Abstract
The association of Epstein-Barr virus (EBV) with various malignancies is well established but the pattern of EBV latent gene expression in these different tumours is variable, reflecting distinct aspects of the virus–cell interaction. These different forms of EBV latency are associated with phenotypic variation and highlight the influence of EBV latent proteins on cell growth and survival. The EBV latent proteins have distinct functions associated with the maintenance of EBV infection and the control of various signalling and transcriptional pathways that facilitate the proliferation and survival of infected cells. Understanding the function of these EBV latent proteins will not only provide insight into the mechanisms governing fundamental cell processes but will also identify targets for novel treatment.
Keywords: Epstein-Barr virus, EBV latent genes
Previous chapters in this volume have considered the role of Epstein-Barr virus (EBV) in the aetiology of nasopharyngeal carcinoma and Hodgkin's disease, but this is not the whole story. EBV was originally discovered in the tumour cells of Burkitt's lymphoma, an undifferentiated B cell tumour, and is also implicated in the development of immunoblastic B cell lymphomas in immunocompromised patients.1 These tumours highlight the B lymphotropic nature of EBV, which is further demonstrated by its unique ability to immortalise normal resting B cells in vitro, converting them into permanently growing lymphoblastoid cell lines (LCLs).2 When peripheral blood lymphocytes from chronic virus carriers are placed in culture, the few EBV infected B cells that are present regularly give rise to a spontaneous outgrowth of EBV transformed LCLs, as long as immune T cells are either removed or inhibited by the addition of cyclosporin A to the culture.3 This phenomenon highlights the importance of EBV specific cytotoxic T lymphocytes (CTLs) in controlling EBV induced B cell transformation in vivo. Certain tumours of T cell origin have also been found to harbour EBV. Nasal T cell lymphoma, a tumour that is more common in the Far East, is invariably EBV positive, whereas around 20% of T cell lymphomas arising at other sites (such as the gastrointestinal tract, lung, and lymph nodes) are associated with EBV.1,4 Often EBV infection is found in only a proportion (5–50%) of the tumour cells, implying that EBV infection might have occurred subsequent to tumour development.4 Although the association of EBV with a proportion of gastric adenocarcinomas is now well accepted (see K Takada in this volume), reports of EBV infection in breast5 and cervical6 carcinoma are less convincing and require corroborative studies. Finally, EBV has also been detected in rare smooth muscle tumours (leiomyosarcomas) occurring in immunocompromised young people.7,8 This article will first consider the differing patterns of EBV latent gene expression in virus associated tumours and then review the current understanding of EBV latent gene function.
EBV latency in vitro and in vivo
The best characterised in vitro model of latency is provided by LCLs generated by infecting primary, resting B cells with EBV. Every cell in an LCL carries multiple copies of the viral episome and constitutively expresses a limited set of viral gene products, the so called latent proteins, consisting of six nuclear antigens (EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA leader protein (EBNA-LP)) and three latent membrane proteins (LMP1, LMP2A, and LMP2B).2 Transcripts from the BamHI A region of the viral genome, first identified in nasopharyngeal carcinoma cells, are also detected in LCLs, although the ability of these mRNAs to encode proteins remains controversial.9,10 In addition to the latent proteins, LCLs also show abundant expression of the small non-polyadenylated (and therefore non-coding) RNAs, EBERs 1 and 2; the function of these transcripts is not clear but they are consistently expressed in all forms of latent EBV infection.1,2 The relative positions and orientations of these viral genes are illustrated in fig 1 ▶ below a linearised BamHI restriction map of the viral genome. The different EBNAs are encoded by individual mRNAs generated by differential splicing of the same long “rightward” primary transcript expressed from one of two promoters (Cp and Wp), located close together in the BamHI C and W region of the genome.11 These are illustrated in fig 2 ▶ on the large (172 kb), covalently closed EBV episome. The LMP transcripts are expressed from separate promoters in the BamHI N region of the EBV genome, with the leftward LMP1 and rightward LMP2B mRNAs apparently controlled by the same bidirectional promoter sequence (fig 2 ▶).2,11 This pattern of latent EBV gene expression is referred to as the “latency III” (Lat III) form of EBV infection (fig 3 ▶).
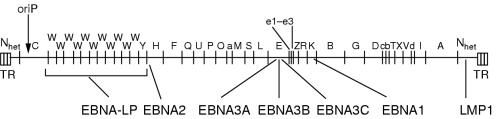
Figure 1.
Location of open reading frames for the Epstein-Barr virus (EBV) latent proteins on the BamHI restriction map of the prototype B95.8 EBV genome. The BamHI fragments are named according to size, with A being the largest. Note that the latent membrane protein 2 (LMP2) proteins are produced from mRNAs that splice across the terminal repeats (TR) in the circularised EBV genome. EBNA, EBV encoded nuclear antigen.
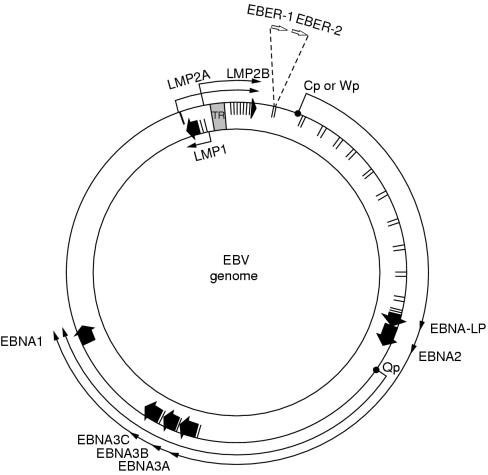
Figure 2.
Location and transcription of the Epstein-Barr virus (EBV) latent genes on the double stranded viral DNA episome. The large solid arrows represent coding exons for each of the latent proteins and the direction in which they are transcribed. EBNA-LP is transcribed from variable numbers of repetitive exons in the BamHI W fragments. Latent membrane protein 2 (LMP2) is composed of multiple exons located either side of the terminal repeat (TR) region, which is formed during the circularisation of the linear DNA to produce the viral episome. The open arrows represent the highly transcribed non-polyadenylated RNAs, EBER1 and EBER2, which are a consistent feature of latent EBV infection. The outer long arrowed line represents EBV transcription in latency type III, where all the EBNAs are transcribed from either the Cp or Wp promoter; the different EBNAs are encoded by individual mRNAs generated by differential splicing of the same long primary transcript. The inner shorter arrowed line represents the EBNA1 transcript originating from the Qp promoter located in the BamHI Q region; this is transcribed in latency types I and II. EBNA, EBV encoded nuclear antigen; LP, leader proteins.
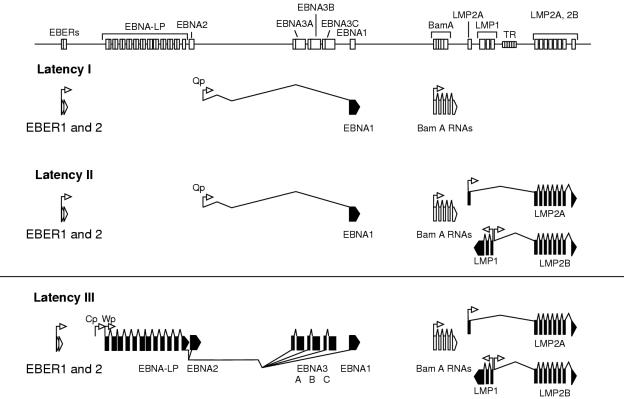
Figure 3.
Epstein-Barr virus (EBV) gene transcription in the three forms of latency. The top panel shows the position of the exons on a linear map of the genome. The lower panels show the direction of transcription from each promoter (arrows) and the splicing structure between the exons. Coding exons are shown in black and non-coding exons in white. EBER, EBV encoded small RNA; EBNA, EBV encoded nuclear antigen; LP, leader protein; LMP, latent membrane protein; TR, terminal repeat.
The consistent pattern of EBV latent protein expression in LCLs is matched by an equally consistent and characteristic cellular phenotype, with a high degree of expression of the B cell activation markers CD23, CD30, CD39, and CD70, and of the cellular adhesion molecules leucocyte function antigen 1 (LFA1; CD11a/18), LFA3 (CD58), and intercellular adhesion molecule 1 (ICAM1; CD54).12,13 That these markers are either absent or expressed at low levels on resting B cells, but are transiently induced to high levels when these cells are activated into short term growth by antigenic or mitogenic stimulation, suggests that EBV induced immortalisation might be elicited through the constitutive activation of the same cellular pathways that drive physiological B cell proliferation. The ability of EBNA2, EBNA3C, and LMP1 to induce LCL-like phenotypic changes when expressed individually in human B cell lines implicates these viral proteins as key effectors of the immortalisation process.14,15
The cell surface phenotype of immunoblastic lymphomas that develop in solid organ or bone marrow transplant recipients resembles that of LCLs, with high expression of the activation antigen CD23 and of the adhesion molecules, LFA1, LFA3, and ICAM1.1,4,16,17 This cellular phenotype is accompanied by a Lat III pattern of EBV latent protein expression, as assessed by immunohistological analysis using monoclonal antibodies to EBNA2 and LMP1 and by immunoblotting analysis with polyvalent human sera.1,4,16 Thus, these lymphomas appear to represent the in vivo counterparts of in vitro immortalised LCLs and, by implication, are likely to be primarily driven by EBV. The LCL-like nature of the immunoblastic B cell lymphomas and their growth in immunosuppressed patients suggests that these tumours are sensitive to EBV specific CTL control. Indeed, the regression of lymphomas has been reported after the relaxation of immunosuppressive treatment in transplant recipients, and recent studies have demonstrated the clinical benefit of adoptive treatment using EBV specific CTLs even in patients with overt lymphoma.1,18 Although a role for secondary genetic events in the pathogenesis of these tumours has been proposed to account for the documented progression of immunoblastic B cell lymphomas from oligoclonality to monoclonality, there is in fact no evidence of consistent genetic or cytogenetic changes in the monoclonal lesions. The likelihood that EBV remains solely responsible for lymphoma growth even after progression to monoclonality is consistent with a phenomenon regularly observed in vitro, where the EBV immortalisation of resting B cells gives rise to an LCL that is initially polyclonal, but which on serial passage becomes dominated by the fastest growing clone. Further support for this view comes from studies of EBV induced B cell lymphomas in animal model systems. Thus, the lymphomas induced in cotton top tamarins within a few weeks of experimental EBV infection,19 and in severe combined immunodeficient (SCID) mice by the inoculation of peripheral blood lymphocytes from healthy EBV carrying individuals,20 can be oligoclonal or monoclonal, yet they all resemble LCLs in their Lat III pattern of EBV latent gene expression and in their cell surface phenotype. The remarkable efficiency of tumour development in both models is strong circumstantial evidence that the development of immunoblastic lymphomas in an immunosuppressed setting in humans only requires EBV induced B cell transformation, with no necessity for secondary genetic change. This clearly sets these particular lesions apart from all other EBV positive malignancies, where viral infection is but one event in a complex multistep oncogenic process.
A second form of EBV infection in B cells, referred to as “latency I” (Lat I), has been identified in Burkitt's lymphoma tumour biopsy cells and in early passage Burkitt's lymphoma cell lines, where abundant EBER transcription is found and EBNA1 is selectively expressed in the absence of the other EBNA and LMP proteins (fig 3 ▶).12,21 The selective expression of EBNA1 involves a different mRNA expressed from a novel EBNA1 promoter (Qp) in the BamHI Q region of the viral genome, which is independent of the Cp or Wp promoters.22,23 In culture, Burkitt's lymphoma cells grow as a carpet of dispersed cells, in contrast to the multicellular aggregates that are seen in LCL cultures.1 Furthermore, Burkitt's lymphoma cells display a distinct cell surface marker phenotype characterised by expression of CD10 (CALLA) and CD77 (BLA), but little or no expression of the cellular activation antigens and adhesion molecules that are regularly found in high amounts in LCLs.21 The Lat I form of latency seen in Burkitt's lymphoma cell lines is not always stably maintained in vitro, and on serial passage a drift to a Lat III pattern of gene expression can be observed concomitant with a change in the cellular phenotype towards that seen in LCLs.21 A similar effect might occur in vivo because recent work has shown that EBNA2 and LMP1 can occasionally be detected in a small proportion of Burkitt's lymphoma cells in biopsy material.24 This indicates that the operational definitions of EBV latencies derived from cell lines in vitro might not readily apply to tumours in vivo.
Another form of EBV latency, Lat II, is characterised by selective expression of the Qp driven EBNA1 mRNA, of the LMP1, LMP2A, and LMP2B transcripts, and of the EBERs (fig 3 ▶).25,26 This form of infection was first identified at the protein level in nasopharyngeal carcinoma biopsies,27,28 but is clearly not restricted to epithelial cells because it is also seen in EBV positive cases of Hodgkin's disease and in certain EBV positive T cell lymphomas.1,29,30 All three forms of EBV latency can be interconverted in somatic cell hybrids between LCLs and either Burkitt's lymphoma cells or certain non-lymphoid lines.31 These transitions are influenced by the cell phenotype of the resultant hybrids, thus emphasising the complex interplay between cellular factors and the resident pattern of EBV latent gene expression. The intricate interaction between the host cell environment and the virus is elegantly depicted in a study in which deregulated c-myc expression introduced into an LCL with regulatable EBNA2 expression resulted in cells with a Burkitt's lymphoma phenotype (growth as single cells, downregulation of activation antigens, and upregulation of CD10) and a Lat I form of EBV latency.32 Table 1 ▶ summarises the different patterns of EBV gene expression in EBV associated tumours.
Table 1.
Viral gene expression in Epstein-Barr virus (EBV) associated tumours
| Tumour | Latent gene expression | Pattern of latency |
| BL, endemic | EBNA1 | I |
| BL, sporadic/AIDS | EBNA1 | I |
| Undifferentiated NPC | EBNA1; LMPs 1, 2 | II |
| Hodgkin's disease | EBNA1; LMPs 1, 2 | II |
| T lymphomas | EBNA1; LMPs 1, 2 | II |
| Immunoblastic lymphomas | EBNAs 1–6; | III |
| LMPs 1, 2 | III |
Note that all EBV positive tumours express the EBER RNAs and the BamHIA transcripts. Preliminary work suggests that another form of latency exists with expression of EBNA1 and LMP2 only, and this is often seen in NPC and gastric adenocarcinomas.
BL, Burkitt's lymphoma; EBNA, EBV encoded nuclear antigen; LMP, latent membrane protein; NPC, nasopharangeal carcinoma.
There are two major types of EBV isolate, originally referred to as A and B and now called types 1 and 2, which appear to be identical over the bulk of the EBV genome but show allelic polymorphism (with 50–80% sequence homology depending on the locus) in a subset of latent genes, namely those encoding EBNA-LP, EBNA2, EBNA3A, EBNA3B, and EBNA3C.1,33–35 A combination of virus isolation and seroepidemiological studies suggests that type 1 virus isolates are predominant (but not exclusively so) in many Western countries, whereas both types are widespread in equatorial Africa, New Guinea, and perhaps certain other regions.36–39 In vitro studies suggest that type 1 isolates are more potent than type 2 in achieving B cell transformation in vitro; the type 2 virus transformed LCLs characteristically show much slower growth, especially in early passage.40 In addition to this broad distinction between EBV types 1 and 2, there is also minor heterogeneity within each virus type, which is most easily detected as variation in the size of the EBNA proteins.1 These differences have been used to trace virus transmission within families and from transplant donors to recipients. The balance of evidence to date suggests that healthy individuals are only infected with one virus type, although this changes in immunologically compromised patients, where multiple EBV types including type 1 and type 2 strains can be detected in the same individual.1
As to the preferential association of EBV strains with virus associated tumours, several studies have shown that the presence of a particular virus strain in the tumour reflects the prevalence of this strain in the same geographical location. For instance, original work demonstrated that around 20% of Burkitt's lymphoma tumours from the endemic areas of Kenya and New Guinea were infected with a type 2 virus and that this reflected the 20% incidence of type 2 virus infection in normal, healthy individuals from these regions.37 More recent work focusing on genetic variation in LMP1 (particularly a 30 bp deletion referred to as delLMP1) has produced confusing data suggesting an association of these alterations with more aggressive disease. However, more extensive analysis in relation to the normal population revealed that the EBV gene polymorphisms (including delLMP1) in virus associated tumours occurred with similar frequency in EBV isolates from healthy virus carriers from the same geographical location.41 Nevertheless, the increased incidence of EBV isolates carrying delLMP1 in the normal Chinese population might be a factor in the increased incidence of nasopharyngeal carcinoma and various T cell tumours in this region.
Function of the EBV latent proteins in cell transformation
The recent use of recombinant EBVs lacking individual latent genes has confirmed the absolute requirement for EBNA2 and LMP1 in the in vitro transformation of B cells and highlighted a role for EBNA-LP, EBNA3A, EBNA3C, and LMP2A in this process.2 These studies confirm that the transformation of B cells by EBV involves the coordinated action of several latent gene functions. With the demonstration of more restricted patterns of EBV gene expression involving LMP1 and LMP2 in nasopharyngeal carcinoma, Hodgkin's disease, and various T cell lymphomas, the function of these membrane proteins has been a focus of much interest.
EBNA1
EBNA1 is a DNA binding nuclear phosphoprotein that has a central role in the maintenance of latent EBV infection.2 It is required for the replication and maintenance of the episomal EBV genome, which is achieved through the binding of EBNA1 to the plasmid origin of viral replication, oriP.2 EBNA1 can also interact with two sites immediately downstream of Qp, the promoter used to drive EBNA1 expression in Lat I and Lat II, thereby negatively regulating its own expression.23 Furthermore, EBNA1 can act as a transcriptional transactivator and has been shown to upregulate Cp and the LMP1 promoter.2 The EBNA1 protein is separated into N-terminal and C-terminal domains by a glycine-glycine-alanine (gly-ala) repeat sequence, which varies in size in different EBV isolates.1,2 This gly-ala repeat domain is a cis-acting inhibitor of major histocompatibility complex (MHC) class I restricted presentation and appears to function by inhibiting antigen processing via the ubiquitin–proteosome pathway.42,43 This effect is also likely to be responsible for the long half life of the EBNA1 protein.43 Directing EBNA1 expression to B cells in transgenic mice results in B cell lymphomas, suggesting that EBNA1 might have a direct role in oncogenesis.44 Previous work has shown that stable EBNA1 expression in epithelial cells requires an undifferentiated cellular environment,45 and that EBNA1 expression can be toxic in certain cell lines (V Robinson et al, unpublished data, 2000). This suggests that EBNA1 might have additional effects beyond its oriP maintenance function, possibly by affecting the origins of replication of cellular genes.
THE OTHER EBNAS
The inability of an EBV strain, P3HR-1, carrying a deletion of the EBNA2 gene and the last two exons of EBNA-LP to transform B cells in vitro was the first indication of the crucial role of the EBNA2 protein in the transformation process.2 The restoration of the EBNA2 gene into P3HR-1 by homologous recombination has unequivocally confirmed the importance of EBNA2 in B cell transformation, and has allowed the functionally relevant domains of the EBNA2 protein to be identified.46,47 EBNA2 is an acidic phosphoprotein that localises in large nuclear granules. An N-terminal polyproline repeat is responsible for the variation in the size of EBNA2 protein in different EBV isolates. EBNA2 is a transcriptional activator of both cellular and viral genes, upregulating the expression of certain B cell antigens, CD21 and CD23, as well as LMP1 and LMP2.2,14,15 EBNA2 also transactivates the viral C promoter (Cp), thereby inducing the switch from Wp to Cp seen early in B cell infection. These EBNA2 responsive promoters have been analysed extensively and have been found to possess a common core sequence (GTGGGAA), which does not directly bind EBNA2. Thus, EBNA2 interacts with a ubiquitous DNA binding protein, RBP-Jκ, and this is partly responsible for targeting EBNA2 to promoters that contain the RBP-Jκ sequence.48–51 Interestingly, the RBP-Jκ homologue in drosophila is involved in signal transduction via the Notch receptor, a pathway that is important in cell fate determination in the fruit fly and has been implicated in the development of T cell tumours in humans.52 Recent work demonstrates that EBNA2 can functionally replace the intracellular region of Notch,53 and activated mouse Notch1 transactivates EBNA2 regulated viral promoters.54,55 The transactivation of genes by EBNA2 also involves PU.1, a transcription factor involved in B cell specific gene transcription, thereby accounting for the ability of EBNA2 to induce LMP1 expression only in B cells. Recent work also demonstrates that an important target of EBNA2 is the c-myc oncogene, and that this effect is important for EBV induced B cell proliferation.56,57
The EBNA3A, EBNA3B, and EBNA3C genes appear to have a common origin and encode hydrophilic nuclear proteins that contain heptad repeats of leucine, isoleucine, or valine, which might act as dimerisation domains.2 Studies with EBV recombinants have shown that EBNA3A and EBNA3C are essential for B cell transformation in vitro, whereas EBNA3B is dispensable.58 Several lines of evidence suggest that members of the EBNA3 family are transcriptional regulators. Thus, EBNA3C can induce the upregulation of both cellular (CD21) and viral (LMP1) gene expression,15,59 repress the Cp promoter,60 and might interact with the retinoblastoma protein (pRb) to promote transformation.61 Although not essential for transformation, EBNA3B has been shown to induce the expression of vimentin and CD40.62 The EBNA3 proteins associate with the RBP-Jκ transcription factor and disrupt its binding to the cognate Jκ sequence and to EBNA2, thus repressing EBNA2 mediated transactivation.58 Recent work shows that EBNA3C interacts with human histone deacetylase 1 (HDAC1) and that this interaction contributes to the transcriptional repression of Cp by RBP-Jκ.63 Thus, the EBNA2 and EBNA3 proteins work together to control RBP-Jκ activity precisely, thereby regulating the expression of cellular and viral promoters containing Jκ cognate sequences.
EBNA-LP is encoded by the leader of each of the EBNA mRNAs and the protein product is of variable size depending on the number of BamHIW repeats contained by a particular EBV isolate.2 Molecular genetic analysis indicates that although not absolutely required for B cell transformation in vitro, EBNA-LP is required for the efficient outgrowth of LCLs.64 Transient transfection of EBNA-LP and EBNA2 into primary B cells induces the G0 to G1 transition as measured by the upregulation of cyclin D2 expression.65 EBNA-LP has been shown to colocalise with pRb in LCLs and in vitro biochemical studies have demonstrated an interaction of EBNA-LP with both pRb and p53.66,67 However, this interaction has not been verified in LCLs and, unlike the situation with the human papillomavirus encoded E6/E7 and adenovirus E1 proteins, EBNA-LP expression appears to have no effect on the regulation of the pRb and p53 pathways. The ability of EBNA-LP to enhance EBNA2 mediated transactivation68,69 suggests a crucial role for EBNA-LP in EBV induced B cell transformation.
LMP1
The expression of LMP1 in several EBV associated tumours including immunoblastic lymphoma, Hodgkin's disease, and nasopharyngeal carcinoma is consistent with its ability to transform rodent fibroblasts, to induce the upregulation of CD40, CD54, and Bcl-2 family members, and to inhibit epithelial differentiation.1,2,70 LMP1 comprises a short 23 amino acid N-terminal cytoplasmic domain, six putative membrane spanning domains that are important for conferring plasma membrane aggregation, and a large 200 amino acid C-terminal cytoplasmic domain.2 Mutational analysis has identified the cytoplasmic C-terminal domain of LMP1 as being important for cell growth transformation and the induction of phenotypic changes.2 Furthermore, a recombinant EBV lacking LMP1 is unable to transform B cells.71 Similarities between the effects of CD40, a member of the tumour necrosis factor (TNF) receptor family, and LMP1 signalling suggest that common biochemical pathways might be activated by these molecules. Both CD40 ligation and transient LMP1 expression are associated with activation of the transcription factor NF-κB, upregulation of CD54, secretion of interleukin 6 (IL-6), induction of the antiapoptotic A20 protein, and growth inhibition.2,72–74 More recent work shows that LMP1 can partially mimic CD40 signals in CD40 knockout mice.75 Thus, it is not surprising that LMP1 has been shown to function as a constitutively activated CD40 by interacting with a common family of TNF receptor associated factors (TRAFs).76 The six transmembrane spanning domains of LMP1 serve to promote aggregation of the protein in the plasma membrane, thereby mimicking the receptor crosslinking effect induced by the interaction of trimeric CD40 ligand with its receptor.77,78 The resultant clustering of the LMP1 cytoplasmic domain stimulates TRAF mediated NF-κB activation, as does clustering of the CD40 cytoplasmic tail induced by ligand binding.79 A TRAF binding motif, PxQxT, is present in a region of LMP1 proximal to the plasma membrane (the so called C-terminal activating region 1 or CTAR1, residues 194–232) and is responsible for mediating TRAF2 dependent activation of NF-κB.73,80–82 TRAF1, TRAF2, TRAF3, and TRAF5 directly associate with LMP1 through this PxQxT domain (TRAF binding domain), whereas TRAF6 does not bind to LMP1.82–86 A more distal region of the LMP1 cytoplasmic tail (CTAR2, residues 351–386) is the dominant NF-κB activating domain of LMP1, and this effect appears to be mediated indirectly via TRAF2; that is, TRAF2 is unable to bind to CTAR2 but dominant negative (RING finger deleted) TRAF2 blocks NF-κB activation from this region.73,80–82 It has recently been shown that TRADD (TNF receptor associated death domain), a death domain protein that interacts with TNF receptor I (TNFRI), directly associates with CTAR2 and probably recruits TRAF2, resulting in NF-κB activation.87 The precise mechanism responsible for TRAF induced NF-κB activation is unknown, but appears to involve a mitogen activated protein (MAP) kinase kinase kinase, NIK, which might function in the phosphorylation of I-κB, leading to the translocation of active NF-κB to the nucleus.88–90 The relative contribution of CTAR1 and CTAR2 to the ability of LMP1 to transform B cells has been examined using recombinant EBV. These studies show that whereas CTAR1 is essential for transformation,87 the last 155 amino acids of the LMP1 C-terminus, which constitute CTAR2, are dispensable, although this region is important for B cell growth at low density and for a reduction in the dependence on paracrine growth factors.91
Several proteins exert a negative regulatory effect on NF-κB activation by either competing for the TRAF binding domain (TRAF3) or by interacting with and inhibiting TRAF2 function (TANK/I-TRAF, A20).73,79,82,92 TRAF mediated NF-κB activation in response to either LMP1 expression or CD40 ligation has been shown to result in the induction of A20 expression and IL-6 secretion, and TRAF3 has been implicated in LMP1 and CD40 induced epithelial cell growth inhibition.72,73,93,94 CD40 ligation also results in the activation of c-Jun N-terminal kinase (JNK, also known as the stress activated protein kinase (SAPK)) leading to activation of the AP-1 transcription factor family.95,96 Recent work has shown that LMP1 can also activate JNK, and that this effect is mediated by CTAR2 and involves both TRADD and TRAF2.90,96,97 The p38 stress activated kinase is also activated by LMP1, predominantly through CTAR2, and this pathway (via activation of the AP-1 and ATF-2 transcription factors) mediates LMP1 induced upregulation of IL-8 synthesis and secretion.98 Thus, LMP1 expression results in the activation of both the NF-κB and AP-1/ATF-2 transcription factors, although the precise cell signalling pathways responsible for eliciting these effects remain unknown. The ability of the NF-κB pathway to mediate the transcription of antiapoptotic genes,99 and the role of the JNK–p38 pathway in both apoptosis and oncogenic transformation,100 suggests that these effects are responsible for the pleiotropic consequences of LMP1 expression. Other effects of LMP1 include the activation of STAT (signal transducers and activators of transcription) proteins by the interaction of the LMP1 repeat region with Janus kinase 3 (JAK3)101 and activation of the small GTPase, cdc42, with concomitant induction of filopodia,102 but the contribution of these effects to EBV induced transformation remains unknown. Figure 4 ▶ is a summary of LMP1 signalling pathways.
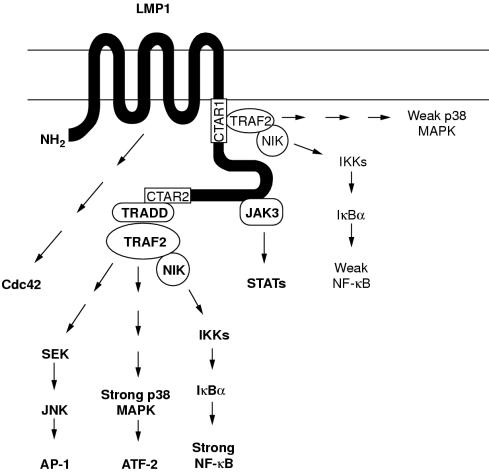
Figure 4.
Schematic representation of the known signalling pathways activated by Epstein-Barr virus (EBV) encoded latent membrane protein 1 (LMP1). The LMP1 cytoplasmic tail contains two functional domains with respect to NF-κB activation (shown as white boxes). The extreme C-terminal domain (CTAR2, aa 352–386) binds TRADD and RIP and is the major mediator of NF-κB, JNK, and p38 signalling in most cell lines. TRAF2, a TRADD interacting protein, regulates CTAR2 induced NF-κB activation via a NIK→IKK→IκBα cascade, but the components of JNK and p38 signalling downstream of TRADD/TRAF2 remain largely unknown. The membrane proximal region (CTAR1, aa 187–231), which is crucial for B cell transformation, interacts weakly with TRAF2 and induces only low amounts of NF-κB and p38 activation. TRAF1 and TRAF3 also bind CTAR1 and might influence TRAF2 mediated signals. The intermediate region between CTAR1 and CTAR2 has been shown to bind JAK3 and activate STAT signalling, whereas the transmembrane domains of LMP1 mediate activation of the small GTPase, Cdc42, leading to cytoskeletal changes. CTAR, C-terminal activating region; IKK, IκB kinase; JAK, Janus kinase; JNK, c-Jun N-terminal kinase; MAPK, mitogen activated protein kinase; NIK, MAPK kinase kinase; RIP, receptor interacting protein; SEK, extracellular signal related kinase (ERK) kinase; STAT, signal transducers and activators of transcription; TRADD, tumour necrosis factor receptor associated death domain; TRAF, tumour necrosis factor receptor associated factor.
The cloning and sequencing of the LMP1 gene from EBV isolates derived from either a Chinese or a Taiwanese nasopharyngeal carcinoma has identified several mutations in comparison with the prototype B95.8 strain, including a point mutation leading to the loss of an XhoI restriction site in the first exon, a 30 bp deletion in the C-terminus immediately upstream of CTAR2, and multiple point mutations.103,104 These so called delLMP1 variants (typified by Cao-LMP1) display increased tumorigenicity in rodent fibroblasts and epithelial cells.104–106 Although initial studies using the polymerase chain reaction suggested that EBV isolates carrying delLMP1 are more frequently detected in lymphoproliferative disorders, a more thorough analysis has shown that the incidence of delLMP1 in EBV associated tumours reflects the frequency with which this isolate in detected in healthy virus carriers from the same geographical region.41 However, given that delLMP1 is the predominant form of LMP1 in Chinese populations it is possible that this variant contributes to the development of nasopharyngeal carcinoma. A recently published functional analysis has revealed that Cao-LMP1 is impaired in its ability to upregulate CD40 and CD54 relative to B95.8-LMP1, even though Cao-LMP1 can induce greater activation of NF-κB than B95.8-LMP1.107 These studies concluded that the 30 bp deletion was not responsible for these differences and that sequences outside CTAR2 were involved. Similar studies using a delLMP1 isolated from a different nasopharyngeal carcinoma (C15) have shown that this LMP1 isolate is also more efficient at activating NF-κB than B95.8-LMP1, with resultant enhanced induction of the epidermal growth factor receptor in the C33A carcinoma cell line; these effects of C15-LMP1 were not a result of the 30 bp deletion.108 We have shown that transient expression of Cao-LMP1 results in JNK activation,96 but our recent studies suggest that Cao-LMP1 is impaired in its ability to induce various phenotypic changes in the SCC12F epithelial cell line.109 Thus, continued study of delLMP1 might help to elucidate the LMP1 signalling pathway, and to assess the contribution of LMP1 sequence variation to the pathogenesis of EBV associated tumours, such as Hodgkin's disease and nasopharyngeal carcinoma.
LMP2
LMP2 is a hydrophobic membrane protein that is transcribed from two different promoters, resulting in the expression of either LMP2A or LMP2B, which differ only in the first exon.2,110 The first exon of LMP2A encodes a 119 amino acid N-terminal cytoplasmic domain, whereas the first exon of LMP2B in non-coding. Both proteins contain 12 identical hydrophobic transmembrane domains and a 27 amino acid cytoplasmic C-domain, and are found in small patches in the plasma membrane of LCLs. Neither LMP2A nor LMP2B is essential for B cell transformation.2,110 The LMP2A N-terminal domain contains eight tyrosine residues, two of which (Y74 and Y85) form an immunoreceptor tyrosine based activation motif (TAM).110,111 When phosphorylated, the TAM present in the B cell receptor (BCR) plays a central role in mediating lymphocyte proliferation and differentiation by the recruitment and activation of the src family of protein tyrosine kinases (PTKs) and the Syk PTK.112 LMP2A can also interact with these PTKs through its phosphorylated TAM and this association appears to regulate PTK activity in a negative manner.110,111 Thus, the LMP2A TAM has been shown to be responsible for blocking BCR stimulated calcium mobilisation, tyrosine phosphorylation, and the activation of the EBV lytic cycle in B cells.110,111,113 More recent work indicates that another tyrosine residue in the LMP2A N-terminal domain (Y112) is also required for the efficient binding of src family PTKs.114 LMP2A is also phosphorylated on serine and threonine residues, and two specific serine residues (S15 and S102) are phosphorylated by MAP kinase in vitro.115 Interestingly, the Erk1 form of MAP kinase was found to interact directly with LMP2A, but the functional relevance of this effect remains unknown.115 The expression of LMP2A in the B cells of transgenic mice abrogates normal B cell development, allowing immunoglobulin negative cells to colonise peripheral lymphoid organs, suggesting that LMP2A can drive the proliferation and survival of B cells in the absence of signalling through the BCR.116 Taken together, these data support a role for LMP2 in modifying the normal B cell development programme to favour the maintenance of EBV latency in the bone marrow and to prevent inappropriate activation of the EBV lytic cycle. A modulatory role for LMP2B in regulating LMP2A function has been suggested.110 The consistent expression of LMP2A in Hodgkin's disease and nasopharyngeal carcinoma suggests an important function for this protein in oncogenesis, but this remains to be shown. A recent report demonstrates the adhesion dependent tyrosine phosphorylation of LMP2A in an epithelial cell line via C-terminal src kinase (Csk), a negative regulator of src kinase activity.117 Our own recent work indicates that LMP2A expression in epithelial cells can affect cell growth and survival as well as augment the signalling capacity of LMP1. The recent demonstration that LMP2A recruits Nedd4-like ubiquitin protein ligases, and that this results in the degradation of LMP2A and Lyn (and probably other signal transduction molecules), provides a novel mechanism whereby LMP2A might modulate receptor signalling.118
Conclusions
Compelling evidence implicates EBV in the pathogenesis of tumours arising in both lymphoid and epithelial tissues. The virus appears to adopt different forms of latent infection in different tumour types, reflecting the complex interplay between EBV and the host cell environment. Another important factor influencing EBV gene expression is the immune response, such that those viral latent proteins to which immunodominant CTL responses are directed, namely the EBNA3 family of proteins, are downregulated in virus associated tumours arising in overtly immunocompetent individuals. EBNA1, an essential protein for the maintenance of EBV infection, which is expressed in all currently known forms of EBV latency, has evolved to evade immunosurveillance by developing a strategy that prevents the protein being processed through the MHC class I pathway. Studies of the function of individual EBV latent genes have highlighted the ability of these proteins to target specific cell signalling pathways. Thus, as is evident from work with proteins encoded by other viruses, an understanding of the functions of EBV latent proteins will not only be relevant to the role of the virus in transformation but will also help to elucidate the mechanisms regulating cell growth, survival, and differentiation. It is hoped that this work will also provide new approaches to treatment. Adoptive transfer of EBV specific CTLs has already been useful in the treatment of immunoblastic B cell lymphomas, and this approach as well as other vaccine strategies are currently being evaluated in patients with Hodgkin's disease or nasopharyngeal carcinoma. The possibility of more direct therapeutic intervention targeting the function of essential EBV latent genes such as EBNA1 and LMP1 is also a possibility. Thus, drugs that inhibit the ability of EBNA1 to bind to oriP or of the TRAFs to interact with LMP1 are likely to be developed. Finally, gene therapy strategies that exploit either the transcriptional regulation of the EBV genome or target the functional effects of EBV latent genes have been described.119,120
Acknowledgments
Work in the authors' laboratory is supported by the Cancer Research Campaign and the Medical Research Council. Dr R Tierney is thanked for help with the figures.
References
- 1.Rickinson AB, Kieff E. Epstein-Barr virus. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology, 3rd ed. Philadelphia: Lipincott-Raven Publishers, 1996:2397–446.
- 2.Kieff E. Epstein-Barr virus and its replication. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology, 3rd ed. Philadelphia: Lipincott-Raven Publishers, 1996:2343–96.
- 3.Rickinson AB, Rowe M, Hart IJ, et al. T-cell-mediated regression of “spontaneous” and of Epstein-Barr virus-induced B cell transformation in vitro: studies with cyclosporin A. Cell Immunol 1984;87:646–58. [DOI] [PubMed] [Google Scholar]
- 4.Niedobitek G, Young LS. Epstein-Barr virus and non-Hodgkin's lymphomas. The non-Hodgkin's lymphomas, 2nd ed. London: Arnold, 1997:309–29.
- 5.Bonnet M, Guinebretiere J-M, Kremmer E, et al. Detection of Epstein-Barr virus in invasive breast cancers. J Natl Cancer Inst 1999;91:1376–81. [DOI] [PubMed] [Google Scholar]
- 6.Sasagawa T, Shimakage M, Nakamura M, et al. Epstein-Barr virus genes expression in cervical intraepithelial neoplasia and invasive cervical cancer: a comparative study with human papillomavirus infection. Hum Pathol 2000;31:318–26. [DOI] [PubMed] [Google Scholar]
- 7.McClain KL, Leach CT, Jenson HB, et al. Association of Epstein-Barr virus with leiomyosarcomas in young people with AIDS. New Engl J Med 1995;332:12–18. [DOI] [PubMed] [Google Scholar]
- 8.Lee ES, Locker J, Nalesnik M, et al. The association of Epstein-Barr virus with smooth-muscle tumors occurring after organ transplantation. New Engl J Med 1995;332:19–25. [DOI] [PubMed] [Google Scholar]
- 9.Brooks LA, Lear AL, Young LS, et al. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J Virol 1993;67:3182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fries KL, Sculley TB, Webster-Cyriaque J, et al. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol 1997;71:2765–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speck SH, Strominger JL. Transcription of Epstein-Barr virus in latently infected, growth-transformed lymphocytes. Advances in Viral Oncology 1989;8:133–50. [Google Scholar]
- 12.Rowe M, Rowe DT, Gregory CD, et al. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J 1987;6:2743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregory CD, Murray RJ, Edwards CF, et al. Downregulation of cell adhesion molecules LFA-3 and ICAM-1 in Epstein-Barr virus-positive Burkitt's lymphoma underlies tumor cell escape from virus-specific T cell surveillance. J Exp Med 1988;167:1811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F, Gregory CD, Rowe M, et al. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci U S A 1987;84:3452–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Gregory C, Sample C, et al. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3c are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol 1990;64:2309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young L, Alfieri C, Hennessy K, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med 1989;321:1080–5. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JA, Hotchin NA, Allday MJ, et al. Immunohistology of Epstein-Barr virus-associated antigens in B cell disorders from immunocompromised individuals. Transplantation 1990;49:944–53. [DOI] [PubMed] [Google Scholar]
- 18.Rooney CM, Smith CA, Brenner MK, et al. Prophylaxis and treatment of Epstein-Barr virus lymphoproliferative disease using genetically modified cytotoxic T lymphocytes. Lancet 1995;345:9–13.7799740 [Google Scholar]
- 19.Cleary ML, Epstein MA, Finerty S. Individual tumors of multifocal Epstein-Barr virus-induced malignant lymphomas in Tamarins arise from different B-cell clones. Science 1985;228:722–4. [DOI] [PubMed] [Google Scholar]
- 20.Rowe M, Young LS, Crocker J, et al. Epstein-Barr virus (EBV)-associated lymphoproliferative disease in the SCID mouse model: implications for the pathogenesis of EBV-positive lymphomas in man. J Exp Med 1991;173:147–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory CD, Rowe M, Rickinson AB. Different Epstein-Barr virus (EBV)–B cell interactions in phenotypically distinct clones of a Burkitt lymphoma cell line. J Gen Virol 1990;71:1481–95. [DOI] [PubMed] [Google Scholar]
- 22.Sample J, Brooks L, Sample C, et al. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is reflected in a novel EBNA-1 mRNA and transcriptional initiation site. Proc Natl Acad Sci U S A 1991;88:6343–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nonkwelo C, Skinner J, Bell A, et al. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol 1996;70:623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedobitek G, Agathanggelou A, Rowe M, et al. Heterogeneous expression of Epstein-Barr virus latent proteins in endemic Burkitt's lymphoma. Blood 1995;86:659–65. [PubMed] [Google Scholar]
- 25.Brooks L, Yao QY, Rickinson AB, et al. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol 1992;66:2689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deacon EM, Pallesen G, Niedobitek G, et al. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med 1993;177:339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahraeus R, Li-Fu H, Ernberg I. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int J Cancer 1988;42:329–38. [DOI] [PubMed] [Google Scholar]
- 28.Young LS, Dawson C, Clark D, et al. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol 1988;69:1051–65. [DOI] [PubMed] [Google Scholar]
- 29.Pallesen G, Hamilton-Dutoit SJ, Rowe M, et al. Expression of Epstein-Barr virus latent gene products in tumor cells of Hodgkin's disease. Lancet 1991;337:320–2. [DOI] [PubMed] [Google Scholar]
- 30.Meijer CJLM, Meijer NM, Jiwa DF, et al. Epstein-Barr virus and human T-cell lymphomas. Semin Cancer Biol 1996;7:191–6. [DOI] [PubMed] [Google Scholar]
- 31.Kerr BM, Lear AL, Rowe M, et al. Three transcriptionally distinct forms of Epstein-Barr virus latency in somatic cell hybrids: cell phenotype dependence of virus promoter usage. Virology 1992;187:189–201. [DOI] [PubMed] [Google Scholar]
- 32.Polack A, Hortnagel K, Pajic A, et al. C-myc activation renders proliferation of Epstein-Barr virus (EBV)-transformed cells independent of EBV nuclear antigen 2 and latent membrane protein 1. Proc Natl Acad Sci U S A 1996;93:10411–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dambaugh T, Hennessy K, Chamnankit L, et al. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc Natl Acad Sci U S A 1984;81:7632–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowe M, Young LS, Cadwallader K, et al. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol 1989;63:1031–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sample J, Young L, Martin B, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol 1990;64:4084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimber U, Adldinger HK, Lenoir GM, et al. Geographical prevalence of two types of Epstein-Barr virus. Virology 1986;154:56–66. [DOI] [PubMed] [Google Scholar]
- 37.Young LS, Yao QY, Rooney CM, et al. New type B isolates of Epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol 1987;68:2853–62. [DOI] [PubMed] [Google Scholar]
- 38.Sixbey JW, Shirley P, Chesney PJ, et al. Detection of a second widespread strain of Epstein-Barr virus. Lancet 1989;ii:761–5. [DOI] [PubMed] [Google Scholar]
- 39.Yao QY, Rowe M, Martin B, et al. The Epstein-Barr virus carrier state: dominance of a single growth-transforming isolate in the blood and in the oropharynx of healthy virus carriers. J Gen Virol 1991;72:1579–90. [DOI] [PubMed] [Google Scholar]
- 40.Rickinson AB, Young LS, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol 1987;61:1310–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khanim F, Yao Q-Y, Niedobitek G, et al. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographical locations. Blood 1996;88:3491–501. [PubMed] [Google Scholar]
- 42.Levitskaya J, Coram M, Levitsky V, et al. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 1995;375:685–8. [DOI] [PubMed] [Google Scholar]
- 43.Levitskaya J, Shapiro A, Leonchiks A, et al. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A 1997;94:12616–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson JB, Bell JL, Levine AJ. Expression of Epstein-Barr virus nuclear antigen-1 induces B cell neoplasia in transgenic mice. EMBO J 1996;15:3117–26. [PMC free article] [PubMed] [Google Scholar]
- 45.Knox PG, Li QX, Rickinson AB, et al. In vitro production of stable Epstein-Barr virus-positive epithelial cell clones which resemble the virus: cell interaction observed in nasopharyngeal carcinoma. Virology 1996;215:40–50. [DOI] [PubMed] [Google Scholar]
- 46.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 1989;340:393–7. [DOI] [PubMed] [Google Scholar]
- 47.Cohen J, Wang F, Mannick J, et al. Epstein-Barr cirus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci U S A 1989;86:9558–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grossman SR, Johannsen E, Tong R, et al. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci U S A 1994;91:7568–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henkel TP, Ling PD, Hayward SD, et al. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 1994;265:92–5. [DOI] [PubMed] [Google Scholar]
- 50.Waltzer L, Logeat F, Brou C, et al. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J 1994;13:5633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science 1995;268:560–3. [DOI] [PubMed] [Google Scholar]
- 52.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science 1995;268:225–32. [DOI] [PubMed] [Google Scholar]
- 53.Sakai T, Taniguchi Y, Tamura K, et al. Functional replacement of the intracellular region of the Notch 1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol 1998;72:6034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofelmayr H, Strobl LJ, Stein C, et al. Activated mouse Notch1 transactivates Epstein-Barr virus nuclear antigen 2-regulated viral promoters. J Virol 1999;73:2770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strobl LJ, Hofelmayr H, Marschall G, et al. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr virus nuclear antigen 2. J Virol 2000;74:1727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaiser C, Laux G, Eick D, et al. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 1999;73:4481–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jayachandra S, Low KG, Thlick A, et al. Three unrelated viral transforming proteins (vIRF, EBNA2 and E1A) induce the MYC oncogene through the interferon-responsive PRF element by using different transcription coadaptors. Proc Natl Acad Sci U S A 1999;96:11566–71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Robertson ES. The Epstein-Barr virus EBNA3 protein family as regulators of transcription. Epstein-Barr Virus Report 1997;4:143–50. [Google Scholar]
- 59.Allday MJ, Farell PJ. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol 1994;68:3491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Radkov SA, Bain M, Farrell PJ, et al. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol 1997;71:8552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker GA, Crook T, Bain M, et al. Epstein-Barr virus nuclear antigen EBNA 3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 1996;13:2541–9. [PubMed] [Google Scholar]
- 62.Silins SL, Sculley TB. Modulation of vimentin, the CD40 activation antigen and Burkitts lymphoma antigen (CD77) by the Epstein-Barr virus nuclear antigen EBNA 4. Virology 1994;202:16–24. [DOI] [PubMed] [Google Scholar]
- 63.Radkov SA, Touitou R, Brehm A, et al. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol 1999;73:5688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allan GJ, Inman GJ, Parker BD, et al. Cell growth effects of Epstein-Barr virus leader protein. J Gen Virol 1992;73:1547–51. [DOI] [PubMed] [Google Scholar]
- 65.Sinclair AJ, Palmero I, Peters G, et al. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J 1994;13:3321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang WQ, Szekely L, Wendel-Hansen V, et al. Co-localization of the retinoblastoma protein and the Epstein-Barr virus-encoded nuclear antigen EBNA-5. Exp Cell Res 1991;197:314–18. [DOI] [PubMed] [Google Scholar]
- 67.Szekely L, Selivanova G, Magnusson KP, et al. EBNA-5, an Epstein-Barr virus-coded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci U S A 1993;90:5455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nitsche F, Bell A, Rickinson AB. Epstein-Barr virus leader protein enhances EBNA2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol 1997;71:6619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol 1997;71:6611–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dawson CW, Rickinson AB, Young LS. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 1990;344:777–80. [DOI] [PubMed] [Google Scholar]
- 71.Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 1993;90:9150–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eliopoulos AG, Dawson CW, Mosialos G, et al. CD40-induced growth inhibition in epithelial cells is mimicked by Epstein-Barr virus-encoded LMP1: involvement of TRAF3 as a common mediator. Oncogene 1996;13:2243–54. [PubMed] [Google Scholar]
- 73.Eliopoulos AG, Stack M, Dawson CW, et al. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL6 production in epithelial cells via and NF-κB pathway involving TNF receptor-associated factors. Oncogene 1997;14:2899–916. [DOI] [PubMed] [Google Scholar]
- 74.Young LS, Eliopoulos AG, Gallagher N, et al. CD40 and epithelial cells: across the great divide. Immunol Today 1998;19:502–6. [DOI] [PubMed] [Google Scholar]
- 75.Uchida J, Yasui T, Takaoka-Shichijo Y, et al. Mimicry of CD40 signals by Epstein-Barr virus LMP1 in B lymphocyte responses. Science 1999;286:300–3. [DOI] [PubMed] [Google Scholar]
- 76.Mosialos G, Birkenbach M, VanArsdale T, et al. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 1995;80:389–99. [DOI] [PubMed] [Google Scholar]
- 77.Gires O, Zimber-Strobl U, Gonnella R, et al. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J 1997;16:6131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Floettmann JE, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region-2 (CTAR-2) maps to the far C-terminus and requires oligomerisation for NF-κB activation. Oncogene 1997;15:1851–8. [DOI] [PubMed] [Google Scholar]
- 79.Rothe M, Sarma V, Dixit VM, et al. TRAF2-mediated activation of NF-κB by TNF receptor 2 and CD40. Science 1995;269:1424–7. [DOI] [PubMed] [Google Scholar]
- 80.Huen DS, Henderson SA, Croom-Carter D, et al. The Epstein-Barr-virus latent membrane protein-1 (LMP1) mediates activation of the NF-κB and cell surface phenotype via 2 effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 1995;10:549–60. [PubMed] [Google Scholar]
- 81.Kaye KM, Devergne O, Harada JN, et al. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci U S A 1996;93:11085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Devergne O, Hatzivassiliou E, Izumi KM, et al. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol Cell Biol 1996;16:7098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J Virol 1997;71:4649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishida T, Tojo T, Aoki T, et al. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc Natl Acad Sci U S A 1996;93:9437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ishida T, Mizushima S, Azuma S, et al. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem 1996;271:28745–8. [DOI] [PubMed] [Google Scholar]
- 86.Devergne O, McFarland EC, Mosialos G, et al. Role of TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol 1998;72:7900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Izumi KM, Kieff E. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci U S A 1997;94:12592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malinin NL, Boldin MP, Kovalenko AV, et al. MAP3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature 1997;385:540–4. [DOI] [PubMed] [Google Scholar]
- 89.Sylla BS, Hung SC, Davidson DM, et al. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-κB through a pathway that includes the NF-κB-inducing kinase and the IκB kinases IKKα and IKKβ. Proc Natl Acad Sci U S A 1998;95:10106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eliopoulos AG, Blake SMS, Floettmann JE, et al. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C-terminus via a mechanism involving TRADD and TRAF2. J Virol 1999;73:1023–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaye KM, Izumi KM, Li H, et al. An Epstein-Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J Virol 1999;73:10525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc Natl Acad Sci U S A 1996;93:6721–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng G, Cleary AM, Ye Z, et al. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science 1995;267:1494–8. [DOI] [PubMed] [Google Scholar]
- 94.Sarma V, Lin Z, Clark L, et al. Activation of the B-cell surface receptor CD40 induces A20, a novel zinc finger protein that inhibits apoptosis. J Biol Chem 1995;270:12343–6. [DOI] [PubMed] [Google Scholar]
- 95.Sutherland CL, Heath AW, Pelech SL, et al. Differential activation of the ERK, JNK, and p38 mitogen-activated protein kinases by CD40 and the B cell antigen receptor. J Immunol 1996;157:3381–90. [PubMed] [Google Scholar]
- 96.Eliopoulos AG, Young LS. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 1998;16:1731–42. [DOI] [PubMed] [Google Scholar]
- 97.Kieser A, Kilger E, Gires O, et al. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J 1997;16:3478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eliopoulos AG, Gallagher NJ, Blake SMS, et al. Activation of the p38 MAPK pathway by Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) co-regulates interleukin-6 and interleukin-8 production. J Biol Chem 1999;274:16085–96. [DOI] [PubMed] [Google Scholar]
- 99.Baichwal VR, Baeuerle PA. Apoptosis: activate NF-κB or die? Curr Biol 1997;7:R94–6. [DOI] [PubMed] [Google Scholar]
- 100.Ip YT, Davis RJ. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol 1998;10:205–19. [DOI] [PubMed] [Google Scholar]
- 101.Gires O, Kohlhuber F, Kilger E, et al. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J 1999;18:3064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Puls A, Eliopoulos AG, Nobes CD, et al. Activation of the small GTPase Cdc42 by the inflammatory cytokines TNFα and IL-1 and by the Epstein-Barr virus transforming protein LMP1. J Cell Sci 1999;112:2983–92. [DOI] [PubMed] [Google Scholar]
- 103.Hu LF, Zabarovsky ER, Chen F. Isolation and sequencing of the Epstein-Barr virus BNLF-1 (LMP1) from a Chinese nasopharyngeal carcinoma. J Gen Virol 1991;72:2399–409. [DOI] [PubMed] [Google Scholar]
- 104.Chen ML, Tsai CN, Liang CL, et al. Cloning and characterization of the latent membrane protein (LMP) of a specific Epstein-Barr virus variant derived from the nasopharyngeal carcinoma in the Taiwanese population. Oncogene 1992;7:2131–40. [PubMed] [Google Scholar]
- 105.Hu LF, Chen F, Zheng X, et al. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 1993;8:1575–83. [PubMed] [Google Scholar]
- 106.Zheng N, Yuan F, Hu L, et al. Effect of B-lymphocyte- and NPC-derived EBV-LMP1 gene expression on in vitro growth and differentiation of human epithelial cells. Int J Cancer 1994;57:747–53. [DOI] [PubMed] [Google Scholar]
- 107.Johnson RJ, Stack M, Hazlewood SA, et al. The 30-base-pair deletion in Chinese variants of the Epstein-Barr virus LMP1 gene is not the major effector of functional differences between variant LMP1 genes in human lymphocytes. J Virol 1998;72:4038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller WE, Cheshire JL, Baldwin AS, et al. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 1998;16:1869–77. [DOI] [PubMed] [Google Scholar]
- 109.Dawson CW, Eliopoulos AG, Blake SM, et al. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology 2000;272:204–17. [DOI] [PubMed] [Google Scholar]
- 110.Longnecker R. Epstein-Barr virus latency: LMP2, a regulator or means for Epstein-Barr virus persistence? Adv Cancer Res 2000;79:176–200. [DOI] [PubMed] [Google Scholar]
- 111.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMPA2 is essential for blocking BCR-mediated signal transduction. J Virol 1997;235:241–51. [DOI] [PubMed] [Google Scholar]
- 112.Cambier JC, Pleiman CM, Clark MR. Signal-transduction by the B-cell antigen receptor and its coreceptors. Annu Rev Immunol 1994;12:457–86. [DOI] [PubMed] [Google Scholar]
- 113.Miller CL, Burkhardt AL, Lee JH. Integral membrane protein 2 (LMP2) of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein tyrosine kinases. Immunity 1995;2:155–66. [DOI] [PubMed] [Google Scholar]
- 114.Freuhling S, Stuart R, Dolwick KM, et al. Tyrosine 112 of latent membrane protein 2A is essential for protein kinase loading and regulation of Epstein-Barr virus latency. J Virol 1998;72:7796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Panousis CG, Rowe DT. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J Virol 1997;71:4752–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Caldwell RG, Wilson JB, Anderson SJ, et al. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998;9:405–11. [DOI] [PubMed] [Google Scholar]
- 117.Scholle F, Longnecker R, Raab-Traub N. Epithelial cell adhesion to extracellular matrix proteins induces tyrosine phosphorylation of the Epstein-Barr virus latent membrane protein 2: a role for C-terminal src kinase. J Virol 1999;73:4767–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ikeda M, Ikeda A, Longan LC, et al. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 2000;268:178–91. [DOI] [PubMed] [Google Scholar]
- 119.Franken M, Estabrooks A, Cavacini L, et al . Epstein-Barr virus-driven gene therapy for EBV-related lymphomas. Nat Med 1996;2:1379–82. [DOI] [PubMed] [Google Scholar]
- 120.Piche A, Kasono K, Johanning F, et al. Phenotypic knock-out of the latent membrane protein 1 of Epstein-Barr virus by an intracellular single-chain antibody. Gene Therapy 1998;5:1171–9. [DOI] [PubMed] [Google Scholar]