Abstract
Apoptosis is the genetically regulated form of cell death that permits the safe disposal of cells at the point in time when they have fulfilled their intended biological function. Examples of apoptosis can be cited throughout the whole of the animal and plant kingdoms. It is a vitally important process during normal development and the adult life of many living organisms. In humans, dysregulation of apoptosis can result in inflammatory, malignant, autoimmune, and neurodegenerative diseases. In addition, infectious agents, including viruses, exploit cellular apoptosis in the host to evade the immune system. This review gives a brief historical perspective of some of the landmark discoveries in apoptosis research. The morphological and biochemical stages of apoptosis are then covered, followed by an overview of how it can be studied in the laboratory. Finally, the implications for therapeutic intervention in disease treatment are discussed.
Keywords: apoptosis, necrosis, tumour necrosis factor, development
Slime mould, nematode worms, butterfly larvae, fruit flies, dwarf tree frogs, chickens, mice, and rats, to name but a few species, have all played their part in defining the importance of “apoptosis” or “programmed cell death”. Apoptosis refers to the genetically regulated form of cell suicide that is morphologically distinct from the degenerate process of necrosis.
In 1858, Virchow described the progressive changes in the appearance of body tissues that occur shortly after death, and it was he who was the first to recognise that more than one form of cell death existed. Necrosis being where “the mortified cell is left in its external form” and “necrobiosis or shrinkage necrosis being where the cell vanishes and can no longer be seen in its previous form”.1 Necrobiosis is what we now recognise as apoptosis.
By 1887, others were investigating the different morphologies of cell death.2 In 1885, Walther Flemming was studying the regression of epithelium in mammalian lymphoid follicles, and it was he who produced the first drawings of what we today would recognise morphologically as apoptosis, and he adopted another term “chromatolysis”.3 The first review describing chromatolysis appeared in 1914 and was written by Ludwig Graper, a German anatomist studying yolk sac shrinkage.4 Almost 40 years later, in 1951 Glucksmann was able to describe “chromatolysis” during embryo development.5 Thirteen years hence, experimental pathologists began in earnest to investigate the ways in which the cells in individual organs die during development and in response to insult or injury.6
Some of the early studies of this process in mammalian tissues centred around the cellular pathology of liver atrophy after restriction of the blood supply by portal vein ligation. Even then, the liver was recognised as a potentially useful organ in this field of study because of its phenomenal capacity to regenerate.6,7 It was essentially from this model that the concept was evolved whereby cell populations could be held in balance not only by proliferation but also by some type of controlled death. Even at that time, many good theoretical reasons for such a mechanism were obvious including, regeneration and repair, maintenance of cellular homeostasis, and control of organ size. Concepts relating to genetic editing for the deletion of defective, mutated, infected, or otherwise functionally effete cells were shortly to follow, and the scientific understanding of regulated cell death slowly began to advance.7,8
Several early studies focused on histopathological observations, and during the mid to late 1960s, a great deal was learnt about cell death at the ultrastructural level using electron microscopy.6 In 1972, a group of pathologists in Edinburgh—Kerr, Wyllie, and Currie—were working in the field and realised that conventional terms such as necrobiosis, shrinkage necrosis, chromatolysis, coagulative necrosis, or ischaemic death were confusing and inadequate to delineate the two forms of cell death.8–10 Thus, it was Kerr, Wyllie, and Currie who proposed, with the assistance of Professor James Cormack (at that time the Professor of Greek at Aberdeen University) the adoption of an unambiguous term “apoptosis” to describe specifically one of these methods of cell death.8 Even today, it seems that pathologists are divided in opinion about the term “necrotic cell death”, and alternative terminology has been proposed recently.11
It was perhaps also Kerr, Wyllie, and Currie who were the first to realise the far reaching implications of apoptotic cell death within the context of human cell biology and disease. Their work in the early 1970s has proved to be a scientific landmark, which to a large extent still upholds the principles that have conceptually dominated research in this field until the present day.8–12
Early histopathological studies gave enough information to describe the division of apoptosis into discrete morphological stages, and to advance hypotheses that have since been substantiated and underpinned by research performed in laboratories throughout the world. During the 1970s, however, it is fair to say that interest in apoptosis research reached a plateau, with many members of the scientific community being unimpressed and uncertain of the potential importance of apoptosis.
The real watershed for apoptosis research did not arrive until the early 1080s, when a succession of papers led to a huge revival of interest. In 1980, Andrew Wyllie produced a paper on glucocorticoid induced apoptosis in cultured lymphocytes, which was possibly responsible for the resurgence of interest.13 Three years later, apoptosis was shown to be linked to the activation of an endonuclease that cleaved DNA into regular sized fragments, which could be demonstrated by gel electrophoresis. Thus, the first biochemical evidence for apoptotic cell death was established.14
From that point forward, there has been almost a decade and a half where we have learned much about the detail of the remarkable and genetically complex process of apoptosis. Powerful genetic manipulation and molecular biology techniques have led to an exponential growth in our knowledge of apoptosis, and we now have a good, albeit incomplete, understanding of the myriad of genes that are involved in the regulation of this process. The race is now on to dissect these complex interactive pathways, in the hope that more detailed knowledge will lead to the design of more specific strategies for the combat of disease.
Today, a current literature search using apoptosis as a keyword will reveal in excess of 29 200 listed manuscripts, and interest in apoptosis research shows no signs of abating. As new apoptotic genes continue to be identified, and new apoptosis related functions are assigned to familiar proteins, our understanding of this complex process will have to remain under constant review for the foreseeable future.
Apoptosis or necrosis: are there really two pathways to death?
It is important to recognise that these terms relate to a “morphological description” of cell death only, and it remains possible that the two processes might be part of a continuum, with morphological necrosis being the ultimate fate even for cells that undergo apoptotic changes initially.15
That the two pathways overlap to some extent is evident because in vivo one type of cell death is often seen accompanying the other and in cultured cells apoptotic cells can, under certain circumstances, assume necrotic morphology.15
It is also important in the broader context to appreciate that regulated cell death occurs across the entire animal and plant kingdom—but for different reasons in humans compared with the slime mould (dichtyostelium).16,17 It therefore follows that during the course of evolution, many different pathways of achieving the same endpoint have developed. That is to say that cellular dismantling and recycling has developed in response to environmental pressures faced by the individual organism. In addition, it should not always be assumed that necrosis has no part to play throughout the normal life cycle of living organisms. For example, trees use dead tissues as an important structural component. In the dichtyostelium slime mould, stalk cell replication, a crucial phase leading to spore dispersal, is arrested via necrosis rather than apoptosis.16 Some very recent and fascinating developmental studies also suggest that when the apoptotic pathway is genetically deleted, the normal developmental cell loss during mouse forelimb formation can still proceed—and it does so predominantly by necrosis.18 Thus, an open mind is still required about the relative contribution of these two types of cell death to development and cellular homeostasis.
Although we still have an incomplete picture of the individually tailored mechanisms of apoptosis or “programmed cell death”, there is now a broad acceptance of its importance to all living organisms. Sufficient genetic and biochemical evidence exists to show that the pathway for regulated cell death in digit formation in the developing mouse embryo—for example, is different to the pathway that has evolved for the modulation of total organ mass or leucocyte numbers, the maintenance of immunological tolerance, the protection against deleterious chromosomal aberrations/mutations, or the elimination of pathogenically infected cells within the whole organism.
In the course of this review we hope to cover in summary what is known about how apoptosis is initiated, how it is regulated, and what can happen when it goes wrong, all within the context of the pathophysiological processes that occur throughout life in humans.
The stages of apoptotic cell death
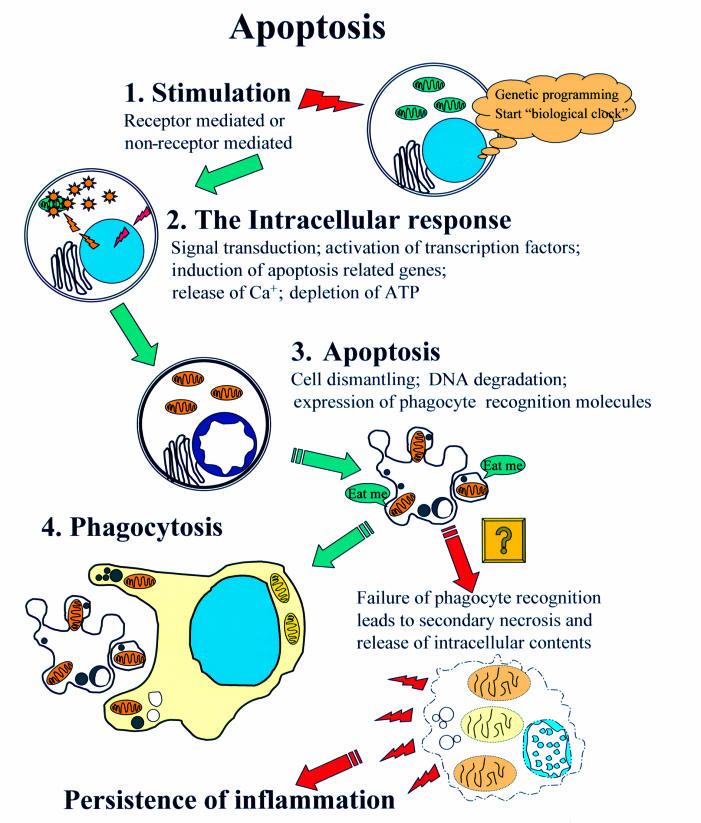
Apoptosis can be thought of as a series of progressive steps that lead desirably to the efficient dismantling of the cell in a manner that minimises the risk of leakage of potentially harmful intracellular contents into the surrounding microenvironment, where it could otherwise have harmful cytotoxic side effects (fig 1 ▶).
Figure 1.
The stages of apoptosis. This diagram illustrates the four basic stages of the apoptotic pathway. Once the cell has reached stage two and the caspase pathway has been activated it is believed that the process is irreversible and the cell cannot be rescued. After stage three, if the cell is in close enough proximity to a phagocytic neighbour and is displaying the right molecular signature it is engulfed and broken down within the phagocyte. If the apoptotic cell is not recognised it will eventually assume necrotic morphology, so called “secondary necrosis”.
STAGE 1: TRIGGERING APOPTOSIS-MECHANISMS OF STIMULATION
It seems likely that all living cells have evolved the genetic capability to undergo apoptosis spontaneously. Although the mechanisms that lead to the starting of the “biological clock” are not understood, it seems likely that alterations in the environmental conditions that the cell experiences can start, accelerate, or slow down the process.19,20
To enter into the apoptotic cycle the cell first encounters a signal to activate the relevant genetic machinery. A good example of physiologically important apoptosis in adult life is the life cycle of the mature neutrophil. When it leaves the bone marrow and enters the peripheral circulation, it is terminally differentiated and incapable of further division. Should it not be recruited to an inflammatory lesion (whereupon it would release its potent and highly cytotoxic contents via degranulation), it sequester in the lung, where the senescent neutrophils undergo apoptosis. Here, they are normally phagocytosed by resident alveolar macrophages.21–23 This mechanism ensures the safe disposal of these otherwise harmful cells. When this system is stressed as a result of overwhelming toxic injury or infection, massive systemic damage results (septic shock syndrome).
In the lymph nodes, cells that are not exposed to frequent antigenic stimulation undergo clonal deletion via apoptosis. This is a very good example of how withdrawal of factors can lead to apoptotic cell death.24 This mechanism is of crucial importance in the recognition of self and non-self, and maintenance of immunological tolerance.25 Breakdown of this regulatory pathway results in a range of autoimmune and malignant conditions.26 There is now also good evidence that entry into the proliferative cycle is a crucial step in sensitising some, but not all, cells to apoptotic signals.26
There are stimuli that originate from outside the cell that can advance or delay apoptotic death. This stage of the process can be triggered by agents that can penetrate the cell directly, and modulate the apoptotic cascade in the absence of specific cell surface receptors. Examples of such agents include heat shock/stress factors,25 free radicals,27 ultraviolet radiation,28 numerous drugs and synthetic peptides,29 toxins,30 and potent lymphocyte enzymes (the granzymes).31 Other mechanisms are dependant on expression of appropriate cell surface receptors.
Receptor mediated apoptosis has been demonstrated for numerous growth factors (for example, transforming growth factor β32 and cytokines, including tumour necrosis factor (TNF)).33 Of these molecular families, the TNF like molecules are a family of ligands that exist as soluble or cell bound forms and which are known to have a major role in the modulation of cell survival/apoptosis in cells of many lineages.34,35 The TNFs act via a large family of receptors expressed on the surface of the target cell, known as the TNF receptor superfamily. The TNF receptor family comprises at least 19 members, which are grouped together based on structural homology, but which in reality have a wide array of biological effects that sometimes extend beyond the regulation of apoptosis.35,36 At least four of them can trigger apoptosis directly, and although not containing an intracellular death domain themselves, several others do have the potential to interact with and activate other apoptosis inducing receptor–ligand pairs. In addition, it seems that some family members, which are incapable of signalling because they lack a cytoplasmic tail, can function as “decoy receptors”. At present, little is known about the true biological role of many of these receptor family members, beyond their sequence homology and distribution throughout tissues. Detailed discussion of individual receptors is clearly beyond the scope of this review, and the reader is referred to other articles for further information.34–36
Expression of some of the TNF receptors appears to be restricted to specific cell types whereas others (for example, Fas (CD95/Apo 1)) are distributed across a wide range of cell types, and are thought to have a central role in the triggering of apoptosis.37–39 This prototypic family member illustrates how many receptor mediated apoptotic pathways can be activated.
STAGE 2: INTRACELLULAR SIGNALLING AFTER RECEPTOR LIGAND BINDING
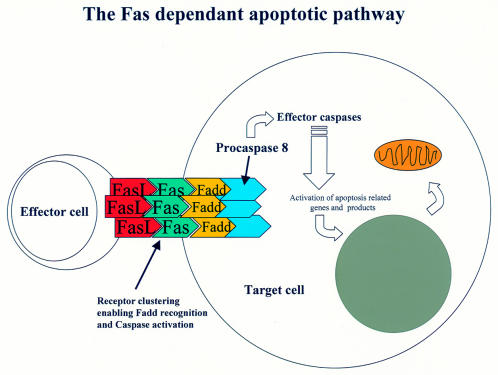
The TNF receptor superfamily members are all typical type 1 membrane spanning glycoproteins, with the N-terminus on the outside of the cell containing anywhere between one and six ligand binding domains. Within the cell, the C-terminus typically (for the death transducing receptors see fig 2 ▶) contains a region of about 60–70 residues, which upon activation with a trimeric ligand, forms a cluster and a complex beneath the cell membrane with a number of cytosolic proteins to form the active “death domain” (for example, Fas forms the Fas associated death domain (FADD)). This receptor–ligand complex then becomes the recognition molecule for a precursor enzyme, procaspase 8.40
Figure 2.
Apoptotic cell death mediated by the Fas receptor pathway. Effector cells (for example, activated T lymphocytes) present the Fas ligand (FasL) in a conformation that permits crosslinking and aggregation of the Fas receptor on the target cell. The clustering of Fas is the first step in activating the intracellular apoptotic cascade. Formation of a complex, the Fas associated death domain (FADD), then results in binding and activation of procaspase 8 and the caspase cascade.
The caspase family of enzymes contains cysteine proteinases that mediate a whole range of intracellular events, and a number of agents act by modulating caspase expression and activity directly, bypassing the receptor pathway41 (table 1 ▶). There are presently 13 known caspase family members, which form an intracellular proteolytic cascade that modulates many cellular events in the apoptotic pathway, including activation of transcription factors.42 Transcription factors are regulatory proteins that bind to specific initiation sites on DNA, and in turn modulate expression of genes that regulate production of pro-apoptotic and antiapoptotic factors. Such important apoptotic genes include the bcl-2 family of mitochondrial proteins,43 cytochrome c,44 the tumour suppressor genes p53 and p21(WAF 1), and many others.45 In a highly complex and as yet poorly understood way, it is the balance between these pro-apoptotic and antiapoptotic factors that controls the fate of the cell (table 1 ▶).46
Table 1.
Genes and molecules that regulate apoptosis
| Gene/molecule | Effects |
| Bcl-2 subfamily (Ced9 in Caenorhabditis elegans) | |
| Bcl-2 | Promotes survival |
| Bcl-xl | Promotes survival |
| Bcl-w | Promotes survival |
| Bcl-xs | Promotes death |
| Bax family | |
| Bax | Promotes death |
| Bak | Promotes death |
| Bok | Promotes death |
| BH3 subfamily | |
| Bad | Promotes death |
| Bik | Promotes death |
| Bid | Promotes death |
| Blk | Promotes death |
| HRK | Promotes death |
| BimL | Promotes death |
| Egl (C elegans) | Promotes death |
| Caspases 1–13* (Ced3 in C elegans) | These interleukin 1β converting enzyme (ICE) like cysteine proteinases form a central part of the apoptotic cascade |
| Apaf-1 (Ced4 in C elegans) | These adaptor molecules link caspase (Ced3) activation and Bcl-2 (Ced9) expression. Their functions in promoting death or survival remain to be defined |
| p53 family of tumour supressor genes | p53 is necessary for apoptosis induced by agents that cause DNA damage. Growth arrest occurs by activation of p21. p53 can also affect the expression of bcl-2 and bax directly. p21/WAF1 inhibits cyclin dependent kinases and prevents progression through the cell cycle |
| Nitric oxide | This molecule prevents apoptosis by altering bcl-2 expression and by nitrosylation of caspases |
| Cytochrome c | This molecule induces apoptosis via caspase activation |
This table summarises the major families of regulatory genes that are activated when a cell undergoes apoptosis. The list is by no means exhaustive but gives representative family members.
The products of the bcl-2 gene subfamily are mitochondrial proteins that can exist as homodimers or heterodimers made up of different pairings. Their functions are still unclear but changes in the ratio of their expression has profound effects on cell survival.
*Inhibitors of ICE have also been described recently.
There is recent evidence suggesting that not all apoptotic cell death requires caspase activation. However, little is currently known about caspase independent death and its pathophysiological relevance awaits clarification.18
Much of what is currently known about the molecular and genetic regulation of apoptosis has been the result of work carried out on the nematode worm, Caenorhabditis elegans, where the Ced (cell death abnormal) and Egl gene family members have been cloned and sequenced, and to which an array of apoptotic functions have been ascribed. Because many of these genes are highly conserved, mammalian homologues of the Ced genes have now been found (table 1 ▶).46
STAGE 3: APOPTOSIS-THE MORPHOLOGICAL AND BIOCHEMICAL CHANGES
Release of intracellular calcium and depletion of ATP accompany the ongoing process.47 In contrast to necrosis, there is no mitochondrial swelling and the cell begins to shrink with the plasma membrane remaining intact. Chromatin begins to condense and marginate towards the nuclear membrane periphery (fig 1 ▶). Mitochondrial activity increases with the synthesis and release of apoptosis regulatory proteins such as the bcl-2 family members, cytochrome c, and nitric oxide.47 The activated caspase cascade continues to initiate the induction of other apoptotic genes, including death promoters such as p53 or p21.
Mutations in one of these tumour suppressor genes (p53), in particular, is thought account for a large proportion of malignancies in human disease,26 and p53 is a major target for transforming viral oncoproteins.48 Other important pro-apoptotic genes include those that encode the endonuclease that degrades the DNA into 50 kb fragments, the biochemical hallmark of apoptosis in most, although not all, cell types.49 Genes are also activated that lead to expression of other cell surface phagocyte recognition molecules, such as intercellular adhesion molecule 3 (ICAM 3).50 A crucial step appears to be the loss of membrane asymmetry, leading to the exposure of anionic phospholipids and phosphatidylserine (PS). These are thought to be the molecular signals recognised by neighbouring phagocytic cells.51 Eventually, the plasma membrane of the cell begins to bud off encapsulating and packaging lysosomes, mitochondria, chromatin fragments (known also as Councilman bodies), and other degenerate organelles including the Golgi/endoplasmic reticulum (figs 1 and 3 ▶ ▶).
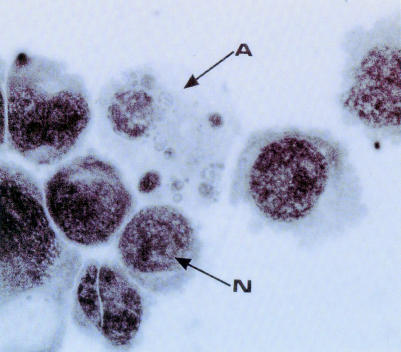
Figure 3.
The morphology of apoptosis. This photomicrograph shows a culture of promonocytic U937 cells, some of which show classic morphological features of apoptosis (arrow A). The cell marked N shows the normal morphology of a viable neighbour for comparison.
STAGE 4: PHAGOCYTIC RECOGNITION OF APOPTOTIC BODIES
The importance of this part of the process to normal tissue homeostasis and during the resolution of the inflammatory response is now well recognised.51,52 Hypothetically, defects in phagocyte recognition of apoptotic cells may lead to persistence of inflammatory disease, which is exacerbated because apoptotic cells eventually assume necrotic morphology (so called secondary necrosis). This idea has gained credence because it seems a particularly attractive explanation for persistent inflammation and tissue damage. As yet, there is little histopathological or clinical proof that such a mechanism occurs in vivo, but this field of work is continuing to advance.
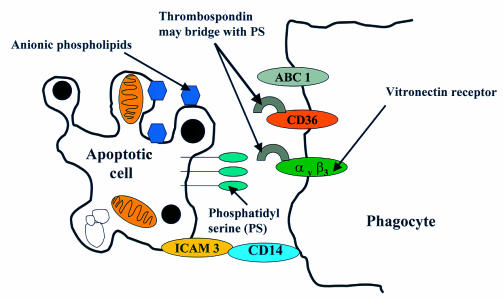
The point at which apoptotic cells are recognised by phagocytic cells is not really known, although it is probably at quite an early stage, because in tissues where cell turnover is high, the frequency of mature apoptotic bodies can be relatively low.53 Research into the mechanisms involved in phagocytic recognition are still at an early stage. Several molecules on the surface of the phagocyte and the apoptotic cell have been identified as important mediators of the process (fig 4 ▶), most of which have turned out to be cell surface molecules that have been familiar to immunologists and biochemists for quite a few years.54–56 Research in this area is currently focused on the professional macrophages of leucocytic origin, and much less is known about phagocytosis of apoptotic cells in other systems. Key molecules on the phagocyte include the class A scavenger receptors macrosialin and ABC 1, the vitronectin receptor, and the class B scavenger receptors CD36 and CD14 (a glycophosphatidylinositol anchored protein which is the lipopolysaccharide receptor). These molecules interact with several counter-receptors on the apoptotic cell including anionic phospholipids, PS, and ICAM 3. On the phagocytic cell, the vitronectin receptor (αvβ3 integrin) and CD36 both have the ability to bind a bridging molecule thrombospondin. Although the partner for thrombospondin bridging on the apoptotic cell has yet to be defined, likely candidates are PS and ICAM 3, although the latter has been shown recently to partner CD14.50,53
Figure 4.
Phagocytic recognition of an apoptotic cell. At present, little is known about the interactions between apoptotic cells and phagocytes, but a series of molecules seem to be important including phosphatidylserine (PS), anionic phospholipids, and intracellular adhesion molecule 3 (ICAM 3). The hierarchical nature of these interactions is not yet known but recent evidence suggests that ICAM 3 partners CD14 on the macrophage. An important pairing also seems to be between PS and the vitronectin receptor or CD36. This probably occurs via thrombospondin, which can avidly bind both molecules on the phagocyte.
The fact that the vitronectin receptor and CD36 can signal via tyrosine kinase implies that these molecules in particular might be of crucial importance in the subsequent activation of the signalling pathway in the phagocytic cell, about which virtually nothing is currently known.
How can apoptosis be studied?
MOLECULAR GENETICS
Today's apoptotic research is becoming dominated be the power of modern molecular biology; in particular, the use of transgenic animal models to test the function of particular genes. There are well known caveats to these models, the most important of which is that, as the number of interactive gene products in any particular system increases, so does the likelihood of redundancy. In other words, when a gene is deleted and no biological effect is observed it is possible that other pathways have compensated for the defect. That being said, transgenics do continue to be a powerful analytical approach to the analysis of gene function. In relation to apoptotic cell death, transgenic animals where death receptor genes, including Fas and CD40 or their cognate ligand encoding genes, have been deleted have given important clues as to the importance of these molecules for the regulation of apoptosis in several cell types.57,58 The Fas ligand “knockout animals” suffer from lymphoproliferative disease and liver hyperplasia. CD40–ligand knockout animals are also severely immunocompromised. Interestingly, CD40–ligand knockout animals are also susceptible to the development of cholangiocarcinoma, have liver hyperplasia, and fail to clear virally infected cells effectively.59,60 All these observations underpin the systemic importance of an effective functional apoptotic pathway .
In addition, the ability to modify individual cell types, either by gene deletion or inducing or inserting pro-apoptotic and antiapoptotic genes, has contributed greatly to our understanding of the role of individual molecules in apoptosis. For example, manipulating hepatocytes to overexpress the survival factor gene bcl-2 makes the cells resistant to Fas mediated apoptosis.61 Molecular analysis of apoptotic gene mutations has also provided some tantalising clues about how malignant transformation can occur. Aggressive tumour growth has been associated with mutations or the aberrant expression of several apoptotic genes, including bcl-2 family members, p53, Fas,62 and CD40.63 An understanding of how these genes function offers great potential for therapeutic intervention.
CELL AND MOLECULAR BIOLOGY
A great deal of information can also be gained from cell biology, molecular biology, and biochemical approaches to the study of cells in tissue culture, or ex vivo in tissue sections. These approaches fall into categories that relate to either the morphological or biochemical assessment of apoptosis. There are no single specific markers of this process, and it is widely accepted that several different techniques should be carried out. As stand alone techniques go, the gold standard method for morphological assessment is electron microscopy, where the ultrastructural changes that typify apoptosis can be seen in individual cells.64
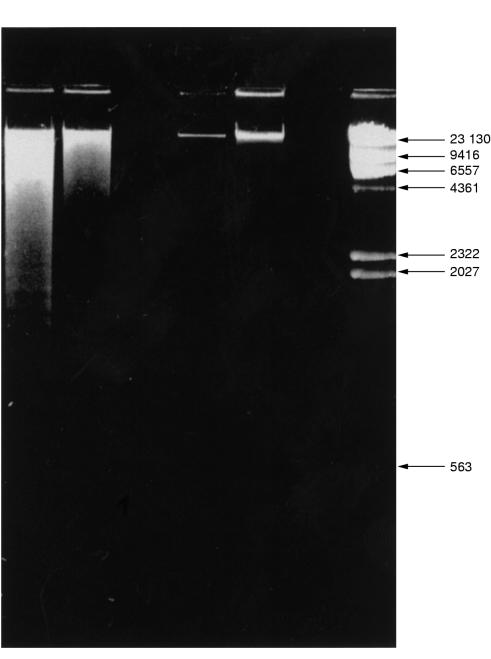
Quantification of apoptosis requires other approaches. Electrophoretic analysis of DNA, the so called “DNA laddering technique”, allows confirmation of the presence of degraded DNA, but in most cases does not permit quantitative assessment of the degree of apoptosis (fig 5 ▶). This method is only really appropriate for isolated pure populations of cells. Assessing cell and nuclear morphology microscopically in combination with an immunohistochemical technique for detection of DNA strand breaks (which occur at high frequency in apoptotic cells) is a widely used and generally accepted approach.
Figure 5.
Electrophoretic analysis of DNA from apoptotic cells. DNA extracts can be purified, run on an agarose gel, and stained with fluorescent ethidium bromide. DNA from apoptotic cells (aged blood polymorphs in this case) forms a distinctive laddering pattern as a result of endonuclease cleavage (lane 1). Lane 2, DNA isolated from necrotic cells; lanes and 6, no sample; lanes 3 and 4, DNA immediately after isolation (before treatment); lane 7, standard DNA molecular weight markers. Band sizes are in kDa.
There are many apoptosis detection kits now commercially available that are based on DNA fragmentation, most being variations on similar themes. Two frequently used techniques are the in situ DNA end labelling method (ISEL)65 (fig 6 ▶) and the in situ terminal deoxynucleotidyl transferase d uridine triphosphate nick end labelling (TUNEL) method.66 Both techniques exploit the endonuclease dependant cleavage of DNA. The assumption is made that this occurs with much higher frequency in apoptotic cells than under any other circumstances. Both techniques rely on the labelling of the nicked DNA with a chromogen or fluorogen conjugated complex.
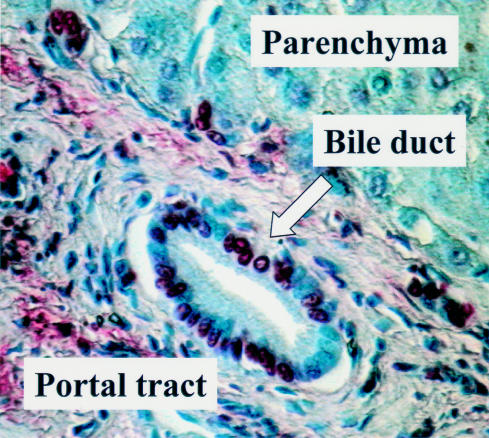
Figure 6.
Identification of apoptotic cells in tissue sections using in situ DNA end labelling (ISEL). This figure shows a section of human liver tissue from a patient with end stage primary biliary cirrhosis. Strongly staining ISEL positive cells (red) can be seen; in this case, in the bile duct within the portal tract.
Other techniques exploit the changes in nuclear morphology and cell membrane biochemistry, and in some circumstances allow for the automated measurement of apoptosis via flow cytometry. These methods rely on various combinations of propidium iodide, which can enter necrotic cells and stain the nucleus, and labelling of the cell surface phosphatidyl serine residues, which are expressed at early stages of apoptosis, with an anticoagulant protein called annexin V, conjugated to a reporter molecule, such as fluorescein or biotin (fig 7 ▶).
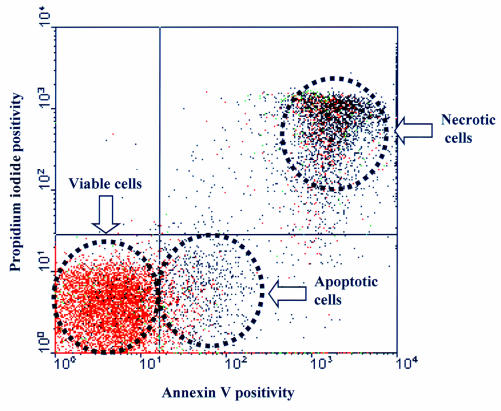
Figure 7.
Quantitative assessment of apoptosis using flow cytometry. This figure shows a typical profile for activated peripheral blood lymphocytes that have been stained with propidium iodide (PI) or annexin V and subjected to fluorescence activated cell sorting. Each dot represents an individual cell. Viable cells stain with neither PI nor annexin V. Apoptotic cells stain with annexin V only, whereas necrotic cells stain with both.
The downstream intracellular events of apoptosis can also be assessed using a variety of cell and molecular biological approaches. Again an abundance of commercially available reagents are readily available. Gene expression and the consequent protein synthesis can be monitored using western immunoblotting (for proteins) and reverse transcriptase PCR (for mRNAs). A wide range of antibodies to receptors, ligands, transcription factors, caspases, and tumour supressor gene products permits localisation of many molecules using immunohistochemistry. There are also several chromogenic substrates, with varying degrees of specificity for the caspase family members, which allow for functional assessment of enzyme activity.
The implications of apoptosis in disease and potential for treatment
The consequences of manipulating apoptosis are potentially beneficial and deleterious. Although we still have much to learn, we already know that the system is extremely complex, and there are several areas where the modulation of apoptotic death could be of benefit in the treatment of disease in the future.
Control of malignant disease: the induction of apoptosis in malignant cells would be desirable.
Delay of premature senescence/neurodegenerative disorders: the prevention or delay of apoptosis in neural cells is a desirable goal.
Regulation of inflammatory disease: the induction of apoptosis and phagocytosis would be desirable for the suppression/and resolution of the inflammatory response.
Treatment of transplant rejection: the prevention of apoptosis in parenchymal cells and the induction of apoptosis for establishing immunological tolerance (cells of the immune system) would both be desirable in this setting.
Regulation of tissue regeneration/repair: induction of apoptosis would be useful to limit fibroblast activity and control scar formation.
The potential for therapeutic intervention in any of these scenarios is theoretically possible, and there are already some studies in each of these settings that suggest short term beneficial effects. The predominant problems with treatments aimed at such a widespread cellular mechanism are the difficulties associated with achieving the necessary degree of specific targeting to the cells or genes of interest. At the moment, strategies for the modulation of apoptosis involving both gene therapy and non-gene therapy approaches are being pursued vigorously worldwide. It remains to be seen which, if any, approach will be sufficiently successful to offer real hope for therapeutic use in the treatment of a disease where dysregulation of cell death is implicated.
References
- 1.Virchow R, Chandler AB, eds. Cellular pathology as based upon physiological and pathological histology. New York: Dover Publications, 1859.
- 2.Weigert C. Uber Croup und Diptheritis. Ein experimenteller und anatomischer Beitrag zur Pathologie der specifischen Entzundungsformen. Virchows Arch Pathol Anat 1877;72:461–501. [Google Scholar]
- 3.Flemming W. Uber die bildung von richtungsfiguren in saugethiereiern beim untergang graaf'scher follikel. Arch Anat Entwgesch 1885:221–4.
- 4.Graper L. Eine neue Anschauung uber physiologische Zellausschaltung. Archive Zellforsch 1914;12:373–94. [Google Scholar]
- 5.Glucksmann A. Cell deaths in normal vertebrate ontogeny. Biol Rev Camb Philos Soc 1951;26:59–86. [DOI] [PubMed] [Google Scholar]
- 6.Majno G, La Gattuta M. Cellular death and necrosis: chemical, physical and morphological changes in rat liver. Virchows Arch A Pathol Anat Histopathol 1960;333:421–65. [DOI] [PubMed] [Google Scholar]
- 7.Kerr JF. Shrinkage necrosis: a distinct mode of cellular death. J Pathol 1971;105:13–20. [DOI] [PubMed] [Google Scholar]
- 8.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 1972;26:239–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr J, Pearn J, eds. Apoptosis: past and future. In: Milestones of Australian medicine. Brisbane: Amphion Press, 1994:153–66.
- 10.Kerr JF, Searle J. A suggested explanation for the paradoxically slow growth rate of basalcell carcinomas that contain numerous mitotic figures. J Pathol 1972;107:41–4. [DOI] [PubMed] [Google Scholar]
- 11.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death [see comments]. Am J Pathol 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 12.Kerr JF, Searle J. The digestion of cellular fragments within phagolysosomes in carcinoma cells. J Pathol 1972;108:55–8. [DOI] [PubMed] [Google Scholar]
- 13.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 1980;284:555-6. [DOI] [PubMed] [Google Scholar]
- 14.Duke RC, Chervenak R, Cohen JJ. Endogenous endonuclease-induced DNA fragmentation: an early event in cell-mediated cytolysis. Proc Natl Acad Sci U S A 1983;80:6361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong Z, Saikumar P, Weinberg JM, et al. Internucleosomal DNA cleavage triggered by plasma membrane damage during necrotic cell death. Involvement of serine but not cysteine proteases. Am J Pathol 1997;151:1205–13. [PMC free article] [PubMed] [Google Scholar]
- 16.Cornillon S, Olie RA, Golstein P. An insertional mutagenesis approach to dictyostelium cell death. Cell Death Differ 1998;5:416–25. [DOI] [PubMed] [Google Scholar]
- 17.Wyllie AH. The genetic regulation of apoptosis. Curr Opin Genet Dev 1995;5:97–104. [DOI] [PubMed] [Google Scholar]
- 18.Chautan M, Chazal G, Cecconi F, et al. Interdigital cell death can occur through a necrotic and caspase-independent pathway [in process citation]. Curr Biol 1999;9:967–70. [DOI] [PubMed] [Google Scholar]
- 19.Raff MC, Barres BA, Burne JF, et al. Programmed cell death and the control of cell survival: lessons from the nervous system. Science 1993;262:695–700. [DOI] [PubMed] [Google Scholar]
- 20.Fraser A, McCarthy N, Evan GI. Biochemistry of cell death. Curr Opin Neurobiol 1996;6:71–80. [DOI] [PubMed] [Google Scholar]
- 21.Weiss SJ. Tissue destruction by neutrophils [see comments]. N Engl J Med 1989;320:365–76. [DOI] [PubMed] [Google Scholar]
- 22.Savill JS, Henson PM, Haslett C. Phagocytosis of aged human neutrophils by macrophages is mediated by a novel “charge-sensitive” recognition mechanism. J Clin Invest 1989;84:1518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigg JM, Savill JS, Sarraf C, et al. Neutrophil apoptosis and clearance from neonatal lungs. Lancet 1991;338:720–2. [DOI] [PubMed] [Google Scholar]
- 24.Allen PD, Bustin SA, Macey MG, et al. Programmed cell death (apoptosis) in immunity and haematological neoplasia. Br J Biomed Sci 1993;50:135–49. [PubMed] [Google Scholar]
- 25.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science 1995;267:1456–62. [DOI] [PubMed] [Google Scholar]
- 26.Evan GI, Brown L, White M, et al. Apoptosis and the cell cycle. Curr Opin Cell Biol 1995;7:825. [DOI] [PubMed] [Google Scholar]
- 27.Buttke TM, Sandstrom PA. Oxidative stress as a mediator of apoptosis [see comments]. Immunol Today 1994;15:7–10. [DOI] [PubMed] [Google Scholar]
- 28.Yamada T, Ohyama H. Radiation-induced interphase death of rat thymocytes is internally programmed (apoptosis). Int J Radiat Biol Relat Stud Phys Chem Med 1988;53:65–75. [DOI] [PubMed] [Google Scholar]
- 29.Schwall RH, Robbins K, Jardieu P, et al. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology 1993;18:347–56. [DOI] [PubMed] [Google Scholar]
- 30.Sachs L, Lotem J. Control of programmed cell death in normal and leukemic cells: new implications for therapy. Blood 1993;82:15–21. [PubMed] [Google Scholar]
- 31.Hayes MP, Berrebi GA, Henkart PA. Induction of target cell DNA release by the cytotoxic T lymphocyte granule protease granzyme A. J Exp Med 1989;170:933–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberhammer F, Fritsch G, Pavelka M, et al. Induction of apoptosis in cultured hepatocytes and in the regressing liver by transforming growth factor-beta 1 occurs without activation of an endonuclease. Toxicol Lett 1992;64–65 (Spec No):701–4. [DOI] [PubMed] [Google Scholar]
- 33.Shinagawa T, Yoshioka K, Kakumu S, et al. Apoptosis in cultured rat hepatocytes: the effects of tumour necrosis factor alpha and interferon gamma. J Pathol 1991;165:247–53. [DOI] [PubMed] [Google Scholar]
- 34.Nagata S, Golstein P. The Fas death factor. Science 1995;267:1449–56. [DOI] [PubMed] [Google Scholar]
- 35.Orlinick JR, Chao W. TNF-related ligands and their receptors. Cell Signal 1998;10:543–51. [DOI] [PubMed] [Google Scholar]
- 36.Smith CA, Farrah T, Goodwin RG. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell 1994;76:959–62. [DOI] [PubMed] [Google Scholar]
- 37.Luo KX, Zhu YF, Zhang LX, et al. In situ investigation of Fas/FasL expression in chronic hepatitis B infection and related liver diseases. J Viral Hepat 1997;4:303–7. [DOI] [PubMed] [Google Scholar]
- 38.Loweth AC, Williams GT, James RF, et al. Human islets of Langerhans express Fas ligand and undergo apoptosis in response to interleukin-1 beta and Fas ligation. Diabetes 1998;47:727–32. [DOI] [PubMed] [Google Scholar]
- 39.Desbarats J, Duke RC, Newell MK. Newly discovered role for Fas ligand in the cell-cycle arrest of CD4+ T cells. Nat Med 1998;4:1377–82. [DOI] [PubMed] [Google Scholar]
- 40.Boldin MP, Goncharov TM, Goltsev YV, et al. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1and TNF receptor-induced cell death. Cell 1996;85:803–15. [DOI] [PubMed] [Google Scholar]
- 41.Buckley CD, Pilling D, Henriquez NV, et al. RGD peptides induce apoptosis by direct caspase-3 activation. Nature 1999;397:534–9. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann K, Bucher P, Tschopp J. The CARD domain: a new apoptotic signalling motif. Trends Biochem Sci 1997;22:155–6. [DOI] [PubMed] [Google Scholar]
- 43.Newmeyer DD, Farschon DM, Reed JC. Cell-free apoptosis in xenopus egg extracts: inhibition by Bcl-2 and requirement for an organelle fraction enriched in mitochondria [see comments]. Cell 1994;79:353–64. [DOI] [PubMed] [Google Scholar]
- 44.Scaife JF. The effect of lethal doses of x-irradiation on the enzymatic activity of mitochondrial cytochrome c. Can J Biochem 1966;44:433–48. [DOI] [PubMed] [Google Scholar]
- 45.Evan G, Littlewood T. A matter of life and cell death. Science 1998;281:1317–22. [DOI] [PubMed] [Google Scholar]
- 46.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science 1998;281:1322–6. [DOI] [PubMed] [Google Scholar]
- 47.Green DR, Reed JC. Mitochondria and apoptosis. Science 1998;281:1309–12. [DOI] [PubMed] [Google Scholar]
- 48.Harrington EA, Fanidi A, Evan GI. Oncogenes and cell death. Curr Opin Genet Dev 1994;4:120–9. [DOI] [PubMed] [Google Scholar]
- 49.Walker NI, Harmon BV, Gobe GC, et al. Patterns of cell death. Methods Achiev Exp Pathol 1988;13:18–54. [PubMed] [Google Scholar]
- 50.Moffatt OD, Devitt A, Bell ED, et al. Macrophage recognition of ICAM-3 on apoptotic leukocytes. J Immunol 1999;162:6800–10. [PubMed] [Google Scholar]
- 51.Savill J. Apoptosis in resolution of inflammation. J Leukoc Biol 1997;61:375–80. [DOI] [PubMed] [Google Scholar]
- 52.Chinnaiyan AM, Dixit VM. The cell-death machine. Curr Biol 1996;6:555–62. [DOI] [PubMed] [Google Scholar]
- 53.Savill J. Phagocytic docking without shocking. Nature 1998;392:442–3. [DOI] [PubMed] [Google Scholar]
- 54.Platt N, Suzuki H, Kurihara Y, et al. Role for the class A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes in vitro. Proc Natl Acad Sci U S A 1996;93:12456–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramprasad MP, Terpstra V, Kondratenko N, et al. Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc Natl Acad Sci U SA 1996;93:14833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luciani MF, Chimini G. The ATP binding cassette transporter ABCl, is required for the engulfment of corpses generated by apoptotic cell death. EMBO J 1996;15:226–35. [PMC free article] [PubMed] [Google Scholar]
- 57.Singer GG, Carrera AC, Marshak-Rothstein A, et al. Apoptosis, Fas and systemic autoimmunity: the MRL-LPR/LPR model. Curr Opin Immunol 1994;6:913–20. [DOI] [PubMed] [Google Scholar]
- 58.Adachi M, Suematsu S, Kondo T, et al. Targeted mutation in the fas gene causes hyperplasia in peripheral lymphoid organs and the liver. Nat Genet 1995;11:294–300. [DOI] [PubMed] [Google Scholar]
- 59.Stephens J, Cosyns M, Jones M, et al. Liver and bile duct pathology following Cryptosporidium parvum infection in immunodeficient mice. Hepatology 1999;30:27–35. [DOI] [PubMed] [Google Scholar]
- 60.Thomsen AR, Nansen A, Christensen JP, et al. CD40 ligand is pivotal to efficient control of virus replication in mice infected with lymphocytic choriomeningitis virus. J Immunol 1998;161:4583–90. [PubMed] [Google Scholar]
- 61.Lacronique V, Mignon A, Fabre M, et al. Bcl-2 protects from lethal hepatic apoptosis induced by an anti-Fas antibody in mice. Nat Med 1996;2:80–6. [DOI] [PubMed] [Google Scholar]
- 62.Wyllie AH. Apoptosis and carcinogenesis. Eur J Cell Biol 1997;73:189–97. [PubMed] [Google Scholar]
- 63.Biancone L, Cantaluppi V, Camussi G. CD40–CD154 interaction in experimental and human disease [review]. Int J Mol Med 1999;3:343–53. [DOI] [PubMed] [Google Scholar]
- 64.Cummings MC, Winterford CM, Walker NI. Apoptosis [see comments]. Am J Surg Pathol 1997;21:88–101. [DOI] [PubMed] [Google Scholar]
- 65.Ansari B, Coates PJ, Greenstein BD, et al. In situ end-labelling detects DNA strand breaks in apoptosis and other physiological and pathological states. J Pathol 1993;170:1–8. [DOI] [PubMed] [Google Scholar]
- 66.Mori C, Nakamura N, Okamoto Y, et al. Cytochemical identification of programmed cell death in the fusing fetal mouse palete by specific labelling of DNA fragments. Anat Embryol (Berl) 1994;190:21–8. [DOI] [PubMed] [Google Scholar]