Abstract
Aims—Human herpesvirus 8 (HHV-8) has been identified in multicentric Castleman's disease and in angioimmunoblastic lymphadenopathies. However, the presence of the virus does not necessarily indicate an aetiological role in these conditions. This study investigates the cell types infected by HHV-8 in Castleman's disease and examines the correlation between HHV-8 and Castleman's disease lymph node angiogenesis.
Methods—Sixteen formalin fixed, paraffin wax embedded samples from patients with Castleman's disease (six multicentric, 10 solitary) were examined for the presence of HHV-8 using the polymerase chain reaction (PCR), non-isotopic in situ hybridisation, PCR in situ hybridisation (PCR-ISH), and real time quantitative TaqMan PCR to HHV-8 open reading frame 26 (ORF-26), and viral (v)-cyclin encoding regions. Vascularity was assessed using CD34, CD31, and factor VIII immunocytochemistry, and lymph nodes were scored as “low” or “high”.
Results—Five multicentric Castleman's disease and two solitary Castleman's disease biopsies were positive for HHV-8. HHV-8 was identified in approximately 10% of intranodal B lymphocytes, in endothelial cells, and in subcapsular spindle cell proliferations. The copy number of HHV-8 was low at 10–50 copies/1000 cells. The highest copy number was in subcapsular spindle cells. There was no correlation between vascularity score and HHV-8 status.
Conclusion—The preferential localisation of HHV-8 in subcapsular spindle cell proliferations (where early intranodal Kaposi's sarcoma initiates) and endothelial cells in Castleman's disease might finally explain the link between intranodal Kaposi's sarcoma and Castleman's disease.
Keywords: Castleman's disease, human herpesvirus 8, Kaposi's sarcoma
Recently, human herpesvirus 8 (HHV-8) sequences have been identified in cases of multicentric Castleman's disease and in angioimmunoblastic lymphadenopathies,1–5 implicating a role for HHV-8 in the pathobiology of these conditions.
Castleman's disease is a morphologically distinct form of lymph node hyperplasia. Microscopically, there are two major categories described.6,7 The first is designated hyaline vascular type (angiofollicular), the second is the plasma cell variant.
Clinically, Castleman's disease is divided into a solitary and a multicentric type. The solitary form presents as a mass, commonly located in the mediastinum, but also found in the neck, lung, mesentery, axillary lymph nodes, peritoneum, soft tissues, and nasopharynx. Microscopically, over 90% of these cases are of the hyaline vascular type and the remainder are of the plasma cell type.
The multicentric or systemic form of Castleman's disease is nearly always of the plasma cell type. This presents as a generalised lymphadenopathy, which might involve the spleen.8–10 This is an atypical lymphoproliferative disorder, showing a polygonal lymphoid proliferation with vascular hyperplasia. The prognosis of systemic Castleman's disease is poor, with the disease tending to persist for months or years. Patients usually succumb to renal and pulmonary complications. Furthermore, some patients have coexistent Kaposi's sarcoma (18–20% of patients).11 Others develop large cell lymphomas of the immunoblastic type. Evidence of clonal rearrangement of immunoglobulin and T cell receptor genes has been found in some patients with systemic Castleman's disease, together with Epstein-Barr virus (EBV) genomes. However, no such features have been described in the solitary form of the disease.12–15 It is clear from these data that multicentric Castleman's disease differs from the classic localised type, and might eventually evolve into clonal lymphoproliferation.
In some cases of Castleman's disease of the hyaline vascular type, there might be an undue prominence of stromal elements.16 These types of lesions can occasionally acquire the features of an angiomatous hamartoma or a cellular spindle cell proliferation resembling Kaposi's sarcoma or haemangiopericytoma.17 Conceivably, some of these vascular proliferative lesions might result from the secretion of angiogenic factors by lymphoid elements. Interestingly, cutaneous and nodal forms of Kaposi's sarcoma have been reported to be associated with multicentric Castleman's disease.1
In light of previous findings of HHV-8 in multicentric Castleman's disease and angioimmunoblastic lymphadenopathy, we investigated the prevalence of HHV-8 in multicentric and solitary Castleman's disease, calculated viral copy numbers in such lesions, and identified the cell types infected by HHV-8. In addition, we examined any possible links between the degree of angiogenesis in Castleman's disease lymph nodes and the presence of HHV-8.
Materials and methods
Sixteen biopsies from lymph nodes of patients affected with Castleman's disease were examined for the presence of HHV-8. These biopsies were from three human immunodeficiency virus (HIV) positive and 13 HIV negative patients (table 1 ▶). Ten control reactive lymph nodes were also examined.
Table 1.
Epidemiology of Castleman's disease (CD) cases
| Case | Subtype of CD | Age | Sex | HIV status | HHV-8 status |
| 1 | MCD-h/p* | 30 | F | Negative | Positive |
| 2 | MCD-hv | ? | M | Negative | Positive |
| 3 | SCD-hv | 32 | M | Negative | Positive |
| 4 | MCD-h/p* | 35 | F | Positive | Positive |
| 5 | MCD-h/p* | 30 | M | Positive | Positive |
| 6 | MCD-hv | 46 | F | Negative | Negative |
| 7 | SCD-pc | 20 | M | Negative | Negative |
| 8 | SCD-pc | ? | F | Negative | Negative |
| 9 | SCD-pc | 47 | M | Negative | Negative |
| 10 | SCD-pc | ? | M | Positive | Positive |
| 11 | MCD-pc | 47 | F | Negative | Positive |
| 12 | SCD-pc | ? | M | Negative | Negative |
| 13 | SCD-hv | 39 | F | Negative | Negative |
| 14 | SCD-hv | 29 | M | Negative | Negative |
| 15 | SCD-hv | ? | M | Negative | Negative |
| 16 | SCD-hv | 50 | M | Negative | Negative |
F, female; HHV-8, human herpesvirus 8; HIV, human immunodeficiency virus; h/p, hyaline vascular and plasma cell type admixed; hv, hyaline vascular type; M, male; MCD, multicentric CD; pc, plasma cell type; SCD, solitary CD.
*Predominantly hyaline vascular in type.
HISTOLOGICAL AND IMMUNOCYTOCHEMISTRY ANALYSIS
To investigate any possible link between the presence of HHV-8 and the type of Castleman's disease or the amount of angiogenesis, the blood vessels in all Castleman's disease biopsies were stained for factor VIII, CD34, and CD31, and vascularity was scored as “low” or “high”. A biopsy score was attributed according to the number of vascular “hits” using an intercept eye piece graticule. Scoring was performed by three independent pathologists. Immunocytochemical detection was achieved using the streptavidin horseradish peroxidase (HRP) method (Dako), according to the manufacturer's recommendations.
IMMUNOCYTOCHEMISTRY FOR VASCULAR MARKERS (CD31, CD34, AND FACTOR VIII)
CD31
Sections were dewaxed in xylene and taken to water, through a series of graded alcohols. Endogenous tissue peroxidase was blocked in a solution of 3% (wt/vol) H2O2 in methanol for 25 minutes. Pretreatment was achieved using either a pressure cooking technique or protease treatment for 20 minutes.
After antigen unmasking, an anti-CD31 antibody (JC 70) diluted 1/30 in Tris buffered saline (TBS) was applied to the tissue section and allowed to interact for 30 minutes. Two washes in TBS for five minutes each were then performed. The secondary antibody (biotinylated rabbit antimouse) (Dako, Copenhagen, Denmark) diluted 1/400 in TBS (pH 7.6) was then incubated with the tissue section at room temperature. Immunocytochemical detection was achieved as described above.
CD34
No pretreatment was required for CD34 staining. The primary antibody dilution consisted of anti-CD34 (Q bend) diluted 1/100 in TBS (pH 7.6). Immunocytochemical detection was achieved as above.
Factor VIII
Pretreatment with either pressure cooking or trypsin digestion for 20 minutes was required for factor VIII staining. Neat antifactor VIII (F8B6) was used as the primary antibody. Immunocytochemical detection was achieved as above.
DETECTION OF HHV-8 BY SOLUTION PHASE POLYMERASE CHAIN REACTION (PCR)
DNA was extracted from three or four 5 μm sections of paraffin wax embedded biopsies by treatment with xylene, digestion with proteinase K, extraction with phenol/chloroform, and alcohol precipitation. The quality of the extracted DNA was controlled by PCR amplification of β actin gene fragment using the following primers: forward primer, 5′-TCACCCACACTGTGCCCATCTACGA-3′ and reverse primer, 5′-CAGCGGAACCG CTCATTGCCAATGG-3′.
For HHV-8 detection, KS4 primer (5′-AGCACTCGCAGGGCAGTACG-3′) and KS5 primer (5′-GACTCTTCGCTGATGAA CTGG-3′), derived from the putative minor capsid protein (homologue of open reading frame 26 (ORF-26) of herpesvirus saimiri) were used and 100 ng of starting DNA template was amplified for 25 cycles. To increase the sensitivity and verify the specificity of the amplicon obtained with these primers, 2 μl of amplified product was reamplified (for 25 cycles) using internal primers KS1 (5′-AGCCGAAAGGATTCCACCAT-3′) and KS2 (5′TCCGTGTTGTCTACGTCCAG -3′).18
All positive cases were confirmed by amplification with non-overlapping primers derived from the major capsid protein of HHV-8 (ORF-25): outer primers, 5′-AGGCAACGT CAGATGTGAC-3′ and 5′-GAAATTACCC ACGAGATCGC-3′; inner primers, 5′-CATG GGAGTACATTGTCAGGACCTC-3′ and 5′-GGAATTATCTCGCAGGTTGCC-3′.18
MEASUREMENT OF HHV-8 VIRAL LOAD IN CASTLEMAN'S DISEASE
HHV-8 viral load in Castleman's disease biopsies was assessed by real time TaqMan PCR using the 7700 sequence detector (PE Biosystems, Warrington, Cheshire, UK) to amplify segments within the ORF-26 and the viral (v)-cyclin encoding region of the HHV-8 genome. This newly described technique uses the 5′→3′ endonucleolytic activity of AmpliTaq DNA polymerase.19–21 The technique enables the direct detection of the PCR product by the specific release of a fluorescent reporter molecule during the PCR reaction.22 TaqMan PCR uses a primer pair (as in conventional solution phase PCR) and an internal oligoprobe, called a TaqMan probe. The TaqMan probe consists of an oligonucleotide 20–30 bases in length with a 5′ reporter dye, a 3′ quencher dye, and a 3′ blocking phosphate. The fluorescence reporter dye—for example, FAM (6-carboxy-fluoroscein), is covalently linked to the 5′ end of the oligonucleotide probe. TET (tetradichloro-6-carboxy-fluoroscein) and HEX (hexachloro-6carboxy-fluoroscein) can also be used as fluorescent reporter dyes in this system. Each of these reporters is quenched by TAMRA (6-carboxy-tetramethyl-rhodamine), which is attached by a LAN (linker arm modified nucleotide) to the 3′ end of the probe. The probe is chemically phosphorylated at its 3′ end, which prevents probe extension during PCR applications. When the probe is intact (linearised), the proximity of the reporter dye to the quencher dye results in direct suppression of the fluorescence from the reporter dye by Forster type energy transfer.23,24 During PCR, if—and only if—the target of interest is present, the probe will specifically anneal between the forward primer and the reverse primer. Owing to the nucleolytic activity of the AmpliTaq DNA polymerase, the probe is cleaved between the reporter and the quencher sequence only if the probe is hybridised to its target. The release of the fluorescent reporter only occurs if target specific amplification occurs, obviating the need to confirm the amplicon after amplification. The sensitivity of the TaqMan PCR is 1–2.5 viral copies in 106 contaminating human DNA sequences (J O'Leary et al, 1999, unpublished data). For the detection of HHV-8 triplicate samples of 100 ng of starting DNA template were used. Magnesium chloride titration assays were performed initially to identify the optimal MgCl2 concentration for the reaction. Primers and TaqMan probe were designed using the “nearest neighbour method”, which determines the optimal annealing temperature.18 The amplification mix consisted of l× PCR buffer II, 4 mM MgCl2, 200 μM dNTPs (dCTP, dATP, dGTP), 400 μM dUTP, 300 nM forward and reverse primers, 200 nM TaqMan probe, 0.025 U/μl AmpliTaq Gold DNA polymerase, and AmpErase UNG 0.01 U/μl (to prevent PCR cross contamination). The following primers were used: KS1 ORF-26, 5′-AGCCGAAGGATTCCACCAT-3′; KS2 ORF-26, 5′-TCCGTGTTGTCTA CGTCCAG-3′; TaqMan probe ORF-26, 5′-F-CGCTATTCTGCAGCAGCTGTTGG TGT ACCA-T-3′ (where F = FAM and T = TAMRA); v-cyclin 1, 5′-ACCAGTT CACTTTGCTATGCC-3′; v-cyclin 2, 5′-GC TTTTGTAATCAGGGTGTTGAC-3′; and TaqMan probe v-cyclin, 5′-F-CAGACTC CTTTTCCCGCCAAGAACTTATAG-T-3′ (where F = FAM and T = TAMRA).
The quality of amplifiable DNA was assessed by β actin PCR using the following primers: forward, 5′-TCACCCACACTGTG CCCATCTACGA-3′; reverse, 5′-CAGCGGA ACCGCTCATTGCCAATGG-3′; and TaqMan probe, 5′-F-ATGCCCTCCCCCATG CCATCCTGCGT-T-3′ (where F = FAM and T = TAMRA).
Controls without template were also performed in triplicate for ORF-26, v-cyclin, and β actin amplifications.
The following thermocycling conditions were applied: 50°C for two minutes (activation of UNG), 95°C for 10 minutes (activation of AmpliTaq Gold), 95°C for 15 seconds (denaturation), and 60°C for one minute (combined annealing and denaturation) for 40 cycles. Specific product was detected using the PE Biosystems 7700 sequence detector for real time PCR.
PCR IN SITU HYBRIDISATION (PCR-ISH) IDENTIFICATION OF HHV-8 IN CASTLEMAN'S DISEASE
PCR-ISH was used to identify the precise cell types infected in Castleman's disease.25 All PCR-ISH assays were performed in triplicate. An EBV positive post-transplant lymphoproliferative disorder specimen was also included to exclude crossreactivity with EBV.
Representative 5 μm sections from solution phase and TaqMan positive cases were cut on to APES coated slides specifically designed for PCR-ISH (PE Biosystems). Tonsil tissue was used as a negative control. The tissue was first dewaxed in xylene and cleaned with alcohol, followed by digestion in proteinase K (0.2–0.5 mg/ml) for 10–15 minutes at 37°C. Endogenous peroxidase activity was blocked by incubation in a 3% hydrogen peroxide solution (0.1% sodium azide). Slides were then washed in high performance liquid chromatography (HPLC) water and air dried.
PCR reagents used were as follows: 1 μM each primer, 200 μM dNTPs, 4.5 mM MgCl2, l× PCR buffer II (PE Biosystems) and 10 U AmpliTaq IS in a reaction volume of 25–50 μl. HHV-8 inner primers as above were used. PCR reactions were performed in the GeneAmp in situ PCR system 1000 (PE Biosystems). Slides were assembled on a dedicated assembly unit held at 70°C. A “hot start” modification was performed by heating the reaction solution to 70°C before the addition of AmpliTaq IS. The PCR reaction mix was placed over the tissue section, which was subsequently overlaid by an Ampli Cover disc (silicon rubber) and clip. Two negative controls were performed on each slide—one without PCR reagents and one with omission of AmpliTaq IS and/or primers. The following cycling conditions were used: 94°C for one minute, 94°C for 50 seconds, and 55°C for 55 seconds, for 40 cycles, followed by soaking to 4°C.
After amplification, the Ampli Cover discs and clips were removed and slides fixed in 100% ethanol for five minutes. Hybridisation was carried out with a 5′-end biotin labelled oligonucleotide probe (5′-TGTTGGTGTA CCACATCTACTCCAAAATAT-3′).
The probe was applied at a concentration of 5 pmol/100 μl hybridisation buffer, composed of 5% dextran sulphate, 2× saline sodium citrate (SSC), and 10% formamide. Slides were denatured at 94°C for 15 minutes and then hybridised for four to 12 hours at 42°C. Coverslips were removed and sections washed using different stringencies as follows: 4× SSC at 22°C for 15 minutes, 2× SSC at 30°C for 15 minutes, 2× SSC at 42°C for 15 minutes, 2× SSC at 55°C for 15 minutes, l× SSC at 55°C for 15 minutes, or 0.l× SSC at 55°C for 15 minutes, followed by a final wash in TBT (100 mM NaCl, 40 mM Tris, pH 7.2, 3% bovine serum albumin (BSA), and 0.05% Triton X-100) for 10 minutes. The intensity of the signal was dependent on the stringency of the post-hybridisation wash used.
A three step detection technique was used. Monoclonal antibiotin (Dako; diluted 1/40) was followed by biotinylated rabbit antimouse F(ab′)2 (Dako; diluted 1/200), and finally avidin/peroxidase (Dako; diluted 1/50 in 5% dried skimmed milk solution). The product was detected by AEC chromagen (Histostain-SP kit; Zymed Lab Inc, San Francisco, USA) with careful monitoring of signal development (five to 15 minutes). Simultaneous immunohistochemistry was not performed because vascular spaces were easily identified morphologically.
Control amplifications and hybridisations were carried out using a pair of β globin primers (5′-ACACAACTGTGTTCACTAGC-3′ and 5′-CAACTTCATCCACGTTCACC-3′) and a biotinylated β globin oligonucleotide probe (5′-GACTCCTGAGGAGAAGTCT GCCGTTACTGC-3′), as well as a biotinylated human papilloma virus 16 (HPV-16) 30 mer oligonucleotide probe (5′-TATTAAC TGTCAAAAGCCACTGTGTCCTGA-3′).
Labelled primer driven in situ amplification was performed on selected cases. The KS-1 primer was a 5′-end labelled 30 mer oligoprobe (sequence as for the PCR-ISH oligonucleotide probe) and we used the same reaction conditions as for PCR-ISH. After amplification the slides were held at 55°C. Stringency washes and detection steps were as for PCR-ISH.
Positive control material for in cell PCR reactions included BC-1 and BC-3 cells (lymphoblastoid cell lines containing HHV-8) and Kaposi's sarcoma biopsies. Negative tissues and cell line controls included myocardium and SiHa cells.
Results
Seven of 16 Castleman's disease (five multicentre and two solitary) cases yielded HHV-8 amplicons. Tables 1 and 2 ▶ ▶ give results for HHV-8 and Castleman's disease and relate the vascularity count to HHV-8 positivity. HHV-8 was found by PCR and TaqMan PCR in Castleman's disease of all types (but predominantly multicentric Castleman's disease) and in HIV positive and negative individuals. Three HIV associated Castleman's disease cases were positive for HHV-8, with two of these patients having coexistent multicentric Castleman's disease and cutaneous Kaposi's sarcoma. In one of these biopsies, there was evidence of microscopic Kaposi's sarcoma within the lymph node affected by Castleman's disease. Four of 13 HIV negative patients with Castleman's disease were positive for HHV-8, and one of these patients had underlying Kaposi's sarcoma. In total, two of six plasma cell variant Castleman's disease nodes, and five of 10 hyaline vascular Castleman's disease nodes were positive for HHV-8. When scored for angiogenesis, six of 11 high, and one of five low nodes were positive for HHV-8 (table 2 ▶). None of the reactive lymph nodes contained HHV-8 by either solution phase PCR or TaqMan PCR.
Table 2.
Angiogenesis and HHV-8 in Castleman's disease (CD)
| CD subtype | Vessel count | HHV-8 |
| Hyaline vascular | Low | 1/3 |
| High | 4/7 | |
| Plasma cell | Low | 0/2 |
| High | 2/4 |
HHV-8, human herpesvirus 8.
Using PCR-ISH under high and low stringency post-hybridisation washing conditions, HHV-8 amplicons were detected predominantly in lymphoid cells and occasionally in endothelial cells (in two cases) within germinal centres (figs 1–4 ▶ ▶ ▶ ▶). PCR-ISH analysis of a solution phase PCR HHV-8 negative lymph node did not reveal any amplicons (fig 1 ▶).
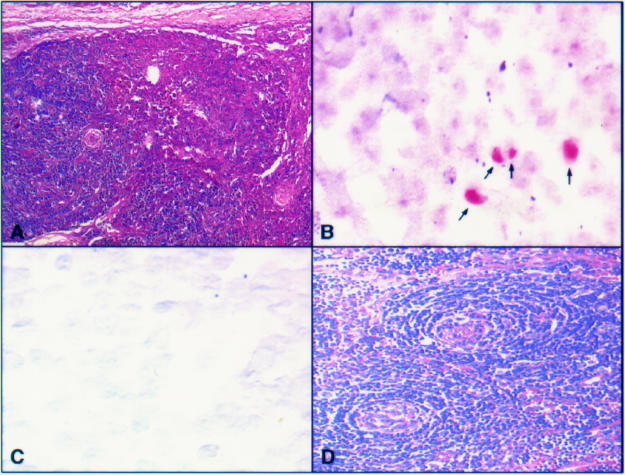
Figure 1.
(A) Haematoxylin and eosin stained section of a plasma cell variant Castleman's disease node in a human immunodeficiency virus (HIV) positive patient with a high vascular score and early intranodal Kaposi's sarcoma. (B) Human herpesvirus 8 (HHV-8) amplicons in lymphoid cells in the same patient identified using polymerase chain reaction in situ hybridisation (PCR-ISH). In addition, HHV-8 was identified in the early Kaposi's sarcoma lesion. (C) Hyaline vascular type Castleman's disease in an HIV negative patient with a high vessel count also negative for HHV-8 on solution phase PCR and negative here on PCR-ISH. (D) Haematoxylin and eosin section illustrating the high vessel count in this sample.
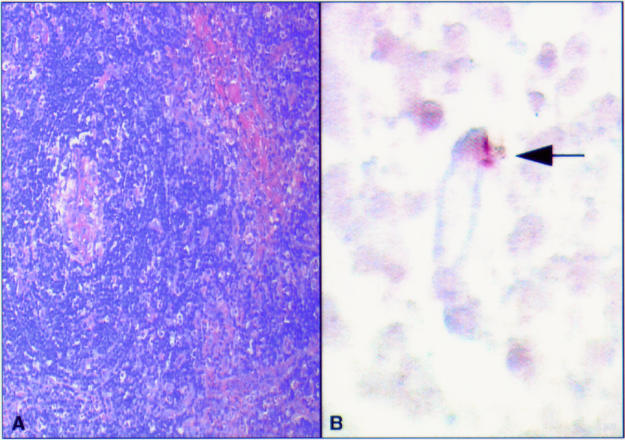
Figure 2.
(A) Haematoxylin and eosin stained section of a hyaline vascular human immunodeficiency virus (HIV) negative Castleman's disease specimen showing a central vessel. (B) Polymerase chain reaction in situ hybridisation (PCR-ISH) assay on the same area identifying human herpesvirus 8 (HHV-8) in an endothelial cell (indicated by an arrow).
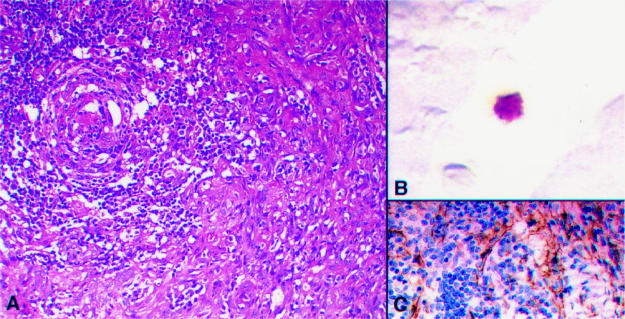
Figure 3.
(A) Haematoxylin and eosin stained section of multicentric Castleman's disease in a human immunodeficiency virus (HIV) positive patient showing a high vascular score. (B) A human herpesvirus 8 (HHV-8) infected lymphoid cell in a subcapsular space. (C) Vascular CD31 staining in the same node.
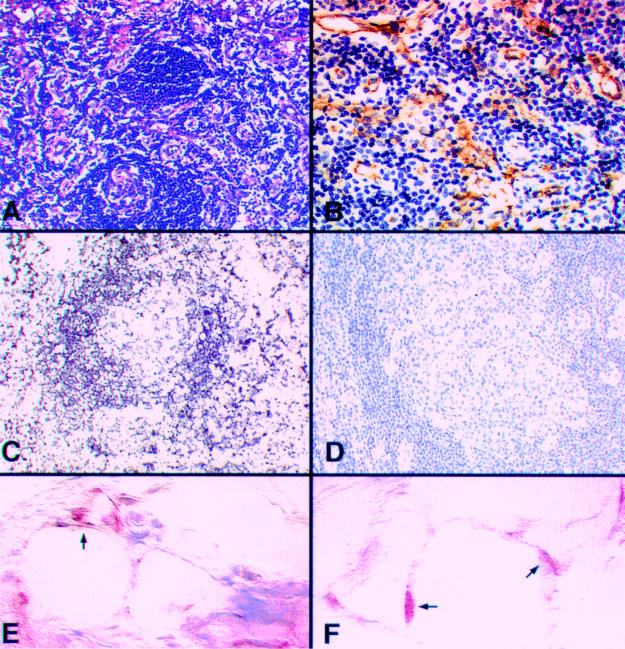
Figure 4.
(A) Haematoxylin and eosin stained section showing a hyaline vascular type Castleman's disease specimen with a high vessel count, as demonstrated by CD34 staining (B). (C) β Globin gene polymerase chain reaction in situ hybridisation (PCR-ISH) on the same specimen and (D) a parallel negative control. (E) And (F) human herpes virus 8 (HHV-8) amplicons (indicated by arrows) in spindle cells in a subcapsular location in a Castleman's disease lymph node, after microdissection of lymphoid cells. Note the suboptimal morphology after proteolytic digestion.
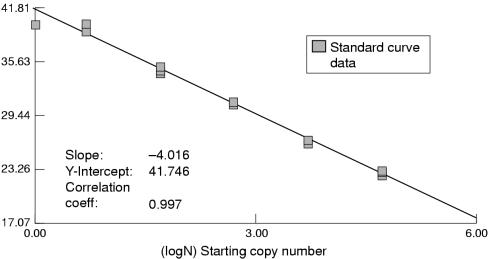
HHV-8 amplicons were detected in lymphoid cells in B cell rich areas of lymph nodes (predominantly germinal centres) and in a single lymphoid cell in an afferent subcapsular lymphatic (fig 3 ▶). In any one node, the number of positive lymphoid cells was less than 10–15% of the nodal lymphoid population. The HHV-8 copy number in each 1000 cells in the lymph node as assessed by TaqMan PCR was 10–50. Corrected for by PCR-ISH results, this represents one to three copies of HHV-8 DNA for each infected endothelial and lymphoid cell. The efficiency of TaqMan PCR can be assessed by the slope of the standard curve (generated by positive plasmid controls containing the gene of interest; fig 5 ▶). The slope here is −4.016, with a correlation coefficient of 0.997. This represents 100% efficient PCR (given the slope of the curve is 1/log(l + E), where E is the efficiency of the PCR).
Figure 5.
Standard curve for viral (v)-cyclin TaqMan PCR using plasmid standards containing the human herpes virus 8 (HHV-8) v-cyclin gene. The slope of the curve is −4.016, with a correlation coefficient of 0.997, illustrating high reproducibility of the assay.
In some cases (three), HHV-8 amplicons were found preferentially in focal areas of spindle cell proliferations beneath the capsule of the node (fig 4 ▶). To confirm the precise cell infected by HHV-8 in these areas, we microdissected out lymphoid cells. Figure 4E and F ▶ shows the localisation of HHV-8 amplicons in spindle like cells in these areas. This occurred in all of the three cases, which incidentally were scored as high vascularity. This result was reproducible on repeated investigation. One case in particular showed very weak in situ signals when the probe alone was applied, but after amplification there was a qualitative and quantitative difference in the intensity of the signal and the number of cells containing HHV-8 amplicons. We believe that this might be the result of a relatively high copy number of HHV-8 DNA (approximately 20–50 copies as assessed by microdissection assisted TaqMan PCR) in some subcapsular spindle cells; in this case, at a copy number sufficient to allow detection by the oligoprobe. However, high stringency washes abolished all signals.
Discrete endothelial amplicon localisation was also identified in two cases (fig 2 ▶) and the results were reproducible on a parallel section of the node.
Discussion
In 1995, Soulier and colleagues found HHV-8 DNA sequences in 14 of 14 cases of HIV associated multicentric Castleman's disease (which included five cases without detectable Kaposi's sarcoma). In addition, HHV-8 was detected in seven of 17 cases of multicentric Castleman's disease in HIV negative patients, including one patient with cutaneous Kaposi's sarcoma.1 HHV-8 DNA sequences were also found in lymph nodes and peripheral blood samples from these patients. In Soulier's control cohort of 34 non-multicentric Castleman's disease reactive lymph nodes (follicular and/or interfollicular hyperplasia) in HIV negative patients, HHV-8 was detected in only one case. These results suggest that HHV-8 might also play a role in the pathogenesis of multicentric Castleman's disease, especially in HIV infected patients. Alternatively, the findings might represent viral passenger positivity. These results have subsequently been confirmed by other investigators.2,3 In addition, HHV-8 DNA sequences have been identified in a related condition, angioimmunoblastic lymphadenopathy.4
In our study, HHV-8 was identified in seven of 16 cases of Castleman's disease. Interestingly, five of six multicentric cases were positive for HHV-8. The presence of HHV-8 in the Castleman's disease samples tested is not linked to a histological subtype of Castleman's disease and neither is it always seen in highly vascular cases (χ2 = 0.55873; <p < 0.50), although it is seen more often in multicentric Castleman's disease. Although one of the HIV negative, HHV-8 positive patients was considered not to have an increased risk of developing Kaposi's sarcoma (that is, did not have multicentric Castleman's disease), there is in fact an increased incidence of Kaposi's sarcoma in individuals with any type of Castleman's disease.26 The presence of HHV-8 in only a proportion of patients with Castleman's disease (43%), as described here, might suggest a reactivation of latent HHV-8 infection in such patients. The cytokine rich microenvironment of these lesions, as well as of other angiogenic lymphoproliferations, could be particularly favourable to HHV-8 replication, or to the host cells which harbour this virus. However, it is also possible that HHV-8 can help to induce angiogenesis in Castleman's disease—for example, through the production of HHV-8 viral interleukin 6 (vIL-6). Preliminary data from our own group (using peptide nucleic acid (PNA-ISH) vIL-6 probes) suggests that this might be an important mechanistic action of HHV-8 in patients with Castleman's disease. Indeed, using TaqMan PCR, HHV-8 vIL-6 transcripts are relatively abundant in Castleman's disease (J O'Leary et al, 1999, unpublished data).
PCR-ISH conveniently localises the cells infected by HHV-8 in Castleman's disease. Of importance, is the finding of HHV-8 in lymphoid cells, which is as predicted from peripheral blood assays.27 However, the localisation of HHV-8 amplicons to endothelial cells (fig 2 ▶) and to subcapsular spindle cell areas (fig 4 ▶), where a high HHV-8 copy number is seen, raises important pathobiological questions.
In lymph nodes, early Kaposi's sarcoma is usually seen in the subcapsular and trabecular sinuses, with advanced Kaposi's sarcoma effacing the lymph node architecture.26 If this is the case, then the localisation of HHV-8 amplicons at this site might not occur by chance alone, and might lead to the following sequence of events. Circulating infected HHV-8 B cells enter the lymph node through afferent lymphatics (fig 3 ▶). Here, virginal mesenchymal cells and/or endothelial cells are infected and assume a spindle cell morphology (akin to Kaposi's sarcoma development). With permissive infection, spindle cell proliferation proceeds and small foci of spindle cells appear in the subcapsular space of the node. Two events can then follow: HHV-8 is disseminated throughout the node, thereby contributing to increased vascularity (via v-IL-6 production) and Castleman's disease, or the virus may remain localised and support focal subcapsular spindle cell proliferation, leading to the development of Kaposi's sarcoma.28
The questions that now remain are: (1) are Castleman's disease and Kaposi's sarcoma inextricably linked by a viral factor, that factor being HHV-8; and (2) are some forms of Castleman's disease early intranodal Kaposi's sarcoma, as has been suggested by some clinical studies?17,29
Acknowledgments
The authors wish to acknowledge the support of the cancer research advancement group (Irish Cancer Society).
References
- 1.Soulier J, Grollet L, Oksenhendler E, et al. Kaposi's sarcoma associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 1995;86:1276–80. [PubMed] [Google Scholar]
- 2.Gessain A, Sudaka A, Briere J, et al. Kaposi's sarcoma-associated herpesvirus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association with non-human immunodeficiency virus-infected patients? Blood 1996;87:414–16. [PubMed] [Google Scholar]
- 3.Karcher DS, Alkan S. Herpes-like DNA sequences, AIDS-related tumors, and Castleman's disease. N Engl J Med 1995;333:797–8. [DOI] [PubMed] [Google Scholar]
- 4.Luppi M, Barozzi P, Maiorana A, et al. Human herpesvirus 8 DNA sequences in human immunodeficiency virus-negative angioimmunoblastic lymphadenopathy and benign lymphadenopathy with giant center hyperplasia and increased vascularity. Blood 1996;87:3903–9. [PubMed] [Google Scholar]
- 5.Frizzera G. Castleman's disease and related disorders. Semin Diagn Pathol 1988;5:346–64. [PubMed] [Google Scholar]
- 6.Keller AR, Hochholzer L, Castleman B. Hyaline-vascular and plasma-cell types of giant lymph node hyperplasia of mediastinum and other locations. Cancer 1972;29:670–83. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen DT, Diamond LW, Hansmann ML, et al. Castleman's disease. Differences in follicular dendritic network in the hyaline vascular and plasma cell variants. Histopathology 1994;24:437–43. [DOI] [PubMed] [Google Scholar]
- 8.Frizzera G, Banks PM, Massarelli G, et al. A systemic lymphoproliferative disorder with morphologic features of Castleman's disease. Pathological findings in 15 patients. Am J Surg Pathol 1983;7:211–31. [DOI] [PubMed] [Google Scholar]
- 9.Frizzera G, Peterson BA, Bayrd ED, et al. A systemic lymphoproliferative disorder with morphologic features of Castleman's disease. Clinical findings and clinicopathologic correlations in 15 patients. J Clin Oncol 1985;3:1202–16. [DOI] [PubMed] [Google Scholar]
- 10.Weisenburger DD, Nathwani BN, Winberg CD, et al. Multicentric angiofollicular lymph node hyperplasia. A clinicopathologic study of 16 cases. Hum Pathol 1985;16:162–72. [DOI] [PubMed] [Google Scholar]
- 11.Peterson BA, Frizzera G. Multicentric Castleman's disease. Semin Oncol 1993;20:636–47. [PubMed] [Google Scholar]
- 12.Hall PA, Donaghy M, Cotter FE, et al. An immunohistological and genotypic study of the plasma cell form of Castleman's disease. Histopathology 1989;14:333–46. [DOI] [PubMed] [Google Scholar]
- 13.Hanson CA, Frizzera G, Patton DF, et al. Clonal rearrangement for immunoglobulin and T-cell receptor genes in systemic Castleman's disease. Association with Epstein-Barr virus. Am J Pathol 1988;131:84–91. [PMC free article] [PubMed] [Google Scholar]
- 14.Ohyashiki JH, Ohyashiki K, Kawakubo K, et al. Molecular genetic, cytogenetic, and immunophenotypic analyses in Castleman's disease of the plasma cell type. Am J Clin Pathol 1994;101:290–5. [DOI] [PubMed] [Google Scholar]
- 15.Radaszkiewicz T, Hansmann ML, Lennert K. Monoclonality and polyclonality of plasma cells in Castleman's disease of the plasma cell variant. Histopathology 1989;14:11–24. [DOI] [PubMed] [Google Scholar]
- 16.Danon AD, Krishnan J, Frizzera G. Morpho-immunophenotypic diversity of Castleman's disease, hyaline-vascular type. With emphasis on a stroma-rich variant and a new pathogenetic hypothesis. Virchows Arch A Pathol Anat Histopathol 1993;423:369–82. [DOI] [PubMed] [Google Scholar]
- 17.Rywlin AM, Rosen 1, Cabello B. Coexistence of Castleman's disease and Kaposi's sarcoma. Report of a case and speculation. Am J Dermatopathol 1983;5:277–81. [DOI] [PubMed] [Google Scholar]
- 18.Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res 1990;18:6409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawyer FC, Stoffel S, Saiki RK, et al. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from the extreme thermophile, Thermus aquaticus. J Biol Chem 1989;264:6427–37. [PubMed] [Google Scholar]
- 20.Holland PM, Abramson RD, Watson R, et al. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A 1991;88:7276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyamichev V, Brow MAD, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science 1993;260:778–83. [DOI] [PubMed] [Google Scholar]
- 22.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res 1993;21:3761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forster VTH. Zwischennolekulare Energie-Wanderung und Fluoreszenz. Annals of Physics (Leipzig) 1948;2:55–75. [Google Scholar]
- 24.Lakowicz JR. Energy transfer. In: Principles of fluorescent spectroscopy. New York: Plenum Press, 1983:303–39.
- 25.Boshoff C, Schulz TF, Kennedy MM, et al. Kaposi's sarcoma associated herpes virus (KSHV) infects endothelial and spindle cells in KS. Nat Med 1995;1:1274–8. [DOI] [PubMed] [Google Scholar]
- 26.Rosai J. Ackerman's surgical pathology, 8th ed. St Louis: Mosby, 1996.
- 27.Whitby D, Howard M, Tenant-Flowers M, et al. Detection of KSHV in peripheral blood of HIV-infected individuals predicts progression to Kaposi's sarcoma. Lancet 1995;346:799–802. [DOI] [PubMed] [Google Scholar]
- 28.Parravinci C, Corbellino M, Paulli M, et al. Expression of a virus-derived cytokine KSHV vIL-6 in HIV seronegative Castleman's disease. Am J Pathol 1997;151:1517–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler E, Beer R. Multicentric giant lymph node hyperplasia clinically stimulating angioimmunoblastic lymphadenopathy: associated Kaposi's sarcoma in two of three cases. Isr J Med Sci 1983;19:230–4. [PubMed] [Google Scholar]