therefore, we conclude that the HHV-8 negative patients were probably truly negative at the time of their lymph node diagnosis and might have subsequently acquired HHV-8 during their illness or, as a result of continuing immunosuppression, might have suffered activation of a minority population of infected HHV-8 B cells, with subsequent dissemination of the virus.
In two of the positive samples, PCR-ISH localised the target cell(s) for HHV-8 in AIDS related lymphadenopathy. Figure 1 ▶ shows lymphocyte positivity in a B cell rich area. In addition, this sample clearly demonstrates follicular expansion (fig 1A ▶). The second HHV-8 positive sample shows HHV-8 amplicons in spindle cells of an intranodal KS lesion. In the third sample, the location of HHV-8 DNA was only revealed in one cell, but the results were not reproducible on serial sections of the lymph node.
The final part of our study examined the BCP-1 cell line using labelled primer driven in situ amplification. As shown in fig 3 ▶, amplicons are clearly visible in the nuclei of all BCP-1 cells and are detectable after only five rounds of amplification (with a three step detection technique), supporting a value of approximately 50 copies of HHV-8/cell, estimated from the fact that using a one step technique (detecting 30–40 copies of virus) and a three step detection technique (detecting 2.5–12 copies (the higher value representing usual detection sensitivity)) we are unable to identify signals using non-isotopic in situ hybridisation without amplification.19, 20
In conclusion, the presence of HHV-8 in AIDS related lymphadenopathy is predictive of KS development and probably represents seeding of HHV-8 infected B cells from the peripheral blood. Our findings again support a role for HHV-8 in the pathobiology of KS, independent of immunosuppression.
References
- 1.Umlas J, Federman M, Crawford C, et al. Spindle cell pseudotumor due to Mycobacterium avium-intracellulare in patients with acquired immunodeficiency syndrome (AIDS). Positive staining of mycobacteria for cytoskeleton filaments. Am J Surg Pathol 1991;15:1181–7. [DOI] [PubMed] [Google Scholar]
- 2.Baroni CD, Uccini S. The lymphadenopathy of HIV infection. Am J Clin Pathol 1993;99:397–401. [DOI] [PubMed] [Google Scholar]
- 3.Said JW. AIDS-related lymphadenopathies. Semin Diagn Pathol 1988;5:365–73. [PubMed] [Google Scholar]
- 4.Burns BF, Wood GS, Dorfman RF. The varied histopathology of lymphadenopathy in the homosexual male. Am J Surg Pathol 1985;9:287–97. [DOI] [PubMed] [Google Scholar]
- 5.Wood GS, Carda CF, Dorfman RF, et al. The immunohistology of follicle lysis in lymph node biopsies from homosexual men. Blood 1985;66:1092–7. [PubMed] [Google Scholar]
- 6.Schuurman H-J, Kluin PM, Gmelig Meijling FHJ, et al. Lymphocyte status of lymph node and blood in acquired immunodeficiency syndrome (AIDS) and AIDS-related complex disease. J Pathol 1985;147:269–80. [DOI] [PubMed] [Google Scholar]
- 7.O'Hara CJ, Groopman JE, Federman M. The ultrastructural and immunohistochemical demonstration of viral particles in lymph nodes from human immunodeficiency virus-related and non-human immunodeficiency virus-related lymphadenopathy syndromes. Hum Pathol 1988;19:545–9. [DOI] [PubMed] [Google Scholar]
- 8.Harris NL. Hypervascular follicular hyperplasia and Kaposi's sarcoma in patients at risk from AIDS. N Engl J Med 1984;310:462–3. [DOI] [PubMed] [Google Scholar]
- 9.Abrams DI. Lymphadenopathy syndrome in male homosexuals. Advances in Host Defence Mechanisms 1985;5:75–97. [Google Scholar]
- 10.Renne R, Zhong W, Herndier B, et al. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 1996;2: 342–6. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy M, Lucas SB, Russell-Jones R, et al. KSHV in female Kaposi's sarcoma. J Pathol 1997;183:447–52. [DOI] [PubMed] [Google Scholar]
- 12.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 1995;86:1276–80. [PubMed] [Google Scholar]
- 13.Gessain A, Sudaka A, Briere J, et al. Kaposi's sarcoma-associated herpesvirus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association with non-human immunodeficiency virus-infected patients? Blood 1996;87:414–16. [PubMed] [Google Scholar]
- 14.Karcher DS, Alkan S. Herpes-like DNA sequences, AIDS-related tumors, and Castleman's disease. N Engl J Med 1995;333:797–8. [DOI] [PubMed] [Google Scholar]
- 15.Luppi M, Barozzi P, Maiorana A, et al. Human herpesvirus 8 DNA sequences in human immunodeficiency virus-negative angioimmunoblastic lymphadenopathy and benign lymphadenopathy with giant center hyperplasia and increased vascularity. Blood 1996;87:3903–9. [PubMed] [Google Scholar]
- 16.Whitby D, Howard M, Tenant-Flowers M, et al. Detection of KSHV in peripheral blood of HIV-infected individuals predicts progression to Kaposi's sarcoma. Lancet 1995;346:799–802. [DOI] [PubMed] [Google Scholar]
- 17.Bigoni B, Dolcetti R, deLellis L, et al. Human herpesvirus 8 is present in the lymphoid system of healthy persons and reactivates in the course of AIDS. J Infect Dis 1996;173:542–9. [DOI] [PubMed] [Google Scholar]
- 18.Corbellino M, Poirel L, Bestetti G, et al. Restricted tissue distribution of extralesional Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS patients with Kaposi's sarcoma. AIDS Res Retroviruses 1996;8:651–7. [DOI] [PubMed] [Google Scholar]
- 19.Herrington CS, Burns J, Graham AK, et al. Interphase cytogenetics using biotin and digoxigenin-labelled probes: I. Relative sensitivity of both reporter molecules for HPV 16 detection in CaSki cells. J Clin Pathol 1989;41:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrington CS, Graham AK, McGee JO'D. Interphase cytogenetics using biotin and digoxigenin labelled probes: III. Increased sensitivity and flexibility for detecting HPV in cervical biopsy specimens and cell lines. J Clin Pathol 1991;44:33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]