Abstract
Aims—To study the distribution of Epstein-Barr virus (EBV) variants containing mutations in the latent membrane protein 1 (LMP-1) oncogene and promoter in EBV associated Hodgkin's disease and infectious mononucleosis compared with previous findings in asymptomatic EBV carriers.
Methods—Sequence analysis of the EBV LMP-1 promoter and gene in isolates from Danish patients with Hodgkin's disease (n = 61) and infectious mononucleosis (n = 10).
Results—Viruses (previously designated group D) that contain two mutations in the activating transcription factor/cAMP response element (ATF/CRE) in the LMP-1 promoter, which are known to decrease promoter activity greatly, were significantly less frequent in Hodgkin's disease than in both infectious mononucleosis (p = 0.0081) and asymptomatic EBV carriers (p = 0.0084). In some cases, the LMP-1 gene contained mutations in a recently identified cytotoxic T cell (CTL) epitope. Most viral isolates contained mutations shown to increase nuclear factor κB (NF-κB) activation and had one of two newly identified C-terminal activation regions 3 (CTAR-3) deleted. The exon 1 Xho-I restriction site in the LMP-1 gene could be lost through a range of different mutations.
Conclusions—These findings indicate selection pressure against EBV strains with weak LMP-1 promoter activity in Hodgkin's disease and thus provide further strong circumstantial evidence for the pathogenic role of EBV (and LMP-1) in this disease. Mutation of the CTL epitope suggests immune selection of EBV strains. Many EBV isolates contain functionally important mutations in the LMP-1 gene. Loss of the Xho-I restriction site should not be used as a marker of specific LMP-1 variants.
Keywords: Epstein-Barr virus, Hodgkin's disease, latent membrane protein 1, promoter mutations
The Epstein-Barr virus (EBV) is not only one of the most widespread human viruses but, somewhat paradoxically, it is also linked to a range of neoplasms.1 These include various B and T cell non-Hodgkin's lymphomas, Hodgkin's disease, and several lymphoepithelioma-like carcinomas, of which nasopharyngeal carcinoma is the archetype. The association of EBV with these tumours, and the oncogenic potential of EBV in vitro, is well documented.2 However, evidence that the virus plays a decisive role in the pathogenesis of EBV associated tumours in vivo is less clear cut.
The identification of tumour specific EBV strains would provide important confirmatory evidence for the oncogenic role of EBV, particularly if structural changes affecting the transcription or function of key viral genes could be demonstrated. Latent membrane protein 1 (LMP-1) is the only EBV gene with known oncogenic potential to be expressed in tumour cells in Hodgkin's disease and nasopharyngeal carcinoma3–10 and, therefore, much attention has been directed at identifying tumour specific sequence variations within this gene. Two nasopharyngeal carcinoma derived LMP-1 variants (CAO and C1510) show increased experimental tumorigenicity compared with the EBV prototype B95.8,11–13 apparently associated at least in part with a 30 bp deletion in the C-terminus of the LMP-1 gene.14 In the original reports, the loss of a Xho-I restriction site in exon 1 of the CAO-LMP-1 and C1510-LMP-1 genes was used as a marker of this new LMP-1 variant.12,13,15 Loss of this restriction site and/or detection of the 30 bp deletion, or partial sequence analysis of the LMP-1 gene, are widely accepted criteria for defining this variant, and have been used in numerous studies to describe its frequency in different EBV associated diseases.12,13,16–23 However, recent reports suggest that the prevalence of the 30 bp deletion variant of LMP-1 in EBV positive tumours is not increased compared with normal controls from the background population in the same geographical region.24–26
In contrast to the N-terminus and the C-terminus of the LMP-1 protein, the LMP-1 promoter and the C-terminal part of the transmembrane domain (amino acid positions 90–190) of the protein have attracted little attention.25 However, our recent survey of EBV strains in asymptomatic seropositive Europeans showed that about 20% of isolates (which we designate the group D variant) contained mutations in the LMP-1 promoter, most of which were identical to those found in the CAO-LMP-1 and C1510-LMP-1 promoters.25 The functional importance of these mutations in CAO, C1510, and our group D isolates is not known. However, two of the promoter mutations (GA → CT; −44 to −45, relative to the transcription start site) found in these isolates have been shown to decrease LMP-1 promoter activity up to nine times.14,27 These two mutations are located in the activating transcription factor/cAMP response element (ATF/CRE) (−38 to −45, relative to the transcription start site), shown by Sjöblom et al to be one of the limiting factors that determine the final degree of LMP-1 promoter activity.28 Thus, approximately 20% of European wild-type EBV isolates from individuals without EBV associated diseases contain an LMP-1 gene with a weak promoter.25 Neoplastic cells in Hodgkin's disease show strong LMP-1 expression—probably the highest found in any type of EBV associated tumour.3 If a high degree of LMP-1 expression is a prerequisite for developing EBV associated Hodgkin's disease, one might expect there to be selection pressure against EBV variants with weak LMP-1 promoter function. We have tested this hypothesis by examining the distribution of LMP-1 variants in patients with EBV associated Hodgkin's disease compared with those with infectious mononucleosis and healthy controls, based on sequence analysis of the LMP-1 gene and promoter.
A recent study of the C-terminal of the transmembrane domain has shown that amino acid substitutions in this region, found in the C15-LMP-1 variant, result in a twofold increase in LMP-1 induced nuclear factor κB (NF-κB) activation, indicating that the area is of functional importance.29 Furthermore, Khanna et al recently showed that this region of the LMP-1 protein contains a human major histocompatibility complex A2 (HLA-A2) restricted cytotoxic T cell (CTL) epitope, which elicited a strong CTL response.30 We have therefore sequenced the whole LMP-1 gene in the EBV isolates from our patients with Hodgkin's disease and infectious mononucleosis to investigate the frequency of mutations in regions of possible importance.
Methods
Tissue biopsies from 61 patients with Hodgkin's disease and 10 with infectious mononucleosis were selected from the archives of the Institute of Pathology. In some cases, frozen samples were available, in others formalin fixed paraffin wax embedded tissues were studied. Forty of the Hodgkin's disease and nine of the infectious mononucleosis cases were included in our previous study of sequence variation around the LMP-1 30 bp deletion.18 Diagnosis was based on standard clinical, morphological, immunohistological, and when appropriate, serological criteria. Immunohistological staining for LMP-1 was performed on all cases using standard procedures for frozen and paraffin wax embedded tissue.3,31
POLYMERASE CHAIN REACTION (PCR)
DNA for PCR was prepared as reported previously18 and the PCR was carried out using an automated thermal cycler (Perkin Elmer Cetus, Norwalk, California, USA). Cases were EBV subtyped by analysis of the EBV nuclear antigen 2 (EBNA-2) coding region as described previously.18 EBV cell lines B95.8 and AG876 carrying EBNA-2A and EBNA-2B, respectively, were used as controls. Cases were screened for the 30 bp deletion in the LMP-1 gene using the primers and PCR procedure published previously.16,18 The Xho-I restriction site polymorphism (position 169428–23) in the LMP-1 gene was analysed using primer pair LMP1/LMP-PRO2, and digestion with the Xho-I restriction enzyme (Pharmacia Biotech, Weiterstadt, Germany), as described previously.25 The LMP-1 gene repeat region (position 168555–400) was analysed using primer pair LMP-repeat5/LMP-repeat3. PCR products from the repeat region of isolates in which the whole LMP-1 gene had previously been sequenced and from cases sequenced in this study were used as controls.25 The size of the PCR products was verified after electrophoresis in Visigel separation matrix (Stratagene, La Jolla, California, USA) stained with ethidium bromide.
In our previous study on EBV isolates from individuals without EBV associated disease, we found that six of the seven LMP-1 variants that had lost the Xho-I restriction site contained a specific pattern of mutations in the LMP-1 promoter, whereas none of the isolates that had retained the Xho-I restriction site had important LMP-1 promoter mutations.25 Therefore, all cases of Hodgkin's disease in which the EBV isolate had lost the Xho-I restriction site were selected for sequence analysis. In addition, we sequenced 15 Hodgkin's disease isolates without the 30 bp deletion, 12 with the 30 bp deletion, and one with a unique 12 bp deletion. All 10 infectious mononucleosis cases were included in the sequence analysis. The LMP-1 promoter, positions 169720–475 (−205 to +40, relative to the transcription start site), was amplified and sequenced using the primer set Pr1/Pr2.25 A larger part of the LMP-1 promoter was sequenced in selected cases with the primer set Pr0/Pr2.25 The whole LMP-1 gene was sequenced using the primer sets LMP-116/LMP69051 (ATGTTAGATCCCTTAAACCAAGTAAGCA), LMP9134 (CCC CAGTCACCCTCCTACTCATC GC)/ LMP 8602 (TGAGAGAGCAGAG TGGGGGTCCGTGCC), LMP8723 (TCCCTCCCGCA CCCTCAACAAGC)/LMP8286 (ACGLMPGCCG CCACCGTCTCATC), and LMP9/ LMP11.16 Table 1 ▶ shows which parts of the LMP-1 gene and promoter were sequenced in the individual cases.
Table 1.
Sequenced parts of the latent membrane protein 1 (LMP-1) gene and promoter in Epstein-Barr virus (EBV) isolates from patients with Hodgkin's disease
| Number of cases sequenced | ||||
| Parts sequenced | +30 bp deletion | −30 bp deletion | −Xho-I | Total |
| 1 | 12 | 15 | 6 | 33 |
| 2 | 7 | 8 | 6 | 21 |
| 3 | 6 | 7 | 4 | 17 |
| 4 | 3 | 4 | 4 | 11 |
| 5 | 12 | 15 | 6 | 33 |
Parts sequenced, the parts of the LMP-1 gene and promoter sequenced in the individual cases. Numbers 1, 2, 3, 4, and 5 stand for primer pairs Pr1/Pr2, LMP1/LMP69051, LMP134/LMP602, LMP8723/LMP8286, and LMP9/LMP11, respectively.
+30 bp deletion, cases with the 30 bp deletion.
−30 bp deletion, cases without the 30 bp deletion.
−Xho-I, cases that had lost the Xho-I restriction site in exon 1 of the LMP-1 gene.
An EBV isolate with a 12 bp deletion was also sequenced. This case is not shown in the table.
When using the primer pairs LMP-1/LMP69051, LMP-1/Pro-2, and LMP9/LMP11 a standard PCR mix was used (Perkin Elmer Cetus) with 200 mM of each dNTP, 6 mM Mg2+ and 1 U Amply-Taq polymerase. With the primer pairs Pro-0/Pro-2 and Pro-1/Pro-2 0.2 mM of deaza-GTP (Pharmacia Biotech) was added. For primer pairs LMP9134/LMP8602, LMP8723/LMP8286, and LMP-repeat5/LMP-repeat3, the standard PCR buffer was replaced with a mercaptoethanol buffer and 5 ml of DMSO was added. For all primer sets, the PCR programme started with a denaturation step at 98°C for five minutes. With the exception of primer set LMP9/LMP11, subsequent steps consisted of five cycles with an annealing temperature of 65°C, 10–20 cycles at 63°C, and 20–25 cycles at 62°C (maximum, 40 cycles). This programme was designed to achieve the highest possible ratio between specific product and background (especially primer dimers) in the individual cases. In all cycles, the denaturation temperature was 94°C and the extension temperature was 74°C. The Taq polymerase was added after the initial denaturation. The PCR conditions for primer set LMP9/LMP11 have been described previously.18
Sequencing of the PCR product amplified with the primer pair LMP-1/LMP69051 was performed as cycle sequencing in an ABI Catalyst 800 Molecular Biology Lab Station (Applied Biosystems, Foster City, California, USA), using M13 (−21) and M13 reverse tagged primers and a DNA dye primer sequencing kit (Perkin Elmer) under standard conditions. Sequencing with the other primers used biotinylated primers and a DNA dye terminator sequencing kit (Perkin Elmer Cetus) as described previously.18 Bidirectional sequencing was performed on the semiautomated ABI 373 sequencer and analysed with the Seq Ed software (Applied Biosystems, Foster City, California, USA), as described elsewhere.18
Results
In all cases, LMP-1 expression was demonstrated in Hodgkin and Reed-Sternberg tumour cells in Hodgkin's disease, and in blast cells in infectious mononucleosis. EBV subtyping was possible in 58 of 61 Hodgkin's disease and 10 of 10 infectious mononucleosis cases. The EBV subtype 1 was found in 54 of the Hodgkin's disease cases and in each of the 10 infectious mononucleosis cases. Four Hodgkin's disease cases contained EBV subtype 2.
The LMP-1 gene in 60 of 61 Hodgkin's disease and all 10 infectious mononucleosis isolates could be analysed satisfactorily for the loss of the Xho-I restriction site, configuration of the repeat region, and the presence of the 30 bp deletion. In six of the 61 Hodgkin's disease cases the Xho-I restriction site was lost; none of these cases contained the 30 bp deletion. In 24 Hodgkin's disease cases the repeat region was identical to that found in B95.8, consisting of three perfect 33 bp repeats and a fourth with an insertion of a 15 bp “unique” sequence.11 The remaining 36 cases had a repeat region consisting of three to six perfect 33 bp repeats, suggesting that the 15 bp unique sequence was deleted in these cases. Sequence analysis of the region confirmed this result (see below). In 16 Hodgkin's disease cases the 30 bp deletion could be detected. In one Hodgkin's disease case the EBV isolate contained a unique 12 bp deletion. The Xho-I restriction site was lost in four of the EBV isolates from patients with infectious mononucleosis, one of these containing the 30 bp deletion. Two infectious mononucleosis isolates had a repeat region identical to B95.8 and eight isolates contained a repeat region consisting of two to six perfect 33 bp repeats. The 30 bp deletion was found in four isolates, including one that had also lost the Xho-I restriction site. All the EBV subtype 2 isolates contained the 30 bp deletion.
In our previous study of EBV isolates from individuals without EBV associated disease, we found that European LMP-1 variants could be allocated to one of four groups based on sequence variation in the LMP-1 gene and promoter compared with B95.8.11,25 We designated these groups A–D and this terminology will be used throughout our present paper.25 Group A consists of isolates similar to B95.8 except for six single base mutations in the LMP-1 gene. Group B includes isolates containing a repeat region different from B95.8, together with 19 single base mutations in the gene. Group C contains isolates showing the 30 bp deletion and 44 single base mutations in the gene. Groups A–C contained one identical mutation in the LMP-1 promoter (C → T; position 169673). Group D consists of isolates that have lost the Xho-I restriction site, do not contain the 30 bp deletion, but show 66 single base mutations in the gene and 33 single base mutations in the sequenced part of the promoter.
SEQUENCE ANALYSIS OF THE LMP-1 PROMOTER
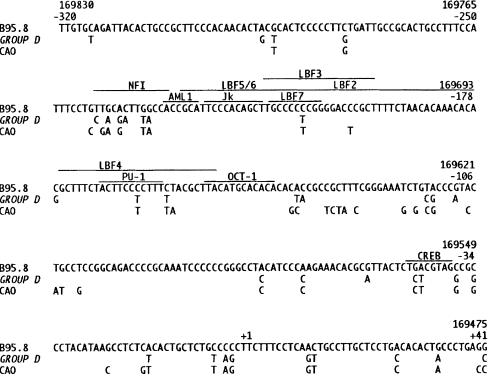
Results are summarised in fig 1 ▶. Sequencing was satisfactory in all 10 infectious mononucleosis and all 34 Hodgkin's disease isolates. Analysis of isolates that had lost the Xho-I restriction site revealed that one of six Hodgkin's disease viruses and three of four infectious mononucleosis viruses (the three isolates without the 30 bp deletion) were characterised by the presence of an identical pattern of 23 single base mutations in the promoter when using PR1/PR2. In HDc68, which was sequenced further upstream with PR0/PR2, this was increased to 33 single base mutations (fig 1 ▶). No unique mutations were found in individual isolates. This set of single base mutations was identical to that previously designated group D.25 In the remaining 33 Hodgkin's disease cases and seven infectious mononucleosis cases, a single base mutation was consistently found (C → T; position 169673) compared with B95.8. In addition, a total of 30 single base mutations were found in the 33 Hodgkin's disease and seven infectious mononucleosis isolates. However, only two of the 30 single base mutations were found in more than one case: G → C, position 168785 (two patients with Hodgkin's disease) and G → C, position 169589 (three patients with Hodgkin's disease).
Figure 1.
The promoter region of the latent membrane protein 1 (LMP-1) gene, from position −320 to +41, relative to the transcription start site. The sequence of B95.8 is shown in full.11 For group D and CAO only mutated nucleotides are shown.15 The sequences of groups A–C are not shown because these isolates only contained one single base mutation compared with B95.8. The recognition sequences and/or binding sites of the transcription factors are marked with a line.28,32–34 CREB indicates the activating transcription factor/cAMP response element (ATF/CRE) (−38 to −45), which binds ATF-1/CREB-1 heterodimers.28
SEQUENCE ANALYSIS OF THE LMP-1 GENE
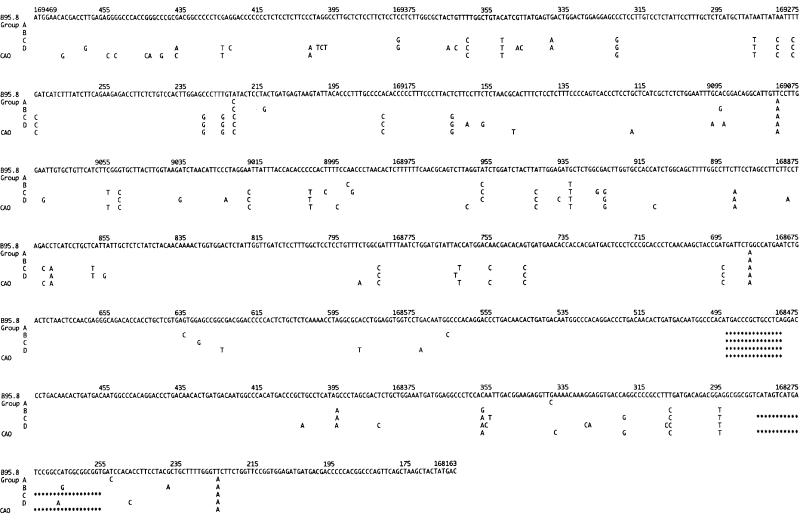
The results are summarised in fig 2 ▶.
Figure 2.
This figure shows the patterns of mutations found in the latent membrane protein 1 (LMP-1) gene in the sequenced cases that could be allocated to one of the four groups (groups A–D) compared with the DNA sequence of the Epstein-Barr virus (EBV) strain B95.811 and CAO.15 The nucleotide sequence of B95.8 is shown in full. For groups A–D and CAO only mutated nucleotides are shown. The CAO-LMP-1 gene contains seven 33 bp repeats and our group B/C isolates contained from three to seven 33 bp repeats in the repeat region. In this figure only four are shown. The 15 bp deletion in the repeat region and the 30 bp deletion in the C-terminus are indicated by asterisks.
Isolates without either the 30 bp deletion or loss of the Xho-I restriction site
Eleven of the 15 Hodgkin's disease and two of the three infectious mononucleosis isolates showed group A sequences in the sequenced parts of the LMP-1 gene, with the exception of occasional individual unique mutations (data not shown). One infectious mononucleosis (IMc8) and three Hodgkin's disease isolates showed group B mutations in the sequenced parts. The infectious mononucleosis case (IMc8) contained four additional unique mutations in the C-terminus (168318, G → C; 168317, G → A; 168278, G → A; 168277, A → C). One Hodgkin's disease isolate (HDc48) contained an intermediate pattern of single base mutations including three of the group B mutations (positions 168992, 168957, and 168687), one of the group C/D mutations (position 168943; L126 → F), and two unique mutations (T → C at position 168814 and G → C at position 168661).
Isolates containing the 30 bp deletion
Ten of the 12 Hodgkin's disease isolates containing the 30 bp deletion showed a group C pattern of mutations in the sequenced parts of the LMP-1 gene. One isolate (HDc115) contained a group A pattern of mutations and the remaining one (HDc66) had a group B pattern of single base mutations in the LMP-1 gene and a group C mutation at position 169339. Three of the four infectious mononucleosis isolates containing the 30 bp deletion showed the group C pattern of single base mutations in sequenced parts of the LMP-1 gene. The fourth infectious mononucleosis isolate (IMc10) showed 44 single base mutations in the LMP-1 gene, which were identical to the group C pattern. However, this isolate had an additional single base mutation (A → C) at position 169424 (fig 3 ▶), which resulted in loss of the Xho-I restriction site.
Figure 3.
The amino acid sequence of the B95.8 latent membrane protein 1 (LMP-1) is shown in full.11 For our group A–D variants and CAO-LMP-1 only mutated amino acids are shown.15 The CAO-LMP-1 gene contains seven 11 amino acid repeats and our group B/C isolates contained from three to seven 11 amino acid repeats in the repeat region. In this figure only four are shown. Amino acid substitutions in the transmembrane domain suggested by Miller and colleagues29 to increase NF-κB activation are at positions 86, 106, 122, 128, and 129. The HLA-A2 restricted cytotoxic T cell epitopes are marked with a double line: the cytotoxic T cell epitope eliciting a strong responce at amino acids 125 to 133 and the cytotoxic T-cell epitope eliciting a weak response at amino acids 159 to 167.30 The C-terminal activation regions 1 (CTAR-1) and CTAR-2, and the recently identified CTAR-3, are indicated by a single line.35–40 The five amino acid (15 bp) deletion in the repeat region and the 10 amino acid (30 bp) deletion in the C-terminus are marked by asterisks.
The Hodgkin's disease case with the 12 bp deletion (HDc115) showed a group B configuration in all three sequenced regions except for one of the group C/D mutations (position 168943; L126 → F)
Isolates that had lost the Xho-I restriction site
The three infectious mononucleosis isolates and the single Hodgkin's disease isolate that showed an identical pattern of single base mutations in the promoter (see above) had 66 identical single base mutations in the LMP-1 gene (fig 2 ▶). In all four isolates, the Xho-I restriction site was found to be lost as a result of a G → T mutation (position 169425). Unlike the other isolates, IMc12 did not have a G → C mutation at position 169399, but had G → C instead of G → T at position 169398, thus changing amino acid 26 from G → A instead of G → V. IMc12 also had a T → C mutation at position 169375, which did not affect the amino acid sequence. IMc7 and IMc12 had an A → G mutation at position 169294, changing amino acid 62 from M to V. HDc68 and IMc7 had an C → A mutation at position 168192, changing amino acid 377 from H to P. IMc3 and IMc12 had an A → G mutation at position 168191, changing amino acid 377 from H to R.
Five of the six Hodgkin's disease isolates that had lost the Xho-I restriction site had done so because of mutations other than G → T (position 169425). In two cases, loss of the Xho-I restriction site was the result of a C → G mutation (position 169426) and a G → C mutation (position 169423; HDc120 and HDc3), and in three cases (HDc87, HDc59, and HDc44) loss was the result of a G → C mutation (position 169423; fig 3 ▶). Apart from this, the single base mutation pattern was identical to group A in one case (HDc3), the other four isolates (HDc87, HDc59, HDc44, and HDc120) containing many of the group B single base mutations.
Sequence analysis with the primer set LMP8723/LMP8286 covering the repeat region showed that all LMP-1 variants with a repeat region differing from B95.8 were deleted for the 15 bp unique sequence located within the repeat region (fig 2 ▶). Thus, based on the combined results of sequence analysis and gel electrophoresis, the EBV isolate in 36 of the 61 Hodgkin's disease cases and eight of the 10 infectious mononucleosis cases contained this deletion. All the sequenced isolates with a repeat region identical to B95.8 had a group A pattern of single base mutations, including the one with the 30 bp deletion.
Table 2 ▶ shows the allocation of the sequenced cases to groups A–D, the results of Xho-I restriction enzyme digestion, detection of the 30 bp deletion, and the configuration of the repeat region.
Table 2.
Characterisation of the latent membrane protein 1 (LMP-1) gene and promoter in Epstein-Barr virus (EBV) isolates from patients with Hodgkin's disease (HD) and infectious mononucleosis (IM)
| Group A–D | Xho-I | Repeat region | 30 bp deletion | Number of cases | |
| HD | A | + | 4+ | – | 9 |
| A | + | 3, 6 | – | 2 | |
| A* | – | 4+ | – | 1 | |
| A* | + | 4+ | + | 1 | |
| B | + | 4–6 | – | 3 | |
| B* | – | 4, 5 | – | 4 | |
| B* | + | 5 | + | 1 | |
| B* | + | 5 | 12 bp | 1 | |
| C | + | 3–6 | + | 10 | |
| D | – | 3 | – | 1 | |
| UG | + | 4+ | – | 1 | |
| IM | A | + | 4+ | – | 2 |
| B | + | 6 | – | 1 | |
| C | + | 5, 6 | + | 3 | |
| D | – | 2–4 | – | 3 | |
| UG | – | 5 | + | 1 |
Group A–D, grouping of the isolates based on sequence analysis of the LMP-1 gene and part of the promoter of sequenced isolates (groups A–D as defined in the result section).
Xho-I, presence (+) or loss (–) of the Xho-I restriction site.
Repeat region, the number of perfect 33 bp repeats in the LMP-1 repeat region; + indicates a repeat region identical to the prototype EBV strain B95.8.11
30 bp deletion, absence (–) or presence (+) of the 30 bp deletion (del-LMP-1).
*These cases contained additional mutations affecting the Xho-I restriction site or contained the 30 bp deletion (one case with a unique 12 bp deletion) but otherwise had a pattern of single base mutations similar to group A or group B.
UG, ungrouped cases.
Statistical analysis
Statistical testing showed the prevalence of the group D LMP-1 variant to be significantly reduced in Hodgkin's disease tumour tissues (one of 60), both compared with infectious mononucleosis lesions (three of 10; p = 0.0081; Fisher's exact test) and with isolates from asymptomatic donors (six of 34; p = 0.0084 Fisher's exact test).25 No difference in group D prevalence was found when comparing isolates from patients with infectious mononucleosis with those from asymptomatic donors (p = 0.4; Fisher's exact test).
Discussion
In EBV associated Hodgkin's disease, LMP-1 is the only EBV gene with a known oncogenic potential to be expressed.3,10 If this gene plays a role in the pathogenesis of Hodgkin's disease, selection pressure can be expected on variants containing mutations affecting LMP-1 function.
Previous studies have indicated that the degree of LMP-1 expression is in itself crucial to the function of the protein. Hu et al found that only transfected Rhec-1 cells expressing high amounts of LMP-1 were tumorigenic in severe combined immunodeficient (SCID) mice, whereas Rhec-1 cells expressing low amounts of LMP-1 were not.13 Miller et al have reported that the expression of the epidermal growth factor receptor (EGFR) was induced in the presence of tumour necrosis factor receptor (TNFR) associated factors (TRAF3-C) in cells expressing high amounts of LMP-1, but was inhibited by TRAF3-C expression when LMP-1 concentrations were low.41 Thus, mutations changing the activity of the LMP-1 promoter might be of importance to the function of the protein.
We showed previously that a substantial minority of healthy European seropositive donors (six of 34) carry wild-type viral isolates (which we designated group D variants) characterised by numerous single base mutations in both the LMP-1 promoter and gene.25 Most of the promoter mutations in these isolates are identical to those originally described in CAO-LMP-1 and C1510-LMP-1.12,15 Two of these promoter mutations (GA → CT; −44 to −45, relative to the transcription start site) found in our group D isolates, CAO and C1510, are located in the ATF/CRE site (−38 to −45, relative to the transcription start site; fig 1 ▶).28 Li et al and Chen et al have reported that in the C1510-LMP-1 promoter these two mutations can reduce promoter activity as much as nine times in luciferase assays,14,27 indicating that the ATF/CRE site is crucial to the activity of the LMP-1 promoter. Recently, Sjöblom et al have confirmed the importance of this site by showing that ATF-1 and CREB-1 (CRE binding protein 1) heterodimers bind to the ATF/CRE site and activate the LMP-1 promoter independently of EBNA-2. They concluded that this is one of the limiting factors that determine the final LMP-1 promoter activity.28
In our study, we found that only one of 61 EBV positive European patients with Hodgkin's disease had a group D isolate, whereas this variant was found in three of 10 patients with infectious mononucleosis. Thus, the frequency of group D variants is significantly lower in Hodgkin's disease than in either infectious mononucleosis (p = 0.0081; Fisher's exact test) or asymptomatic donors (six of 34 cases25; p = 0.0084; Fishers exact test). In Hodgkin's disease, the tumour cells consistently express high amounts of LMP-1 and we believe that the functional difference in promoter activity between group D and other variants provides a plausible explanation for the apparent selection against group D variants in Hodgkin's disease found in our study. This observation provides further strong indirect evidence that EBV (and LMP-1) plays a part in the pathogenesis of EBV associated Hodgkin's disease.
The LMP-1 promoter in group D isolates, CAO and C1510, contains many other single base mutations, some of which are located at binding sites or recognition sequences of other transcription factors (fig 1 ▶).32–34 The functional relevance of these mutations needs further investigation. The frequency of LMP-1 promoter mutations in isolates from healthy carriers and EBV associated diseases in other parts of the world has not been reported. Whether or not Asian wild-type EBV isolates carry LMP-1 promoter mutations identical to CAO-LMP-1 remains to be investigated.
In contrast to the LMP-1 promoter, numerous studies have reported on sequence variation in the LMP-1 gene. Our sequence analysis of the LMP-1 gene shows that in all group A–D isolates the two C-terminal activation regions 1 (CTAR-1; residues 194–232) and CTAR-2 (residues 351–386),35–39 which have been shown to be crucial for signalling of the protein, are highly conserved. No variant contains mutations in the core sequence required for NF-κB activation in either CTAR-1 (PQQAT; position 204 to 208) or CTAR-2 (PVQLSY; position 379 to 384),39,42 or the core sequence for the C-Jun N-terminal kinase (JNK) pathway (PVQLSYYD; position 379 to 386).43 In CTAR-1, only a single non-conservative amino acid change is found (G → S; position 212). However, this is seen in all of the group A–D isolates, suggesting that this might be the prototype sequence (fig 2 ▶). The 30 bp deletion overlaps CTAR-2 and has been found to increase the oncogenicity of the protein,14 but not to affect NF-κB signalling.44 Apart from this, non-conservative amino acid changes in this region have only been found in the group D variant shown in this study to be under-represented in Hodgkin's disease.
The fact that most single base mutations affecting the amino acid sequence of the LMP-1 protein are located outside the two major effector regions raises the question of whether these mutations might just be non-functional polymorphisms. Three recent papers indicate that this is not the case.
Gires and co-workers have reported that B95.8-LMP-1 contains a third activation region (CTAR-3) interacting with Janus kinase 3 (JAK-3), which activates signal transducer and activator (STAT) transcription factors.40 CTAR-3 was mapped to a PXXPXP consensus motif. The B95.8 LMP-1 contains two such motifs, one of which constitutes a unique sequence in the repeat region (PHDPLP; position 275 to 280), the other being located immediately after the repeat region (PHDPLP; position 302 to 307; fig 3 ▶). The results of our present study and a previous study on EBV isolates from individuals without EBV associated disease show that the PHDPLP sequence within the repeat region (position 275 to 280) is only conserved in EBV group A isolates with a repeat region identical to B95.8. In all other isolates the HDPLP sequence is deleted. This is also true for the CAO-LMP-1 and the C1510-LMP-1 proteins. Thus, about 60% of European wild-type EBV isolates from individuals without EBV associated disease and from patients with Hodgkin's disease, and about 80% from patients with infectious mononucleosis, have only retained one of the two motifs interacting with JAK-3. The functional importance of these findings needs further investigation.
Khanna et al have identified two peptide epitopes in LMP-1 (prototype B95.8) recognised by HLA-A2 restricted cytotoxic T cells (CTLs).30 One of these (YLLEMLWRL; amino acid position 125 to 133) induced a strong CTL response and efficient lysis of LMP-1 expressing HLA-A2 positive cells. Interestingly, our sequence analyses show that the group B, C, and D variants contain one, three, and three amino acid changes, respectively, (CAO-LMP-1 containing two) in the epitope eliciting the strong CTL response and none in the other epitope (fig 3 ▶). The substitution of leucine with phenylalanine at anchor position two found in groups C and D (the ungrouped variant HDc48 and the 12 bp deletion variant HDc115 also contained this substitution) is of special interest, because screening of peptides presented by the HLA-A2 molecule has shown that the dominant residue at this position is usually leucine, sometimes methionine, and rarely isoleucine, but never phenylalanine.45,46 Therefore, it is highly unlikely that peptides containing this substitution can be presented to CTLs by the HLA-A2 molecule.45,46 The effects of the other amino acid substitutions are less clear cut and this area needs further investigation. The gene frequency of HLA-A2 in the Danish population is about 53%. In our present study, substitution of leucine with phenylalanine was found in about 35% of patients with Hodgkin's disease and 60% with infectious mononucleosis, compared with 45% of EBV carriers in our previous study.25 This suggests that although LMP-1 only constitutes a minor target component for the total virus specific CTL response,2 this substitution could reflect immune selection to avoid the host CTL response.
Miller et al have reported that five amino acid changes in the transmembrane domain of LMP-1 (Ile85 → Leu; Phe106 → Tyr; Ile122 → Leu; Glu128 → Asp; and Met129 → Ile) are responsible for enhanced NF-κB activation and EGFR induction by the LMP-1 protein in the EBV isolate C15 (derived from a Mediterranean case of nasopharyngeal carcinoma) compared with B95.8-LMP-1.29 They concluded that these mutations found in an EBV isolate from a high risk area of nasopharyngeal carcinoma might contribute to the increased risk of this disease in these geographical areas.29 Our present study and a previous study on EBV isolates from individuals without EBV associated disease25 show that the amino acid changes at positions 85 and 106 are found in all group A–D isolates (fig 2 ▶). The amino acid changes at positions 122 and 129 are found in group B–D isolates, which constitute about 60% of all isolates (fig 2 ▶). Thus, four of the five mutations described by Miller et al are found frequently in European EBV isolates from Hodgkin's disease, infectious mononucleosis, and healthy carriers, and are not restricted to high risk areas of nasopharyngeal carcinoma. Only the amino acid change at position 128 is rare in European isolates, being restricted to group D isolates (fig 2 ▶). However, this is a conservative amino acid substitution unlikely to be responsible for the increase in NF-κB activation. Furthermore, our study shows that the transmembrane domain of European LMP-1 variants contains 17 other amino acid changes (particularly in groups C and D; fig 3 ▶). Further studies are needed to investigate the role of these mutations in LMP1 signalling.
The sequence analysis performed in our present study and a previous study on EBV isolates from asymptomatic carriers are the only ones to include the C-terminal part of the transmembrane domain of LMP-1. The frequency of mutations in this HLA-A2 restricted CTL epitope and of the mutations increasing NF-κB activation in wild-type EBV isolates from other geographical areas is therefore not known.
In our previous study on EBV isolates from asymptomatic donors, 19 of 20 could be placed in groups A, B, C, or D after sequencing.25 In Hodgkin's disease, only 25 of the 34 sequenced cases could be directly allocated to one of the four groups, suggesting a higher degree of heterogeneity of the LMP-1 gene in Hodgkin's disease than in asymptomatic donors. However, only one case (HDc48) contained single base mutations, which placed it in an intermediate position between the four groups (table 2 ▶). The other non-grouped cases were isolates in which the Xho-I restriction site was lost or the 30 bp deletion detected (in one case a unique 12 bp deletion) in an LMP-1 gene together with single base mutations similar to group A or group B (table 2 ▶). Only one infectious mononucleosis isolate could not be grouped, but again this was because of a single mutation affecting the Xho-I restriction site. Apart from this mutation, the pattern of single base mutations was that found in group C. Therefore, we believe that groups A–D can be used to describe the variation in the EBV LMP-1 gene and promoter in European Hodgkin's disease and infectious mononucleosis cases.
In previous studies, loss of the Xho-I restriction site, detection of the 30 bp deletion, or partial sequence analysis of the LMP-1 gene have been used as markers of specific LMP-1 variants.12,13,16–24,26 Our present study shows that in European wild-type EBV isolates the Xho-I restriction site can be lost as a result of a range of mutations (fig 3 ▶). Only the mutation G → T (position 169425) is associated with the group D mutational pattern in the LMP-1 promoter and gene. In six of 10 cases in our study, loss of the Xho-I restriction site was the result of other mutations. Thus, the group of isolates characterised by this polymorphism is heterogeneous and the Xho-I restriction site polymorphism is not by itself a good group marker in European EBV variants. In contrast to European LMP-1 variants, loss of the Xho-I restriction site in Asian LMP-1 variants seems always to be the result of the G → T (position 169425) mutation. However, loss of this restriction site has been reported in different Chinese LMP-1 variants, and should therefore not be used as a marker of specific Asian LMP-1 variants.26 In contrast to the Xho-I restriction site, our sequence analysis shows that the 30 bp deletion is a adequate marker of the group C pattern (10 of 12 isolates) of single base mutations.
In conclusion, our study shows that EBV isolates with a variant of the LMP-1 gene (group D), which contains mutations in the functionally important ATF-1/CREB-1 binding site in the LMP-1 promoter, are under-represented in Hodgkin's disease, indicating selection pressure against this variant in Hodgkin's disease. Furthermore, our results show that our previous proposed grouping of LMP-1 variants (groups A–D) can be applied to Hodgkin's disease and that a substantial number of these wild-type variants contain mutations in an HLA-A2 restricted CTL epitope and possibly functionally important mutations in the transmembrane domain of the protein.
Figure 4.
Loss of the Xho-I restriction site was found in six Hodgkin's disease (HD) and four infectious mononucleosis (IM) isolates. Sequence analysis showed four different patterns of single base mutations in the recognition sequence of the Xho-I restriction enzyme. Only one of these (G → T, position 169425 and G → C, position 169423) was associated with the group D pattern of single base mutations in the latent membrane protein 1 (LMP-1) gene and promoter. The DNA sequence of the prototype Eptein-Barr virus (EBV) strain B95.8 is shown from positions 169430 to 169420.11 For study isolates, only the mutated nucleotides are shown. The recognition sequence of the Xho-I restriction enzyme is underlined. The arrow indicates the position where the enzyme cuts.
Acknowledgments
This work was supported by the Danish Cancer Society (Grant 95 100 16) and the Danish Medical Research Campaign (Grant 9503303 28809).
References
- 1.Anagnostopoulos I, Hummel M. Epstein-Barr virus in tumours. Histopathology 1996;29:297–315. [DOI] [PubMed] [Google Scholar]
- 2.Rickinson AB, Kieff E. Epstein-Barr virus. In: Fields BN, Knipe DM, Howley PM, eds. Fields virology. Philadelphia: Lippincott-Raven Publishers, 1996:2397–446.
- 3.Pallesen G, Hamilton-Dutoit SJ, Rowe M, et al. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet 1991;ii:320–2. [DOI] [PubMed] [Google Scholar]
- 4.Young LS, Dawson CW, Clark D, et al. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol 1988;69:1051–65. [DOI] [PubMed] [Google Scholar]
- 5.Rowe M, Pilon MP, Huen DS, et al. Upregulation of BCL-2 by the Epstein-Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-κB activation and to induction of cell surface markers. J Virol 1994;68:5602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng M, Lundgren E. Transient expression of the Epstein-Barr virus LMP-1 gene in human primary B cells induces cellular activation and DNA synthesis. Oncogene 1993;7:1775–82. [PubMed] [Google Scholar]
- 7.Wang D, Liebowits D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 1985;43:831–3. [DOI] [PubMed] [Google Scholar]
- 8.Kaye KM, Izumi KM, Kieff E. Epstein-Barr virus latent membrane protein-1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci U S A 1993;90:9150–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson CW, Rickinson AB, Young LS. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 1990;344:777–80. [DOI] [PubMed] [Google Scholar]
- 10.Fåhraeus R, Rymo L, Rhim JL, et al. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein-Barr virus. Nature 1990;345:447–9. [DOI] [PubMed] [Google Scholar]
- 11.Baer R, Bankier AT, Biggin MD, et al. DNA sequence and expression of the B95.8 Epstein-Barr virus genome. Nature 1984;310:207–11. [DOI] [PubMed] [Google Scholar]
- 12.Chen ML, Tsai CN, Liang CL, et al. Cloning and characterization of the latent membrane protein (LMP) of a specific Epstein-Barr virus variant derived from the nasopharyngeal carcinoma in the Taiwanese population. Oncogene 1992;7:2131–40. [PubMed] [Google Scholar]
- 13.Hu LF, Chen F, Zheng X, et al. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 1993;8:1575–83. [PubMed] [Google Scholar]
- 14.Li SN, Chang YS, Liu ST. Effect of a 10 amino acid deletion on the oncogenic activity of latent membrane protein 1 of Epstein-Barr virus. Oncogene 1996;12:2129–35. [PubMed] [Google Scholar]
- 15.Hu LF, Zabarovsky ER, Chen F, et al. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP-1) from a Chinese nasopharyngeal carcinoma. J Gen Virol 1991;72:2399–2409. [DOI] [PubMed] [Google Scholar]
- 16.Knecht H, Bachmann E, Brousset P, et al. Deletions within the LMP-1 oncogene of Epstein-Barr virus are clustered in Hodgkin's disease and identical to those observed in nasopharyngeal carcinoma. Blood 1993;82:2937–42. [PubMed] [Google Scholar]
- 17.Miller WE, Edwards RH, Walling DM, et al. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J Gen Virol 1994;75:2729–40. [DOI] [PubMed] [Google Scholar]
- 18.Sandvej K, Peh SC, Andresen B, et al. Identification of potential hot-spots in the carboxy-terminal part of the Epstein-Barr virus (EBV) BNLF-1 gene in both malignant and benign EBV associated diseases: high frequency of a 30 bp deletion in Malaysian and Danish peripheral T-cell lymphomas. Blood 1994;84: 4053–60. [PubMed] [Google Scholar]
- 19.Knecht H, Bachmann E, Brousset P, et al. Mutational hot spots within the carboxy terminal region of the LMP1 oncogene of Epstein-Barr virus are frequent in lymphoproliferative disorders. Oncogene 1995;10:523–8. [PubMed] [Google Scholar]
- 20.Santon A, Manzanal AI, Campo E, et al. Deletion in the Epstein-Barr virus latent membrane protein-1 oncogene in Hodgkin's disease. J Clin Pathol 1995;48:184–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YS, Su IJ, Chung PJ, et al. Detection of an Epstein-Barr virus variant in T-cell lymphoma tissues identical to the distinct strain observed in nasopharyngeal carcinoma in the Taiwanese population. Int J Cancer 1995;62:673–7. [DOI] [PubMed] [Google Scholar]
- 22.Cheung ST, Lo KW, Leung SF, et al. Prevalence of LMP-1 deletion variant of Epstein-Barr virus in nasopharyngeal carcinoma and gastric tumors in Hong Kong. Int J Cancer 1996;66:711–12. [DOI] [PubMed] [Google Scholar]
- 23.Chen HL, Lung ML, Chan KH, et al. Tissue distribution of Epstein-Barr virus genotypes. J Virol 1996;70:7301–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khanim F, Yun Q, Nieedobitek G, et al. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood 1996;88:3491–501. [PubMed] [Google Scholar]
- 25.Sandvej K, Gratama JW, Munch M, et al. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild type EBV isolates. Blood 1997;90:323–30. [PubMed] [Google Scholar]
- 26.Sung NS, Edwards RH, Seillier-Moiseiwitsch F, et al. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int J Cancer 1998;76:207–15. [DOI] [PubMed] [Google Scholar]
- 27.Chen ML, Wu RC, Liu ST, et al. Characterization of 5`-upstream sequence of the latent membrane protein 1 (LMP-1) gene of an Epstein-Barr virus identified in nasopharyngeal carcinoma tissues. Virus Res 1995;37:75–84. [DOI] [PubMed] [Google Scholar]
- 28.Sjöblom A, Yang W, Palmquist L, et al. An ATF/CRE element mediates both EBNA2-dependent and EBNA2-independent activation of the Epstein-Barr virus LMP1 gene promoter. J Virol 1998;72:1365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller WE, Cheshire JL, Baldwin AS, et al. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 1998;16:1869–77. [DOI] [PubMed] [Google Scholar]
- 30.Khanna R, Burrows SR, Nicholls J, et al. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur J Immunol 1998;28:451–8. [DOI] [PubMed] [Google Scholar]
- 31.Zhou XG, Hamilton-Dutoit SJ, Yan QH, et al. The association between Epstein-Barr virus and Chinese Hodgkin's disease. Int J Cancer 1993;55:359–63. [DOI] [PubMed] [Google Scholar]
- 32.Johannesen E, Koh E, Mosialos G, et al. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by jk and PU.1. J Virol 1995;69:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laux G, Adam B, Strobl L, et al. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-jk interacts with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J 1994;13:5624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh D, Kieff E. Cis-acting regulatory elements near the Epstein-Barr virus latent-infection membrane protein 1 transcription site. J Virol 1990;64:1855–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huen DS, Henderson SA, Croom-Carter D, et al. The Epstein-Barr virus latent membrane protein-1 (LMP-1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 1995;10:549–55. [PubMed] [Google Scholar]
- 36.Mosialos G, Birkenbach M, Yalamanchili R, et al. The Epstein-Barr virus transforming protein LMP-1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 1995;80:389–99. [DOI] [PubMed] [Google Scholar]
- 37.Devergne O, Hatzivassiliou H, Izumi KM, et al. Association of TRAF 1, TRAF2 and TRAF3 with an Epstein-Barr virus LMP-1 domain important for B-lymphocyte transformation: role in NFκB activation. Mol Cell Biol 1996;16:7098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaye K, Devergne O, Harada J, et al. Tumor necrosis factor receptor associated factor 2 is a mediator of NFκB activation in latent infection membrane protein 1, the Epstein-Barr virus transforming protein. Proc Natl Acad Sci U S A 1996;93:11085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Floettmann JE, Rowe M. Epstein-Barr virus latent membrane protein-1 (LMP-1) C-terminus activation region 2 (CTAR-2) maps to the far C-terminus and requires oligomerisation for NFκB activation. Oncogene 1997;15:1851–8. [DOI] [PubMed] [Google Scholar]
- 40.Gires O, Kohlhuber F, Kilger E, et al. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J 1999;18:3064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller WE, Mosialos G, Kieff E, et al. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NFκB activation. J Virol 1997;71:586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J Virol 1997;71:4649–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eliopoulos AG, Blake SM, Floettmann JE, et al. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J Virol 1999;73:1023–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RJ, Stack M, Hazlewood SA, et al. The 30 bp deletion in Chinese variants of the Epstein-Barr virus LMP1 gene is not the major effector of functional differencies between variant LMP1 genes in human lymphocytes. J Virol 1998;72:4038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falk K, Rötzschke O, StevanoviÉc S, et al. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991;351:290–6. [DOI] [PubMed] [Google Scholar]
- 46.Engelhard VH. Structure of peptides associated with class I and class II MHC molecules. Annu Rev Immunol 1994;12:181–207. [DOI] [PubMed] [Google Scholar]