Abstract
Background/Aims—In Epstein-Barr virus (EBV) positive cell lines that are stably infected, three different promoters are known to direct the transcription of EBV nuclear antigen 1 (EBNA1). These are located in the BamHI-C, BamHI-Q, and BamHI-F regions of the viral genome (Cp, Qp, and Fp, respectively). Fp is activated upon induction of the viral lytic cycle. The aim of this study was to investigate the activity of Fp in EBV associated diseases.
Methods—Using reverse transcriptase polymerase chain reaction, a qualitative analysis of EBNA1 promoter usage in various EBV associated diseases was performed.
Results—Fp driven transcription was detected in the context of primary infection and/or lytic replication; at least a portion of the Fp driven transcripts encoded EBNA1. Qp driven EBNA1 transcripts were detected in most samples across the range of disorders tested. Cp driven EBNA1 transcripts were detected in the context of immune suppression and in samples containing EBV positive (non-neoplastic) lymphoid cells.
Conclusions—These results confirm the previously proposed “housekeeping” function of the Qp promoter.
Keywords: Epstein-Barr virus, Epstein-Barr virus nuclear antigen 1, BamHI-F region
Epstein-Barr virus (EBV) is associated with several lymphoid and epithelial malignancies.1 These are endemic Burkitt's lymphomas, nasal natural killer (NK)/T cell lymphomas, post-transplant lymphoproliferative disorders, AIDS related lymphomas, nasopharyngeal carcinomas, approximately 40% of cases of Hodgkin's disease, and 8–10% of gastric carcinomas.2 In these malignancies, viral replication rarely occurs. Instead, the virus is latently present within the neoplastic cells (reviewed in Kieff3). The viral episome is maintained by the expression of EBV nuclear antigen 1 (EBNA1),4 which interacts with the host cell DNA and with the origin of replication (oriP) in the viral genome.
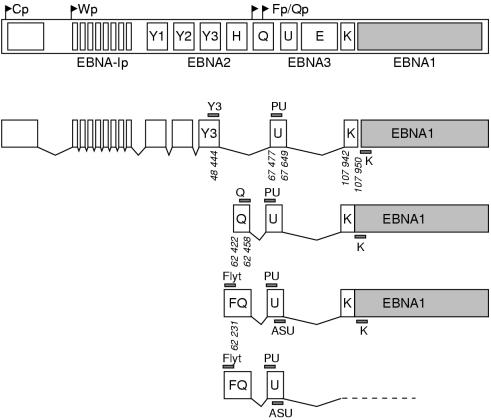
The transcription of EBNA1 can be driven by promoters located in the BamHI-W or BamHI-C fragments of the EBV genome (Wp and Cp, respectively; fig 1 ▶). Transcription from Cp/Wp results in long polycistronic mRNAs encoding not only EBNA1 but also one or more of the other EBNAs.6 This situation results in the extensive gene expression pattern (latency type III) that is found in lymphoblastoid cell lines (LCLs) and lymphomas of the immunocompromised: EBNA1, EBNA2, EBNA3a, EBNA3b, EBNA3c, and EBNA4; together with the latent membrane proteins (LMPs) LMP1, LMP2a, and LMP2b; the small non-coding RNAs, EBER-1 and EBER-2; and rightward transcripts driven from the BamHI-A region of the viral genome (BARTs).
Figure 1.
Schematic representation of Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) transcripts derived from the four different promoters. Large open boxes represent exons; small shaded bars represent reverse transcriptase polymerase chain reaction (RT-PCR) primers. Genomic coordinates given are those of the B95-8 prototype strain.5
Alternatively, EBNA1 can be transcribed from a promoter in the BamHI Q fragment of the viral genome (Qp; fig 1 ▶). This is a TATA-less promoter that resembles housekeeping gene promoters and thus guarantees EBNA1 expression in all EBV positive cells.7 In Burkitt's lymphomas, Hodgkin's disease, nasopharyngeal carcinomas, gastric carcinomas, and non-Hodgkin lymphomas of immunocompetent patients, Cp and Wp are inactive as a result of methylation8 and EBNA1 transcription is Qp driven.7, 9 This results in the expression of EBNA1 but none of the other EBNAs. In addition, EBERs and BARTs are detected in Burkitt's lymphomas (latency type I). Hodgkin's disease, nasopharyngeal carcinomas, gastric carcinomas, and non-Hodgkin lymphomas express EBNA1, EBERs, BARTs, and LMPs (latency type II).
Recently, another promoter for EBNA1 transcription was discovered. This promoter (Fp; fig 1 ▶) is localised 100–200 bp upstream of Qp,9–11 and was found to be active in cell lines after the induction of the viral lytic cycle.9, 11, 12 Originally, Fp was erroneously thought to be the promoter used for EBNA1 transcription during type I latency, but this misinterpretation was based on experiments in which part of the cells underwent spontaneous activation of the lytic cycle.13, 14 Recent studies have shown that Fp and Qp driven transcripts can be distinguished in reverse transcriptase polymerase chain reaction (RT-PCR) assays simply by using specific primers.7, 10, 15 However, these studies were all performed on EBV positive cell lines, whereas the presence and importance of Fp driven EBNA1 transcripts in EBV associated malignancies remained unknown. Therefore, we have designed an RT-PCR assay using a forward primer (Flyt) in combination with an EBNA1 specific antisense primer as described previously (table 1 ▶; primer K),16, 17 or with an antisense primer located in the U exon (table 1 ▶; fig 1 ▶). This primer enables the distinction between Fp and Qp driven transcripts using total RNA from clinical material and cell lines without the need for nested PCRs. Transcripts derived from Cp and Qp can be detected using the Y3 and Q sense primers, respectively, in combination with the U or K antisense primers (table 1 ▶).16
Table 1.
Oligonucleotide primers and probes used in RT-PCR analysis
| Target transcript | Oligo | B95.8 genomic coordinates | Sequence |
| EBNA1* | Y3 (s) | 48397–48416 | TGGCGTGTGACGTGGTGTAA |
| Q (s) | 62440–62457 | GTGCGCTACCGGATGGCG | |
| Flyt (s) | 62336–62355 | GACCACTGAGGGAGTGTTCC | |
| K (as) | 107986–107967 | CATTTCCAGGTCCTGTACCT | |
| ASU (as) | 67610–67589 | TCTACTGGCGGTGTATCATGCG | |
| PU (probe) | 67544–67563 | AGAGAGTAGTCTCAGGGCAT | |
| BARTs | A3 (s) | 157154–157173 | AGAGACCAGGCTGCTAAACA |
| A4 (as) | 159194–159175 | AACCAGCTTTCCTTTCCGAG | |
| AP (probe) | 157359–157378 | AAGACGTTGGAGGCACGCTG | |
| U1A | U1A1 (s) | – | CAGTATGCCAAGACCGACTCAGA |
| U1A2 (as) | – | GGCCCGGCATGTGGTGCATAA | |
| U1A3 (probe) | – | AGAAGAGGAAGCCCAAGAGCCA |
*All EBNA116 primers except Flyt, and all BART primers18 were described previously. We also used BLZF1 and BHRF1 specific primers as described previously.19
as, antisense; BARTs, BamH1A region driven transcripts; EBNA, Epstein-Barr virus nuclear antigen; RT-PCR, reverse transcription polymerase chain reaction; s, sense.
Materials and methods
SELECTION OF CLINICAL MATERIAL
All neoplasms tested in our study (table 2 ▶), with the exception of three nodal peripheral T cell lymphomas not otherwise specified, were associated with EBV (EBER1/2 RNA in situ hybridisation performed as described previously20 showed that most neoplastic cells contained EBV). The three nodal peripheral T cell lymphomas not otherwise specified were considered not “EBV associated”; that is, samples contained EBV positive cells but these were relatively few and non-neoplastic. The definition of EBV associated lymphomas was discussed previously.21 C15 is an nasopharyngeal carcinoma derived xenotransplant propagated in mice.22 The JY lymphoblastoid cell line23 was used as a positive control; the EBV negative Burkitt's lymphoma cell line Ramos was used as a negative control.
Table 2.
Analysis of EBNA1 promoter usage in Epstein-Barr virus (EBV) associated disorders
| Sample type | Qp | Cp | Fp* | BARTs |
| In vitro systems | ||||
| JY cells23 | + | + | + | + |
| C15 tumours22 | + | – | + | + |
| In vivo disorders | ||||
| Infectious mononucleosis (n = 1) | 1/1 | 1/1 | 1/1 | 1/1 |
| Reactive node, EBV positive cells (n = 1) | 1/1 | 0/1 | ND | 1/1 |
| Nasopharyngeal carcinoma (n = 15) | 15/15 | 5/15 | 0/5 | 15/15 |
| Lymphomas in patients without overt immunodeficiency | ||||
| Hodgkin's disease (n = 19) | 14/19 | 4/19 | 0/13 | 19/19 |
| Burkitt's lymphoma (n = 4) | 3/4 | 0/2 | 1/2 | 4/4 |
| Anaplastic large cell lymphoma (n = 1) | 1/1 | 0/1 | 0/1 | 1/1 |
| Nasal T/NK cell lymphoma (n = 6) | 6/6 | 0/5 | 0/2 | 6/6 |
| B non-Hodgkin's lymphoma (n = 4) | 2/4 | 1/4 | ND | 4/4 |
| Nodal PTCL NOS (n = 3) | 2/3 | 1/3 | 0/2 | 3/3 |
| Lymphomas in immunocompromised patients | ||||
| AIDS related lymphoma (n = 2) | 2/2 | 1/2 | 0/2 | 2/2 |
| Post-transplant LPD (n = 7) | 6/7 | 7/7 | 3/6 | 7/7 |
*Some cases could not be tested owing to lack of material.
BARTs, BamH1-A region driven transcripts; EBNA1, EBV nuclear antigen 1; LPD, lymphoproliferative disease; ND, not done; NK, natural killer; PTCL NOS, peripheral T cell lymphomas not otherwise specified.
PREPARATION OF RNA
RNA was isolated from 5 × 106 cultured EBV positive JY and EBV negative Ramos cells and from 10 cryosections, 5 μm thick, of biopsy samples using 1 ml of RNAzolTM (Biotecx Laboratories, Houston, Texas, USA). RNA was stored as an isopropanol precipitate at −80°C. Because the detectability of EBNA1 transcripts relies strongly upon the quality of the RNA,17 the integrity of the RNA was checked by gel electrophoresis; the presence of 28S/18S ribosomal bands was used as an indication of high quality RNA. Only samples that showed ribosomal bands on the gel were included in our study. As an additional quality control, RT-PCRs for the housekeeping gene U1A were performed.24 To check for the presence of amplifiable EBV mRNA in the preparations, we performed RT-PCR for BARTs, which are expressed in all types of latency.25
RT-PCR CONDITIONS
Multiprimed reverse transcription followed by single specific RT-PCR was performed as described previously.17 An amount of RNA equivalent to 1.5 cryosections, 5 μm thick, of the biopsies or to 100 000 cultured cells was reverse transcribed in a 20 μl reaction volume containing 10 pmol each of primer K, ASU, A4, and U1A2; subsequently, 1.5 μl of the generated cDNA was subjected to PCR. PCR products were analysed by gel electrophoresis, transferred to nylon filters (Qiabrane; Qiagen, Chatsworth, California, USA) by alkaline blotting, and hybridised with 32P labelled specific oligonucleotide probes (table 1 ▶).16, 18, 19 Films were exposed to these blots for four hours or overnight.
Results
ANALYSIS OF EBNA1 TRANSCRIPTION IN EBV POSITIVE CELL LINES
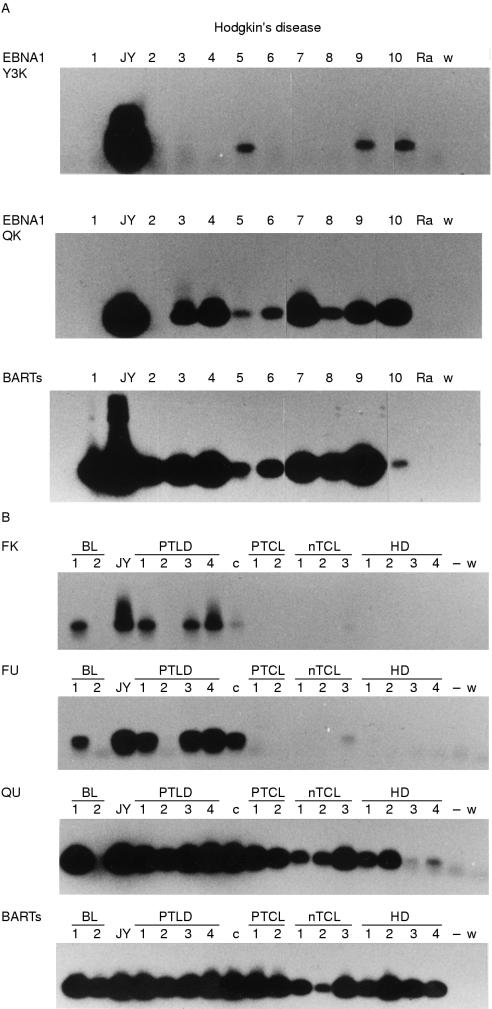
In the JY cell line, Cp/Wp driven EBNA1 transcripts were detected; Qp driven EBNA1 transcripts were detected to a lesser extent (fig 2A ▶, upper and middle panel, respectively). Fp driven transcripts (Fp-U spliced) were also detected (fig 2B ▶, second panel), indicating that at least in a proportion of cultured JY cells, EBV has entered into the lytic state. Moreover, using the Flyt and K primer pair, a signal was detected (fig 2B ▶, upper panel), indicating that (part of) the detected Fp driven transcripts encode EBNA1 in JY cells.
Figure 2.
Reverse transcriptase polymerase chain reaction (RT-PCR) analysis of EBNA1 promoter usage in Epstein-Barr virus (EBV) associated disorders. (A) Analysis of Cp (top) and Qp driven (middle) EBNA1 transcripts and BamHI-A rightward transcripts (BARTs, bottom) in Hodgkin's disease; (B) Analysis of Fp driven transcripts (Fp-U-K spliced, top; and Fp-U spliced, second row), Qp driven transcripts (Q-U spliced, third row) and BARTs in different EBV associated disorders. BL, Burkitt's lymphoma; c, C15 tumour; HD, Hodgkin's disease; JY, lymphoblastoid cell line; nTCL, nodal T cell lymphoma; PTCL, peripheral T cell lymphoma; PTLD, post-transplant lymphoproliferative disorder; Ra/−, EBV negative Ramos cell line; w, water control.
The C15 tumour showed clear expression of Qp driven but not Cp driven EBNA1 transcripts (not shown). After overnight exposure of the PCR blot, a faint signal for Fp driven EBNA1 transcripts (Fp-U-K spliced; fig 2B ▶, upper panel) was seen in the C15 tumour; as in the JY cells and the clinical samples, Fp-U spliced transcripts were also detected (fig 2B ▶, second panel) and this signal was stronger than the Fp-U-K signal.
ANALYSIS OF EBNA1 TRANSCRIPTION IN BIOPSIES FROM EBV ASSOCIATED DISEASES (TABLE 2 ▶)
Fp-U spliced transcripts
Fp-U spliced transcripts were only detected in post-transplant lymphoproliferative disorders (half of the cases tested), the infectious mononucleosis sample (not shown), and one of the two Burkitt's lymphoma samples tested (fig 2B ▶, second panel). We also assessed the presence of Fp-U-K spliced transcripts and found signals in the same samples that were positive for Fp-U spliced transcripts (fig 2B ▶, upper panel).
Q-U-K spliced EBNA1 transcripts
Q-U-K spliced EBNA1 transcripts were detected across the range of diseases tested in most samples, with the exception of five of the 19 Hodgkin's disease samples (in which no EBNA1 transcription was detectable), one of four Burkitt's lymphoma samples, two of four B cell non-Hodgkin's disease samples (of which one expressed Cp driven EBNA1 only and the other showed no detectable EBNA1 transcription), and one nodal peripheral T cell lymphoma not otherwise specified sample that contained < 10 EBV positive cells in each section and showed a weak signal for BARTs. Although the Q primer might also detect Fp driven transcripts (fig 1 ▶), the absence of Fp signals in all cases except the abovementioned post-transplant lymphoproliferative disorders, infectious mononucleosis, and Burkitt's lymphoma samples indicates that all signals found with the Q-U and the Q-K primer pairs are truly derived from Qp driven transcripts.
Q-U spliced transcripts
Q-U spliced transcripts were detected in the samples that were also positive for Q-U-K spliced transcripts (fig 2B ▶, third panel). The signals obtained with the Q-U primer pair were stronger than those of the Q-U-K primer pair, which might be because the amplimer generated with the Q-U primer pair is shorter; in fact, one Hodgkin's disease and one Burkitt's lymphoma sample, which were negative for Q-U-K, did show a signal with the Q-U primer pair and this was also true for the nodal peripheral T cell lymphoma that remained negative in the Q-U-K RT-PCR (fig 2B ▶, third panel).
Cp driven EBNA1 transcription was clearly detected in all post-transplant lymphoproliferative disorders, the infectious mononucleosis sample, and one of the two AIDS related lymphomas (not shown). Moreover, in five of 15 nasopharyngeal carcinoma samples a Cp signal was observed, although these signals were relatively weak compared with the positive controls and the post-transplant lymphoproliferative disorders. One B cell non-Hodgkin's lymphoma clearly expressed Cp driven EBNA1 transcripts. In three of the 19 Hodgkin's disease samples a faint signal was observed after overnight exposure; only one of the 19 Hodgkin's disease samples clearly expressed Cp driven EBNA1 transcripts. A faint Cp signal was also observed in one nodal peripheral T cell lymphoma not otherwise specified.
Discussion
The detection of Fp driven EBNA1 transcription in JY cells disagrees with the findings of Schaefer et al,15 who detected Fp-U spliced transcripts but not Fp-U-K spliced transcripts in JY cells. This discrepancy might result from interlaboratory differences in the JY cell culture. Lytic reactivation in some of the JY cells cultured in our laboratory has been demonstrated in a previous study using immunohistochemistry for the BZLF1 lytic activator.26 The detection of Fp driven EBNA1 transcription in the C15 tumour probably reflects viral replication that occurs in nude mice in the absence of a cytotoxic T cell response.
The detection of Fp driven EBNA1 transcription in infectious mononucleosis and in post-transplant lymphoproliferative disorders probably reflects the lytic replication of EBV. Infectious mononucleosis is known as the clinical manifestation of a primary EBV infection.27 Post-transplant lymphoproliferative disorders, like infectious mononucleosis, usually occur in patients not previously exposed to EBV.28 Moreover, the expression of the lytic replication activator BZLF1 was detected previously in infectious mononucleosis29 and post-transplant lymphoproliferative disorders.26 This is also the case for Burkitt's lymphoma30; Burkitt's lymphoma may even contain cells entering the lytic phase,31 which was recently shown to be related to a good response to chemotherapy.32
We determined whether BZLF1 transcription and/or protein synthesis coincided with Fp activity, but we could not find a clear correlation: the infectious mononucleosis sample and the three post-transplant lymphoproliferative disorders that showed Fp activity also showed BZLF1 mRNA (using RT-PCR) and protein (by immunohistochemistry using the BZ1 monoclonal antibody (Dako, Glostrup, Denmark). However, the three post-transplant lymphoproliferative disorders that did not show Fp activity also showed BZLF1 mRNA and protein expression. Moreover, all five nasopharyngeal carcinoma samples that were negative for Fp driven transcripts showed BZLF1 transcription, but BZLF1 protein could not be detected immunohistochemically. It is not known whether BZLF1 protein expression is necessary to induce Fp activity, although it was shown previously11 that the activation of Fp is dependent on de novo protein synthesis after the initiation of the lytic cycle.
The detection of Fp-U-K spliced RT-PCR products indicates that EBNA1 encoding transcripts can be Fp driven in clinical material. The same was shown recently for cell lines.33 However, the Fp-U RT-PCR signals were clearly stronger than the Fp-U-K RT-PCR signals, indicating that a proportion of Fp driven transcripts may encode proteins other than EBNA1. This was also suggested for cell lines in a previous study.33 Alternatively, RT-PCRs with the U antisense primer might proceed more efficiently than those with the K antisense primer, because with the U antisense primer a shorter amplimer is generated.
In a previous study,34 we showed that lymphomas of immunocompromised patients were positive in RT-PCR both with the Y3/K and with the Q/K primer pair, but the latter signals were ascribed to Fp activity because the Q/K primer pair does not enable the distinction between Fp and Qp driven EBNA1 transcripts (fig 1 ▶). In our present study, we show that Qp driven EBNA1 transcription in the absence of Fp activity does occur in the lymphomas of immunocompromised patients.
The ubiquitous expression of Qp driven transcripts indicates that Qp can indeed be considered a “housekeeping” promoter,7 which ensures constant EBNA1 transcription and thus maintenance of the viral episome in the host cells. The absence of detectable Qp transcripts in some of the samples might reflect the number of EBV infected cells, as is probably the case in the sample from the peripheral T cell lymphoma not otherwise specified (hence the weak BART signal found in this sample). Alternatively, specific downregulation of Qp activity by EBNA1 protein might occur.35 Recently, it was shown that this effect can be overcome by E2F36 and, therefore, the ratio between EBNA1 and E2F in EBV associated diseases is likely to determine Qp activity.
The detection of Cp driven EBNA1 transcription in post-transplant lymphoproliferative disorders, infectious mononucleosis, and AIDS related lymphomas is in agreement with the literature, Cp being the promoter that is active during latency type III, which is commonly found in these disorders.37–39 The EBV gene expression pattern that prevails in nasopharangeal carcinoma is latency type II,16, 40 but these tumours are known to contain abundant reactive lymphoid infiltrate. It is not unlikely that EBV infected B cells are present and contribute to the Cp signals observed. The same holds true for nodal peripheral T cell lymphomas not otherwise specified that contain EBV positive cells: we have shown that in these lymphomas the EBV positive cells are mostly (reactive) B cells41 and, in a previous study, Cp driven BHRF1 transcription was shown for this kind of lymphoma.19 Therefore, Cp activity is associated not only with a latency type III expression pattern but also with the presence of EBV positive reactive B cells.
Acknowledgments
This work was supported by grant VU94–749 from the Dutch Cancer Society, and by the European Community, grant 1C18-CT96–0132. The authors thank Ms J Post for excellent technical assistance. We are grateful to Dr van Gorp for kindly providing post-transplantation lymphoproliferative disorder, AIDS related lymphoma, and infectious mononucleosis biopsies, and Professor Griffin for providing Burkitt's lymphoma biopsy samples and C15 tumour material.
References
- 1.Miller G. Epstein-Barr virus: biology, pathogenesis and medical aspects. In: Fields BN, Knipe DM, eds. Virology, 2nd ed, Vol. 2. New York: Raven Press, 1990:1921–58.
- 2.Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol 1992;140:769–74. [PMC free article] [PubMed] [Google Scholar]
- 3.Kieff E. Epstein-Barr virus and its replication. In: Fields BN, Knipe DM, Howley P, et al, eds. Fields virology. Philadelphia: Lippincott-Raven, 1996:2343–96.
- 4.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 1985;313:821–5. [DOI] [PubMed] [Google Scholar]
- 5.Baer R, Bankier AT, Biggin MD, et al. DNA sequence and expression of the B95.8 Epstein-Barr virus genome. Nature 1984;310:207–11. [DOI] [PubMed] [Google Scholar]
- 6.Bodescot M, Brison O, Perricaudet M. An Epstein-Barr virus transcription unit is at least 84 kilobases long. Nucleic Acids Res 1986;14:2611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaefer BC, Strominger JL, Speck SH. Redefining the Epstein-Barr virus encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci U S A 1995;92:10565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaefer BC, Strominger JL, Speck SH. Host-cell determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol Cell Biol 1998;17:364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nonkwelo C, Skinner J, Bell A, et al. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J Virol 1996;70:623–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai CN, Liu ST, Chang YS. Identification of a novel promoter located within the BamHI Q region of the Epstein-Barr virus genome for the EBNA1 gene. DNA Cell Biol 1995;14:767–76. [DOI] [PubMed] [Google Scholar]
- 11.Schaefer BC, Strominger JL, Speck SH. The Epstein-Barr virus BamHI F promoter is an early lytic promoter: lack of correlation with EBNA 1 gene transcription in group 1 Burkitt's lymphoma cell lines. J Virol 1995;69:5039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lear AL, Rowe M, Kurilla MG, et al. The Epstein-Barr virus (EBV) nuclear antigen-1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle. J Virol 1992;66:7461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer BC, Woisetschlaeger M, Strominger JL, et al. Exclusive expression of Epstein-Barr virus nuclear antigen 1 in Burkitt lymphoma arises from a third promoter, distinct from the promoters used in latently infected lymphocytes. Proc Natl Acad Sci U S A 1991;88: 6550–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sample J, Brooks L, Sample C, et al. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci U S A 1991;88:6343–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaefer BC, Strominger JL, Speck SH. A simple reverse transcriptase PCR assay to distinguish EBNA1 gene transcripts associated with type I and II latency from those arising during induction of the viral lytic cycle. J Virol 1996;70:8204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks L, Yao QY, Rickinson AB, et al. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol 1992;66:2689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brink AATP, Oudejans JJ, Jiwa M, et al. Multiprimed cDNA synthesis followed by PCR is the most suitable method for Epstein-Barr virus transcript analysis in small lymphoma biopsies. Mol Cell Probes 1997;11:39–47. [DOI] [PubMed] [Google Scholar]
- 18.Deacon EM, Pallesen G, Niedobitek G, et al. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med 1993;177:339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oudejans JJ, Van den Brule AJC, Jiwa NM, et al. BHRF1, the Epstein-Barr virus (EBV) homologue of the bcl-2 (proto-)oncogene, is transcribed in EBV associated B-cell lymphomas and in reactive lymphocytes. Blood 1995;86:1893–902. [PubMed] [Google Scholar]
- 20.Jiwa NM, Kanavaros P, De Bruin PC, et al. Presence of Epstein-Barr virus harbouring small and intermediate-sized cells in Hodgkin's disease. Is there a relationship with Reed-Sternberg cells? J Pathol 1993;170:129–36. [DOI] [PubMed] [Google Scholar]
- 21.De Bruin PC, Jiwa M, Oudejans JJ, et al. Presence of Epstein-Barr virus in extranodal T-cell lymphomas: differences in relation to site. Blood 1994;83:1612–18. [PubMed] [Google Scholar]
- 22.Busson P, Ganem G, Flores P, et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int J Cancer 1988;42:599–606. [DOI] [PubMed] [Google Scholar]
- 23.Burakoff SJ, Ratnofsky SE, Benacerraf B. Mouse cytolytic T lymphocytes induced by xenogeneic rat stimulator cell exhibit specificity for H-2 complex alloantigens. Proc Natl Acad Sci U S A 1977;74:4572–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bijl J, van Oostveen JW, Kreike M, et al. Expression of HOXC4, -C5 and -C6 in human lymphoid cell lines, leukemias, and in benign and malignant lymphoid tissue. Blood 1996;87:1737–45. [PubMed] [Google Scholar]
- 25.Brooks LA, Lear AL, Young LA, et al. Transcripts from the Epstein-Barr virus BamHIa fragment are detectable in all three forms of virus latency. J Virol 1993;67:3182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oudejans JJ, Jiwa M, van den Brule AJC, et al. Detection of heterogeneous Epstein-Barr virus gene expression patterns within individual post-transplantation lymphoproliferative disorders. Am J Pathol 1995;147:923–33. [PMC free article] [PubMed] [Google Scholar]
- 27.Evans AS. Clinical syndromes associated with EB virus infection. Adv Intern Med 1972;18:77–93. [PubMed] [Google Scholar]
- 28.Harris NL, Ferry JA, Swerdlow SH. Posttransplant lymphoproliferative disorders: summary of society for hematopathology workshop. Semin Diagn Pathol 1997;14:8–14. [PubMed] [Google Scholar]
- 29.Niedobitek G, Agathanggelou A, Herbst H, et al. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J Pathol 1997;182:151–9. [DOI] [PubMed] [Google Scholar]
- 30.Niedobitek G, Agathanggelou A, Rowe M, et al. Heterogenous expression of Epstein-Barr virus latent protein in endemic Burkitt's lymphoma. Blood 1995;86:659–65. [PubMed] [Google Scholar]
- 31.Gutiérrez MI, Bhatia K, Magrath I. Replicative viral DNA in Epstein-Barr virus associated Burkitt's lymphoma biopsies. Leuk Res 1993;17:285–9. [DOI] [PubMed] [Google Scholar]
- 32.Labrecque LG, Xue SA, Kazembe P, et al. Expression of Epstein-Barr virus lytically related genes in African Burkitt's lymphoma: correlation with patient response to therapy. Int J Cancer 1999;81:6–11. [DOI] [PubMed] [Google Scholar]
- 33.Zetterberg H, Stenglein M, Jansson A, et al. Relative levels of EBNA1 gene transcripts from the C/W, F and Q promoters in Epstein-Barr virus-transformed lymphoid cells in latent and lytic stages of infection. J Gen Virol 1999;80:457–65. [DOI] [PubMed] [Google Scholar]
- 34.Oudejans JJ, Dukers DF, Jiwa NM, et al. Expression of Epstein-Barr virus encoded nuclear antigen 1 in benign and malignant tissues harbouring EBV. J Clin Pathol 1996;49:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sample J, Henson DEB, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol 1992;66:4654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davenport MG, Pagano JS. Expression of EBNA-1 mRNA is regulated by cell cycle during Epstein-Barr virus type I latency. J Virol 1999;73:3154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young L, Alfieri C, Hennessy K, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med 1989;321:1080–5. [DOI] [PubMed] [Google Scholar]
- 38.Tierney R, Steven N, Young L, et al. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol 1994;68:7374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacMahon EME, Glass JD, Hayward SD, et al. Epstein-Barr virus in AIDS-related primary central nervous system lymphoma. Lancet 1991;338:969–73. [DOI] [PubMed] [Google Scholar]
- 40.Hitt MM, Allday MJ, Hara T, et al. EBV gene expression in an NPC related tumour. EMBO J 1989;8:2639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brink AATP, ten Berge RL, van den Brule AJC, et al. Epstein-Barr virus is present in neoplastic cytotoxic T cells in extranodal, and predominantly in B cells in nodal T non-Hodgkin lymphomas. J Pathol 2000;191:400–6. [DOI] [PubMed] [Google Scholar]