Abstract
Aims—To investigate the status of chromosome 17 in a series of medulloblastomas using comparative genomic hybridisation (CGH) and fluorescence in situ hybridisation (FISH).
Methods—Frozen tissue and formalin fixed, paraffin wax embedded tissue from 27 medulloblastomas were analysed by CGH and FISH, respectively. CGH ratio profiles for chromosome 17 were compared with the results of FISH, for which loss or gain of 17p or 17q was assessed in two distinct ways using a combination of differentially labelled subtelomeric and centromeric probes and analysing 200 nuclei in each tumour.
Results—CGH revealed imbalances consistent with isochromosome 17q in eight of 27 tumours. Either loss of 17p or gain of 17q was identified in a further nine tumours, whereas 10 tumours were apparently balanced. Using control results from preparations of paraffin wax embedded tonsils, thresholds for the detection of abnormalities by FISH were established, either by determining the dominant pattern of signals in each case, or the mean ratio of subtelomeric to centromeric signals. Results by CGH and FISH were concordant in 21 of 27 tumours. In the remainder, most discrepancies related to methodological differences.
Conclusions—CGH has a role in disclosing common, genome wide chromosomal gains or losses in tumours, the clinical relevance of which can then be studied in large archival series of paraffin wax embedded tumours using FISH.
Keywords: medulloblastoma, fluorescence in situ hybridisation, comparative genomic hybridisation
Medulloblastomas make up more than 90% of cases of central nervous system (CNS) primitive neuroectodermal tumours, which are the most common CNS tumours in childhood.1,2 Advances in surgical and anaesthetic techniques and the introduction of various regimens of adjuvant radiotherapy and chemotherapy have improved the five year survival from 10% to about 50% in the past 30 years. However, further improvement will only follow an increased understanding of the biology of these tumours, which is needed to refine the use of current adjuvant treatments or to suggest new treatments.
The analysis of genetic abnormalities has been one focus of research on the biology of primitive neuroectodermal tumours. Cytogenetic studies of medulloblastoma have identified several non-random chromosomal aberrations in a high proportion of cases.3–6 Of these, isochromosome 17q (i(17q)) is the most common, occurring in a third of successfully karyotyped cases.4,7 Microsatellite analysis has corroborated the existence of chromosome 17 abnormalities by documenting the loss of heterozygosity (LOH) on 17p.8–10
More recently, the development of comparative genomic hybridisation (CGH) has allowed the screening of all tumours for genetic imbalances, not just those that grow and divide in vitro.11,12 This technique has been used in the study of medulloblastomas, and has supported and extended the results of cytogenetic reports on this tumour type.13–15
Whereas CGH serves as an effective screen for chromosomal imbalance across the genome, fluorescence in situ hybridisation (FISH) answers more specific questions, depending on the probes used.16 For example, probes to α-satellite repeat sequences at the centromere have been used to assess chromosome number, whereas unique sequence probes have been used to target specific loci. A combination of such probes with different fluorochromes can be used to assess loss or gain at specific loci relative to chromosome number, and this technique has been used widely in the study of a large variety of tumours.16,17 This type of FISH was initially developed for use with cultured tumour cells or touch preparations of fresh tumours, precluding its use on large archives of formalin fixed, paraffin wax embedded tumours. However, FISH techniques have been modified successfully to include prehybridisation tissue digestion steps and more sensitive probe detection, such that hybridisation can take place in nuclei extracted from paraffin wax embedded, formalin fixed tissue, or sections of such tissue.18,19
By comparing CGH and FISH in the analysis of chromosome 17 in a series of medulloblastomas, we aimed:
To explore the feasibility of performing FISH reliably on preparations of nuclei extracted from formalin fixed medulloblastomas.
To establish appropriate means of interpreting FISH performed in this way.
To provide more information about chromosome 17 imbalance in medulloblastomas, using a combination of these methods.
Materials and methods
TISSUE
All tumour biopsies (n = 27) came from patients (age range, 8 months to 32 years) presenting to the Wessex Neurological Centre in Southampton between 1985 and 1997. In each case, tumour samples for histological examination had been fixed in 10% neutral buffered formalin, then embedded in paraffin wax. Fresh tumour material surplus to diagnostic requirements was stored in liquid nitrogen. All specimens used in our study were obtained before the start of adjuvant treatment, except in two cases (12 and 15). In every case, the fixed and frozen tissues used for FISH and CGH, respectively, were examined histologically to ascertain the degree of contamination with non-neoplastic tissue. In 21 of 27 samples for CGH and 17 of 27 samples for FISH, there was no identifiable non-neoplastic tissue, and only a small amount of such tissue was present in the remainder. Normal tonsils (n = 5) were used as controls for FISH.
COMPARATIVE GENOMIC HYBRIDISATION (CGH)
CGH was performed on frozen tumour tissue as described previously,13 using a protocol modified from that of Kallioniemi et al.20 In summary, genomic DNA was salt extracted21 and directly labelled by nick translation, using fluorescein-12-dUTP and Texas red-5-dUTP (Dupont, Stevenage, UK) for test and reference DNAs, respectively. Labelled DNA from each of the test and reference sources was used in the hybridisation mixtures, which were denatured at 72°C for eight minutes and applied to normal male target metaphase slides (Vysis, Richmond, UK). After hybridisation for three days at 37°C, images were captured, and green to red fluorescence intensity ratios were calculated along the length of each chromosome. Data from five to 10 metaphases were combined to give a mean ratio profile for each chromosome, together with profiles corresponding to 99% confidence intervals (CIs). Gains or losses of material by the tumour were deduced from deviations of the mean profile beyond thresholds set at ratios of 1.1 and 0.9, as long as the 99% CIs also consistently deviated to the same side of the midline for the regions involved, and that there were no such deviations in the control.
NUCLEAR PREPARATIONS AND FISH
Sections (2 × 15 μm) of paraffin wax embedded tissue were dewaxed in xylene. Tissue then underwent three five minute cycles of suspension and centrifugation at 4000 ×g in absolute alcohol, 50% alcohol, and phosphate buffered saline (PBS), followed by digestion with 0.5% pepsin in 0.01 M HCl at 37°C for 120 minutes. Digestion was stopped with fetal calf serum, and the suspension was passed through a 70 μm mesh cell strainer before centrifugation at 250 ×g for 10 minutes. The pellet was resuspended in PBS before further centrifugation at 800 ×g for five minutes. Subsequent resuspension in a variable volume (up to 2 ml) of PBS preceded the making of cytospin nuclear preparations on 3-aminopropyltriethoxysilane coated slides, which were then stored at −20°C until required for FISH.
An unmasking step preceded hybridisation. Preparations of nuclei were warmed by incubation in distilled water at 37°C for 10 minutes. Nuclei were digested using 100 μl of pepsin (4 mg/ml in 0.2 M HCl) at 37°C for 12–16 minutes. The slides were subsequently washed in distilled water and PBS before dehydration through a series of graded alcohols (70%, 80%, and 95%). Digoxigenin labelled 17p or 17q subtelomere probes, each combined with a biotin labelled D17Z1 centromere probe, were brought to 37°C before vortexing and pulse centrifugation. Probe mixture (12 μl) and coverslips were applied to the preparations, the latter sealed with rubber adhesive. Co-denaturation at 75°C for five minutes preceded hybridisation overnight at 37°C in a humidified chamber.
After hybridisation, slides were placed in 2× saline sodium citrate (SSC) at room temperature before two five minute washes at 43°C in 0.1× SSC + 30% formamide (17p probe mix) or 0.5× SSC + 30% formamide (17q probe mix). After these stringency washes, slides were placed in fresh 2× SSC before application of 4× SSC with Tween 20 and dried skimmed milk (4× SSCTM) at 37°C in a humidified chamber. Antibody solutions (in 4× SSCTM) were applied sequentially at 37°C as follows: avidin–Texas red 1/500 (Vector, Peterborough, UK) plus fluorescein isothiocyanate (FITC) labelled antidigoxigenin Fab fragments 1/20 (Boehringer, Mannheim, Germany) for 30 minutes; biotinylated anti-avidin 1/100 (Vector) plus rabbit antisheep immunoglobulin 1/50 (Dako, Ely, UK) for 20 minutes; avidin–Texas red 1/500 (Vector) and FITC labelled swine antirabbit immunoglobulin 1/40 (Dako) for 20 minutes. After each detection step, slides were washed twice in 4× SSCT for two minutes. Slides were mounted in diamidinophenylindole (DAPI) with antifade (Vectashield).
FISH signals were revealed using a Leica fluorescence microscope with a triple band pass filter. Signals in 200 non-overlapping nuclei were counted, and the proportion of cells with a particular signal ratio, representing subtelomeric gain, loss, or balance, was recorded. In addition, the mean ratio of subtelomeric to centromeric signals was calculated using a formula that summed the relative contributions of signal ratios in all 200 cells, such that a balanced population of cells would have a mean signal ratio of 1. For example, a preparation with two subtelomeric and two centromeric signals in 60% of nuclei, one subtelomeric and two centromeric signals in 20%, two subtelomeric and three centromeric signals in 10%, and three subtelomeric and two centromeric signals in 10% would score the following:
2/2 × 0.6 + 1/2 × 0.2 + 2/3 × 0.1 + 3/2 × 0.1 = 0.917. Split signals representing sister chromatids were counted as one.22,23
The reproducibility of the FISH method was tested in 10 randomly chosen tumours, the entire protocol being repeated (with the same scorer) once or, in four cases, twice. The reliability coefficient was calculated (in SPSS) to be 0.94. The interobserver reliability coefficient was also calculated on the patterns of signals in eight tumours, and found to be 0.93.
Results
CGH (fig 1 ▶) and FISH (fig 2 ▶) were undertaken successfully in all 27 cases of medulloblastoma, and tables 1–3 ▶ ▶ ▶ summarise the results of chromosome 17 analysis. For controls performed with each CGH experiment, no imbalances reaching the criteria for inclusion were found on chromosome 17. In the tonsils acting as controls for FISH, the proportion of cells for which the number of centromeric and subtelomeric signals was equal (balanced) ranged from 69% to 80% (mean, 76.6%; SD, 4.4%) for the 17p probe, and 69% to 81% (mean, 77.6%; SD, 5.0%) for 17q. Of the other cells, 97% contained either two signals of one colour plus one signal of the other, or one signal of each colour. Two centromeric signals in each nucleus were detected in a mean of 82% of lymphoid cells, and two subtelomeric signals in 82.4% and 84.6% for 17p and 17q probes, respectively. Based on 200 nuclei in control tissue, FISH signal ratios (subtelomeric to centromeric) ranged from 1.03 to 1.10 (mean, 1.07; SD, 0.027) and 1.06 to 1.14 (mean, 1.10; SD, 0.03) for 17p and 17q, respectively.
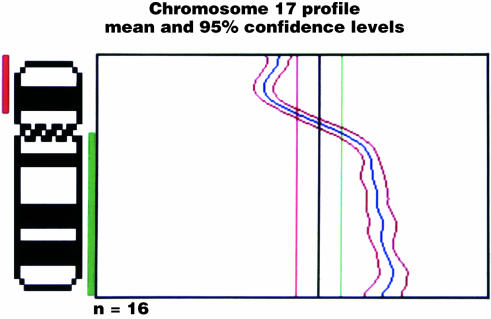
Figure 1.
The comparative genomic hybridisation (CGH) profile of a medulloblastoma with isochromosome 17q. The mean signal (blue) and its 95% confidence intervals deviate across thresholds for gain (green) and loss (red), set at 1.1 and 0.9, respectively.
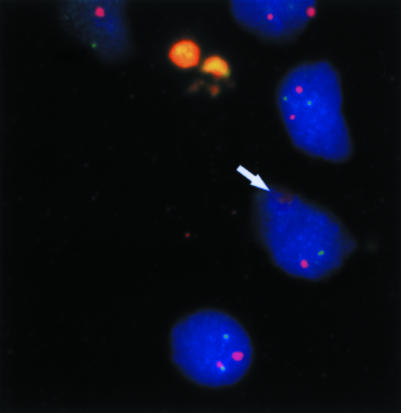
Figure 2.
This unprocessed fluorescence in situ hybridisation (FISH) image shows nuclei from a medulloblastoma outlined by DAPI (blue) against a speckled background of non-specific signals outside nuclei (yellow/orange). Inside the nuclei are centromeric (red) and subtelomeric (green) signals. These number two in all nuclei, but are sometimes out of the plane of focus (arrow), appearing enlarged and faint.
Table 1.
Tumours balanced by fluorescence in situ hybridisation (FISH) and comparative genomic hybridisation (CGH)
| FISH 17p % cells with | FISH 17q % cells with | FISH 17p status | FISH 17q status | CGH | ||||||||||
| Tumour | Loss | Balance | Gain | FISH 17p average signal | Loss | Balance | Gain | FISH 17q average signal | >Threshold | Average signal | >Threshold | Average signal | 17p loss | 17q gain |
| 1 | 15 | 65 | 20 | 1.11 | 13 | 66 | 21 | 1.13 | Balance | Balance | Balance | Balance | No | No |
| 3 | 22 | 61 | 17 | 1.05 | 12 | 62 | 26 | 1.16 | Balance | Balance | Balance | Balance | No | No |
| 4 | 7 | 77 | 16 | 1.12 | 11 | 72 | 17 | 1.10 | Balance | Balance | Balance | Balance | No | No |
| 6 | 12 | 67 | 21 | 1.11 | 7 | 74 | 19 | 1.11 | Balance | Balance | Balance | Balance | No | No |
| 7 | 29 | 59 | 12 | 0.97 | 17 | 63 | 20 | 1.07 | Balance | Balance | Balance | Balance | No | No |
| 11 | 12 | 74 | 14 | 1.08 | 10 | 68 | 22 | 1.15 | Balance | Balance | Balance | Balance | No | No |
| 20 | 16 | 69 | 15 | 1.07 | 10 | 70 | 20 | 1.13 | Balance | Balance | Balance | Balance | No | No |
| 24 | 20 | 68 | 12 | 1.01 | 12 | 68 | 20 | 1.12 | Balance | Balance | Balance | Balance | No | No |
| 25 | 25 | 64 | 11 | 1.00 | 18 | 67 | 15 | 1.04 | Balance | Balance | Balance | Balance | No | No |
Table 2.
Tumours showing loss and/or gain by fluorescence in situ hybridisation (FISH) and comparative genomic hybridisation (CGH)
| FISH 17p % cells with | FISH 17q % cells with | FISH 17p status | FISH 17q status | CGH | ||||||||||
| Tumour | Loss | Balance | Gain | FISH 17p average signal | Loss | Balance | Gain | FISH 17q average signal | >Threshold | Average signal | >Threshold | Average signal | 17p loss | 17q gain |
| 12 | 56 | 40 | 4 | 0.81 | 27 | 50 | 23 | 1.06 | Loss | Balance | Balance | Balance | Yes | No |
| 21 | 66 | 32 | 2 | 0.68 | 16 | 69 | 15 | 1.05 | Loss | Loss | Balance | Balance | Yes | No |
| 27 | 50 | 44 | 6 | 0.80 | 7 | 70 | 23 | 1.16 | Equivocal | Loss | Balance | Balance | Yes | No |
| 5 | 60 | 33 | 7 | 0.77 | 3 | 27 | 70 | 1.43 | Loss | Loss | Gain | Gain* | Yes | Yes§ |
| 10 | 77 | 22 | 1 | 0.62 | 2 | 44 | 54 | 1.39 | Loss | Loss | Equivocal | Gain* | Yes | Yes§ |
| 13 | 65 | 32 | 3 | 0.69 | 6 | 33 | 61 | 1.52 | Loss | Loss | Gain | Gain* | Yes | Yes§ |
| 14 | 67 | 29 | 4 | 0.72 | 5 | 37 | 58 | 1.52 | Loss | Loss | Gain | Gain* | Yes | Yes§ |
| 16 | 57 | 35 | 8 | 0.78 | 2 | 41 | 57 | 1.40 | Loss | Loss | Gain | Gain* | Yes | Yes§ |
| 26 | 68 | 27 | 5 | 0.69 | 8 | 45 | 47 | 1.40 | Loss | Loss | Equivocal | Gain* | Yes | Yes§ |
| 15 | 21 | 62 | 17 | 1.05 | 11 | 41 | 48 | 1.28 | Balance | Balance | Gain | Gain | No | Yes |
| 22 | 56 | 35 | 9 | 0.81 | 19 | 34 | 47 | 1.25 | Loss | Balance | Gain | Gain | No | Yes |
| 23 | 51 | 41 | 8 | 0.85 | 7 | 36 | 57 | 1.29 | Equivocal | Balance | Gain | Gain | No | Yes |
Equivocal indicates that no value is greater or equal to the threshold of 41%, or that more than one value crosses this threshold.
*i(17q) by FISH.
§i(17q) by CGH.
Table 3.
Tumours lacking concordance between fluorescence in situ hybridisation (FISH) and comparative genomic hybridisation (CGH)
| FISH 17p % cells with | FISH 17q % cells with | FISH 17p status | FISH 17q status | CGH | |||||||||||
| Tumour | Loss | Balance | Gain | FISH 17p average signal | Loss | Balance | Gain | FISH 17q average signal | >Threshold | Average signal | >Threshold | Average signal | 17p loss | 17q gain | 17 gain |
| 8 | 46 | 46 | 8 | 0.85 | 18 | 41 | 41 | 1.30 | Equivocal | Balance | Equivocal | Gain | Yes | Yes | No§ |
| 17 | 78 | 20 | 2 | 0.64 | 12 | 35 | 53 | 1.38 | Loss | Loss | Gain | Gain* | No | Yes | No |
| 19 | 72 | 24 | 4 | 0.67 | 7 | 61 | 32 | 1.17 | Loss | Loss | Balance | Balance | Yes | Yes | No§ |
| 2 | 70 | 25 | 5 | 0.75 | 30 | 38 | 32 | 1.08 | Loss | Loss | Equivocal | Balance | No | No | No |
| 9 | 19 | 69 | 12 | 1.02 | 22 | 64 | 14 | 1.02 | Balance | Balance | Balance | Balance | No | Yes | Yes |
| 18 | 27 | 44 | 29 | 1.05 | 16 | 42 | 42 | 1.19 | Balance | Balance | Equivocal | Balance | No | Yes | Yes |
Equivocal indicates that no value is greater or equal to the threshold of 41%, or that more than one value crosses this threshold.
*i(17q) by FISH.
§i(17q) by CGH.
With CGH, chromosome 17 was the most frequently affected by imbalance in this series of medulloblastomas; results relating to other chromosomes have been reported elsewhere.13 Loss of 17p was identified in 11 cases (tables 2 and 3 ▶ ▶), and was associated with reciprocal gain of 17q in eight, suggesting the presence of i(17q). Four cases had gain of 17q without loss of 17p, and two more had gain of the whole of chromosome 17, making a total of 14 with gain of all or part of chromosome 17. The remaining 10 cases showed no imbalance with respect to chromosome 17.
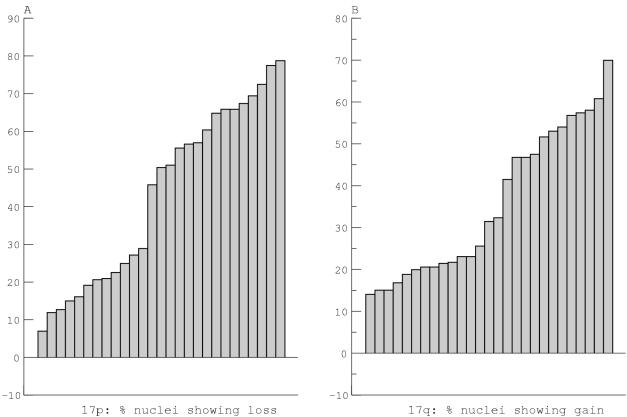
For the FISH analysis, we first categorised tumours according to a threshold set to identify the predominant population of cells. With normal cells and perfect hybridisation, one would expect 100% of nuclei to contain two signals for each probe, and a threshold of 50% would therefore define the predominant population in test cases. However, with fixed archival tissue hybridisation is not perfect, and our control data indicated that a threshold of 41% (half 82%) would be more appropriate. Distributions of the proportions of nuclei with 17p loss and 17q gain (fig 3 ▶) support this approach by demonstrating that this threshold lies on the steepest part of the data curves. For the alternative, averaged signal method of analysis, control data indicated that signal ratios of 0.8 and 1.2 would be appropriate thresholds for the interpretation of losses or gains.
Figure 3.
The series of medulloblastomas is ranked according to the proportion of nuclei showing loss of 17p (A) or gain of 17q (B).
Loss of 17p was identified in 12 tumours (tables 2 and 3 ▶ ▶) by the proportion of nuclei with subtelomeric signal loss, and in 11 by a signal ratio score of ≤ 0.8. FISH detected no significant population of nuclei with gain of 17p. Gain of 17q was identified in eight tumours by the proportion of nuclei with subtelomeric gain, and in 11 by a signal ratio score of ≥ 1.2. Using the signal ratio method of scoring, we found that seven tumours showed both loss of 17p and gain of 17q, suggesting i(17q), and that there was concordance between FISH and CGH results in six of seven cases.
A threshold of 41% proved problematic in a few cases. These were labelled “equivocal” because the threshold was either not reached—for example, tumour 2 (17q)—or crossed by groups of nuclei with different signal profiles. An example of the latter is tumour 8, for which an equal fraction of nuclei showed balance or loss and balance or gain, respectively, when 17p and 17q were examined.
In a comparison of the techniques, FISH results were concordant with CGH in 21 of 27 cases. Of these, tumours 10, 23, 26, and 27 showed one equivocal FISH result, but were concordant in terms of the signal ratio analyses and CGH results. Tumours 12 and 22 showed discrepant 17p results by the two FISH analyses, but could reasonably be counted as examples of 17p loss because their 17p signal ratios were 0.81 and over 50% of nuclei showed loss. We noted that over 41% of nuclei in both these tumours contained three or more centromeric signals, indicating aneuploidy. Of the six of 27 cases where CGH and FISH results differed, tumours 9 and 18 showed gain of the entire chromosome 17 by CGH, but were balanced by FISH analysis, as might be expected in the absence of centromere probes for other chromosomes. Tumour 8 showed equivocal FISH results by both scoring methods, but the findings could be regarded as consistent with the presence of a clonal population with i(17q). Tumours 2, 17, and 19, representing 11% of the series, remain as examples of an inexplicable mismatch between FISH and CGH results.
Discussion
We have successfully used FISH on preparations of nuclei extracted from 27 formalin fixed, paraffin wax embedded medulloblastomas, achieving results that are broadly in line with those demonstrated by CGH on the same tumours. In agreement with other genetic studies of medulloblastomas,6,14,24 our original CGH study showed that abnormalities of chromosome 17, particularly i(17q), are frequently found in these tumours.13 In our present study, we made chromosome 17 the focus of our FISH analysis, and compared signals from differentially labelled probes to the centromeric region of chromosome 17 and to subtelomeric regions of 17p or 17q in the assessment of loss or gain. This approach readily detects the gains and losses indicative of i(17q), and is also suitable for medulloblastomas that show losses on 17p, because microsatellite and FISH studies of multiple loci along the short arm have indicated that 17p loss nearly always involves large deletions that include the subtelomeric region.5,10
We first established control values for chromosome 17 FISH signals in nuclear preparations from five normal tonsils. The results demonstrated that hybridisation efficiency in formalin fixed tissue from paraffin wax blocks cannot match an optimal result obtained with cultured cells or fresh tissue, in which over 90% of normal nuclei would be expected to contain two signals.25,26 However, our results are similar to those reported in studies of a variety of fresh or fixed tissues, in which, because of methodological limitations, two signals were evident in about 80% of normal nuclei.19,23,27–30 Difficulties in adequately and homogeneously exposing DNA in formalin fixed tissue during enzyme digestion are likely to explain the reduced hybridisation efficiency. In such circumstances, there is a fine balance between optimising conditions for DNA exposure and probe hybridisation and the removal of non-specifically bound probe with stringent washes. These factors account for a wider variation in the FISH signals encountered in normal nuclei extracted from archival blocks, even after conditions have been optimised by varying the duration of prehybridisation digestion. Enhancement of probe visualisation using multiple layers in the detection step appears to be a further requirement of reduced hybridisation efficiency in these circumstances.
It is clear that some tumours in our series (for example, tumour 4) showed a distribution of FISH signals that matched control values. In these cases, the proportion of nuclei with balanced centromeric and subtelomeric signals is as high as could be expected from a population of normal nuclei. A similar proportion is also achieved in some tumours (for example, tumour 5), where 17p and 17q probes detect loss and gain, respectively.
In other tumours, one population of nuclei with a particular combination of probe signals did not necessarily predominate. Indeed, the interpretation of FISH results was not clear in several cases. Either variation in combinations of signals was so great that the threshold (41%) was not reached, as was the case for 17q in tumour 2, or the threshold was exceeded in more than one category—for example, tumour 8. Such results can probably be explained by clonal variation in the tumour, given that histological examination showed little contamination with normal cells. In addition, we noted that aneuploidy characterised by an excess of centromeric signals in many nuclei was a feature of several tumours (such as tumours 12, 18, and 22), in which there was no dominant population of nuclei with a single combination of signals.
Results that are not clear cut raise the issue of setting criteria for the categorisation of tumours. In some studies using a single centromeric probe, this has been achieved once control cases have been used to determine the proportion of cells that lies 2 or 3 SD outside the mean error rate for the method.31 With a combination of centromeric and sequence specific probes, genetic loss was simply recorded in two studies on formalin fixed, paraffin wax embedded tissue when the number of sequence specific signals was less than the number of centromeric signals.19,23 A distribution of the proportion of nuclei that showed loss in each series of tumours indicated that thresholds could be set at 40%19 or 50%.23 We chose our threshold to be half (41%) the mean value (82%) of balanced nuclei (2 : 2 signals) detected in the tonsil preparations by the centromeric probes, which gave very similar results to the subtelomeric 17p (82.4%) and 17q (84.6%) probes.
Because of difficulties in defining thresholds, we investigated an alternative method of scoring FISH signals. This summed the relative contribution of signal ratios (subtelomeric to centromeric) across the total population of counted nuclei, such that nuclei with balanced signals would have a score of 1. This method mimics the analysis used in CGH. We set thresholds at 0.8 and 1.2 for loss and gain, respectively. These differed from the CGH thresholds of 0.9 and 1.1, which were set on the basis of separate control results. Mean signal ratios in tonsil preparations deviated from unity, having values of 1.07 (17p) and 1.10 (17q), and we considered alternative, offset thresholds of 0.9 and 1.2 to reflect this. Applying these thresholds improves the concordance between the two methods of categorising FISH results. Thus, tumours 12 and 22 would be labelled as showing 17p loss by both methods. Provided the thresholds lie outside the control mean ± 3 SD, we decided that either approach could be justified.31 Calculating the results in this way reduces the likelihood of identifying abnormalities that are only present in a proportion of the cells, a frequent situation in tumours that show clonal variation, even when there is no normal cell contamination. Although this is also a recognised shortcoming of CGH, this method of analysis identifies abnormalities that are likely to be of most importance in the tumour.
In general, there was good agreement between the results obtained by CGH and FISH. Some discrepancies are easily explained. With CGH, tumours 9 and 18 showed gain of the whole of chromosome 17, but had a balanced profile with FISH. FISH will detect variations in copy number of chromosome 17, whether or not it reflects copy number of the whole genome (ploidy). However, it will not distinguish aneuploidy of chromosome 17 from variations in ploidy, without the concomitant use of probes for other chromosomes, or techniques such as flow cytometry to assess DNA index or ploidy. In contrast, CGH only detects aneuploidy relative to the rest of the genome.
Other discrepancies are not easily explained. Loss of 17p was unequivocally detected in tumours 2 and 17 by FISH, but not by CGH. These differences are unlikely to be the result of the inadequate assessment of FISH signals, in view of the numbers of scored nuclei. The possibility that a small deletion picked up by FISH could be missed by CGH, owing to a limitation in resolution, is not substantiated by data from our earlier study or other reports.5,10,13 Nearly all deletions of 17p in medulloblastomas are large enough to be readily detected by CGH.
Using FISH on nuclei extracted from formalin fixed, paraffin wax embedded tissue and CGH with DNA from frozen tumours, we have analysed the status of chromosome 17 in 27 medulloblastomas. There was good concordance between the results obtained by each method. Although some differences raised interesting issues of interpretation, we believe that CGH has a role in disclosing common, genome wide chromosomal gains or losses in tumours, the importance of which can then be studied in large archival series using FISH. In this way, controversies over such issues as the prognostic relevance of 17p loss in medulloblastomas may be resolved.32–35
Acknowledgments
The authors gratefully acknowledge the support of the Wessex Cancer Trust, the Parthenon Trust, and the Wolfson Foundation. They are also grateful for the assistance of Dr T Revesz, Dr J Darling, and Dr M Luxsuwong of the Institute of Neurology, London, UK; Miss S Beal of the Wessex Regional Genetics Laboratory, Salisbury, UK; Mrs J Buontempo of the Department of Pathology (Neuropathology), Southampton General Hospital; and Dr L Kearney of the Institute of Molecular Medicine, University of Oxford, UK.
References
- 1.Gjerris F, Agerlin N, Borgesen SE, et al. Epidemiology and prognosis in children treated for intracranial tumours in Denmark 1960–1984. Childs Nerv Syst 1998;14:302–11. [DOI] [PubMed] [Google Scholar]
- 2.Packer RJ. Brain tumors in children. Curr Opin Pediatr 1995;7:64–72. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee MB, Armstrong DD, Vogel H, et al. Cytogenetic analysis of 120 primary brain tumours and literature review. Cancer Genet Cytogenet 1997;97:39–53. [DOI] [PubMed] [Google Scholar]
- 4.Biegel JA, Rorke LB, Packer RJ, et al. Isochromosome 17q in primitive neuroectodermal tumours of the central nervous system. Genes Chromosomes Cancer 1989;1:139–47. [DOI] [PubMed] [Google Scholar]
- 5.Biegel JA, Rorke LB, Janss AJ, et al. Isochromosome 17q demonstrated by interphase fluorescence in situ hybridization in primitive neuroectodermal tumours of the central nervous system. Genes Chromosomes Cancer 1995;14:85–96. [DOI] [PubMed] [Google Scholar]
- 6.Bigner SH, McLendon RE, Fuchs H, et al. Chromosomal characteristics of childhood brain tumors. Cancer Genet Cytogenet 1997;97:125–34. [DOI] [PubMed] [Google Scholar]
- 7.Mertens F, Johansson B, Mitelman F. Isochromosomes in neoplasia. Genes Chromosomes Cancer 1994;10:221–30. [DOI] [PubMed] [Google Scholar]
- 8.Blaeker H, Rasheed BK, McLendon RE, et al. Microsatellite analysis of childhood brain tumours. Genes Chromosomes Cancer 1996;15:54–63. [DOI] [PubMed] [Google Scholar]
- 9.Cogen PH, Daneshvar L, Metzger AK, et al. Deletion mapping of the medulloblastoma locus on chromosome 17p. Genomics 1990;8:279–85. [DOI] [PubMed] [Google Scholar]
- 10.Scheurlen WG, Seranski P, Mincheva A, et al. High-resolution deletion mapping of chromosome arm 17p in childhood primitive neuroectodermal tumors reveals a common chromosomal disruption within the Smith-Magenis region, an unstable region in chromosome band 17p11.2. Genes Chromosomes Cancer 1997;18:50–8. [DOI] [PubMed] [Google Scholar]
- 11.du Manoir S, Speicher MR, Joos S, et al. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum Genet 1993;90:590–610. [DOI] [PubMed] [Google Scholar]
- 12.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992;258:818–21. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson JC, Ross FM, Kohler JA, et al. Comparative genomic hybridization and histological variation in primitive neuroectodermal tumours. Br J Cancer 1999;80:1322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reardon DA, Michalkiewicz E, Boyett JM, et al. Extensive genomic abnormalities in childhood medulloblastoma by comparative genomic hybridization. Cancer Res 1997;57:4042–7. [PubMed] [Google Scholar]
- 15.Schütz BR, Scheurlen W, Krauss J, et al. Mapping of chromosomal gains and losses in primitive neuroectodermal tumours by comparative genomic hybridization. Genes Chromosomes Cancer 1996;16:196–203. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher JA. DNA in situ hybridization as an adjunct in tumor diagnosis. Am J Clin Pathol 1999;112(suppl 1):S11–8. [PubMed] [Google Scholar]
- 17.Gray JW, Pinkel D, Brown JM. Fluorescence in situ hybridization in cancer and radiation biology. Radiat Res 1994;137:275–89. [PubMed] [Google Scholar]
- 18.Qian J, Bostwick DG, Takahashi S, et al. Comparison of fluorescence in situ hybridization analysis of isolated nuclei and routine histological sections from paraffin-embedded prostatic adenocarcinoma specimens. Am J Pathol 1996;149:1193–9. [PMC free article] [PubMed] [Google Scholar]
- 19.Sauter G, Deng G, Moch H, et al. Physical deletion of the p53 gene in bladder cancer. Detection by fluorescence in situ hybridization. Am J Pathol 1994;144:756–66. [PMC free article] [PubMed] [Google Scholar]
- 20.Kallioniemi O-P, Kallioniemi A, Piper J, et al. Optimising comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumours. Genes Chromosomes Cancer 1994;10:231–43. [DOI] [PubMed] [Google Scholar]
- 21.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopman AH, Ramaekers FC, Raap AK, et al. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry 1988;89:307–16. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M, Kawashima A, Mai M, et al. Analysis of chromosome 17p13 (p53 locus) alterations in gastric carcinoma cells by dual-color fluorescence in situ hybridization. Am J Pathol 1996;149:1575–84. [PMC free article] [PubMed] [Google Scholar]
- 24.Biegel JA. Genetics of pediatric central nervous system tumors. J Pediatr Hematol Oncol 1997;19:492–501. [DOI] [PubMed] [Google Scholar]
- 25.Arnoldus EP, Noordermeer IA, Peters AC, et al. Interphase cytogenetics reveals somatic pairing of chromosome 17 centromeres in normal human brain tissue, but no trisomy 7 or sex-chromosome loss. Cytogenet Cell Genet 1991;56:214–16. [DOI] [PubMed] [Google Scholar]
- 26.Lomax BL, Kalousek DK, Kuchinka BD, et al. The utilization of interphase cytogenetic analysis for the detection of mosaicism. Hum Genet 1994;93:243–7. [DOI] [PubMed] [Google Scholar]
- 27.Devilee P, Thierry RF, Kievits T, et al. Detection of chromosome aneuploidy in interphase nuclei from human primary breast tumors using chromosome-specific repetitive DNA probes. Cancer Res 1988;48:5825–30. [PubMed] [Google Scholar]
- 28.Ishikawa T, Kobayashi M, Mai M, et al. Amplification of the c-erbB-2 (HER-2/neu) gene in gastric cancer cells. Detection by fluorescence in situ hybridization. Am J Pathol 1997;151:761–8. [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa H, Inazawa J, Misawa S, et al. Detection of an I(17q) chromosome by fluorescent in situ hybridization with a chromosome 17 alpha satellite DNA probe. Cancer Genet Cytogenet 1992;62:140–3. [DOI] [PubMed] [Google Scholar]
- 30.Nederlof PM, van der Flier S, Raap AK, et al. Detection of chromosome aberrations in interphase tumor nuclei by nonradioactive in situ hybridization. Cancer Genet Cytogenet 1989;42:87–98. [DOI] [PubMed] [Google Scholar]
- 31.Roque L, Serpa A, Clode A, et al. Significance of trisomy 7 and 12 in thyroid lesions with follicular differentiation: a cytogenetic and in situ hybridization study. Lab Invest 1999;79:369–78. [PubMed] [Google Scholar]
- 32.Batra SK, McLendon RE, Koo JS, et al. Prognostic implications of chromosome 17p deletions in human medulloblastomas. J Neurooncol 1995;24:39–45. [DOI] [PubMed] [Google Scholar]
- 33.Biegel JA, Janss AJ, Raffel C, et al. Prognostic significance of chromosome 17p deletions in childhood primitive neuroectodermal tumors (medulloblastomas) of the central nervous system. Clin Cancer Res 1997;3:473–8. [PubMed] [Google Scholar]
- 34.Emadian SM, McDonald JD, Gerken SC, et al. Correlation of chromosome 17p loss with clinical outcome in medulloblastoma. Clin Cancer Res 1996;2:1559–64. [PubMed] [Google Scholar]
- 35.Scheurlen WG, Schwabe GC, Joos S, et al. Molecular analysis of childhood primitive neuroectodermal tumors defines markers associated with poor outcome. J Clin Oncol 1998;16:2478–85. [DOI] [PubMed] [Google Scholar]