Abstract
Background—Although a large number of allergens have been characterised, the structural, functional, and biochemical features that these molecules have in common, and that could explain their ability to elicit powerful IgE antibody responses, are still uncertain. Recently, there has been considerable interest in the role of the cysteine protease activity of the house dust mite allergen Der p 1 in biasing the immune response in favour of IgE production.
Aims—To search for remote homologues of Der p 1 with sequences similar to the 30 conserved amino acids surrounding the catalytic cysteine residue (Cys34).
Methods—Potential homologues were analysed by examining their three dimensional structures and multiple sequence alignments using the programs PROPSEARCH, ClustalW, GeneDoc, and Swiss Pdb Viewer.
Results—Diverse allergens (for example, the plant cysteine protease papain, the transport protein lipocalin Mus m 1, and the ragweed allergen Amb a 5) have a similar structural motif; namely, a groove resembling the substrate binding groove of Der p 1. The groove is located inside an α–β motif, between an α helix on one side and an antiparallel β sheet on the other side. A similar common motif (a cysteine stabilised α–β fold) can also be found in some toxins and defensins.
Conclusion—Allergens of diverse sources have a common structural motif, namely a groove located inside an α–β motif, which could potentially serve as a ligand binding site.
Keywords: allergen, Der p 1, protease, three dimensional protein structure
Type I hypersensitivity is the clinical manifestation of an immune response against foreign protein molecules, commonly known as allergens, which are potent inducers of IgE synthesis. Although a large number of allergens have been characterised, we are still uncertain about the structural, functional, or biochemical features that these molecules have in common, and that could explain their ability to elicit powerful IgE antibody responses. It has been suggested that allergenicity might result from an intrinsic biochemical property of the molecule, and on this basis allergens were classified into the following broad biochemical groupings: enzymes and enzyme inhibitors, transport proteins, regulatory proteins, and allergens with biochemical activities that do not fall into the previous categories.1 However, it is difficult to imagine how such relatively diverse biochemical activities could all lead to the stimulation of T cell helper type 2 (Th2) responses and subsequent bias towards the synthesis of IgE.
Thus, the identification of common features among allergens is of great importance to the basic understanding of allergic sensitisation and in the development of effective immunotherapeutic strategies. In recent years, there has been considerable interest in the role of the cysteine protease activity of the house dust mite allergen Der p 1 in subverting the regulatory process controlling IgE synthesis, thereby favouring an allergic outcome.2, 3 A recent study has demonstrated that the grass pollen allergen Phl p 1 also exhibits cysteine protease activity, and that it is a remote homologue of cysteine proteases of the papain family.4 Interestingly, Phl p 1 does not possess the classic catalytic triad found in the enzyme active site of Der p 1 (Cys34, His170, and Asn190).5
In our study, we sought to extend the search for remote homologues of Der p 1 with sequences similar to those surrounding the catalytic cysteine residue (Cys34)5 because this part of the molecule forms the substrate binding groove of cysteine proteases. The function of hydrolytic enzymes, such as cysteine proteases, is defined by their catalytic triad (see above) and the topology of the substrate binding site.6 Therefore, potential homologues were analysed by examining their three dimensional structures and multiple sequence alignments using the programs PROPSEARCH (http://www.infobiosud.univ-montpl.fr/SERVEUR/PROPSEARCH/Presentation.html), ClustalW,7 GeneDoc (www.cris.com/∼ketchup/genedoc.shtml), and Swiss Pdb Viewer.8 Our study has revealed that allergens of diverse sources have a common structural motif; namely, a groove that could potentially serve as a ligand binding site.
Methods
Remote homologues of Der p 1 were searched using the PROPSEARCH program because other programs for protein sequence alignment are not capable of detecting functional or structural homologues, particularly if the sequence identity is below the significance threshold of about 25% (http://www.infobiosud.univ-montpl.fr/SERVEUR/ PROPSEARCH/Presentation.html). The PROPSEARCH program uses the amino acid composition, rather than the order of amino acid residues in a sequence. In addition, 144 chemical properties such as molecular weight, content of bulky residues, content of small residues, average hydrophobicity, average charge, and the content of selected dipeptide groups are calculated from the sequence and used as query vector. The program has been shown to find remote homologues with the same fold, and most often similar function, but with insignificant alignment homology (http://www.infobiosud.univ-montp1.fr/SERVEUR/PROPSEARCH/propsearch.html).
The database was scanned using as template a 30 amino acid long sequence of Der p 1 (Met19–Leu48), which contains the catalytic cysteine residue (Cys34) and the surrounding conserved motifs. This stretch of 30 amino acids participates in the formation of the substrate binding groove of Der p 1, which consists of an α helix on one side and a loop connecting it to an antiparallel β sheet on the other side. To eliminate false positives, potential homologues were compared further with the query motif by performing multiple sequence alignments using the ClustalW program,7 and by checking whether any conserved residues could be found in the query and hit sequences. The chemical properties of the amino acids (negatively and positively charged residues, amide and alcohol residues, aliphatic and aromatic residues, and small and sulphur containing residues) were examined by viewing the sequences using the GeneDoc program (www.cris.com/∼ketchup/genedoc.shtml).
In addition, the three dimensional structures of the query and hit sequences were compared using the coordinates from the PDB9 or HSSP10 databases to identify common fold elements. The coordinates used were as follows: papain (1PPN),11 lipocalin Mus m 1 (1MUP),12 ragweed Amb a 5 (3BBG),13 insect defensin A (1ICA),14 antibiotics (3LEU),15 antifungal protein (1AUN),16 thionin (1AYJ),17 and toxins (1LQQ18 and 2SN319).
Results
REMOTE HOMOLOGUES OF DER P 1
Using the PROPSEARCH program, the hit sequences obtained contained many potential homologues from diverse biochemical groupings (enzymes and enzyme inhibitors, transport proteins, regulatory proteins, and allergens with biochemical activities that do not fall into the previous categories), with toxins and defensins representing the two largest groups (table 1 ▶). No such hit sequences could be obtained for the negative control sequence, containing the first 30 amino acid residues of Der p 1 (Met1–Lys30). Potential homologues of the Der p 1 sequence (Met19–Leu48) were selected for further investigation using three dimensional structure comparison and multiple sequence alignments.
Table 1.
Examples of potential remote homologues of Der p 1 determined by PROPSEARCH and tested by examining their three dimensional structures and multiple sequence alignments
| ID | Description | 3D coordinates |
| Hydrolytic enzymes | ||
| pa21_matbi | Phospholipase A2 isozyme I | 1POB (HSSP) |
| ctrp_penmo | Chymotrypsin (EC 3.4.21.1) | 1AZZ (HSSP) |
| Enzyme inhibitors | ||
| iaac_horvu | α-Amylase/trypsin inhibitor | 1BIP (HSSP) |
| rag1_orysa | Seed allergenic protein RAG 1 precursor | 1HSS (HSSP) |
| Regulatory proteins | ||
| prof_enthi | Profilin | 1YPR (HSSP) |
| Toxins | ||
| scx9_mesma | Toxin BMK-IX precursor | 1LQQ (HSSP) |
| sscx3_ortsc | Neurotoxin OS3 | 1LQQ (HSSP) |
| scx3_butom | Neurotoxin III | 1LQQ (HSSP) |
| scx5_cenno | Toxin CNGT II precursor | 2SN3 (HSSP) |
| Antibacterial/antifungal proteins | ||
| pis1_carpi | Bacteriocin pisciolin 126 | 3LEU (HSSP) |
| thhr_horvu | Antifungal protein (fragment) | 1AUN (HSSP) |
| defi_prote | Insect defensins A and B | 1ICA (PDB) |
| defi_anoga | Defensin precursor | 1ICA (PDB) |
| afp2_rapsa | Cysteine rich antifungal protein | 1AYJ (PDB) |
THREE DIMENSIONAL STRUCTURE COMPARISON
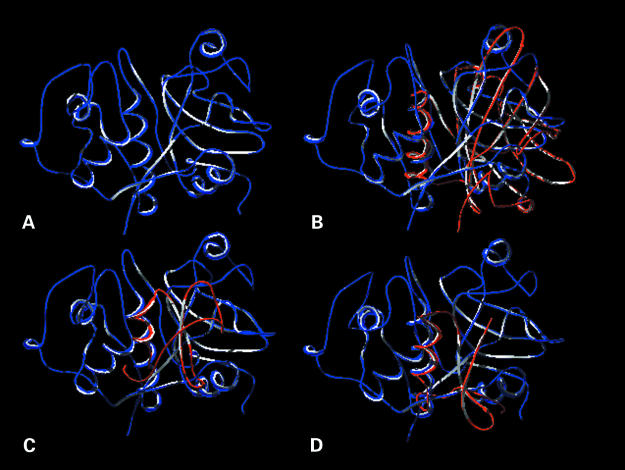
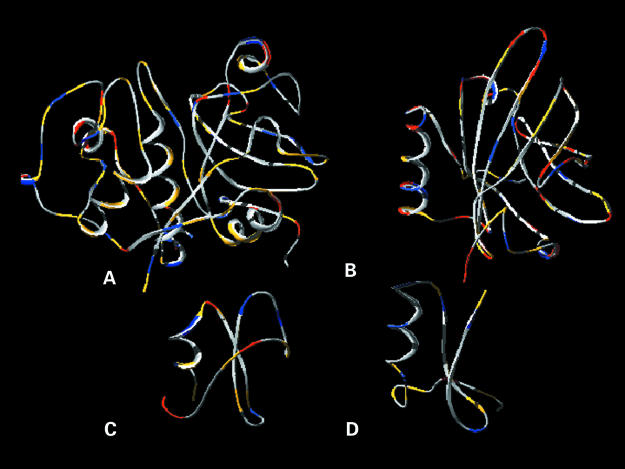
Available PDB files of numerous defensins and toxins, as well as other selected potential remote homologues, were viewed using the program Swiss Pdb Viewer.8 Examination of the three dimensional structures showed that the following potential homologues have a similar structural motif, namely an α–β groove resembling the substrate binding groove of Der p 1 (fig 1 ▶): plant cysteine proteases (such as papain), lipocalin Mus m 1, and ragweed Amb a 5. The defensin 1ICA possesses a similar groove (fig 1 ▶),14 which in toxins and defensins is known as a cysteine stabilised α–β motif (for example, leucocin 3LEU,15 antifungal protein 1AUN,16 thionin 1AYJ,17 and toxins 1LQQ18 and 2SN319 form a similar groove). Furthermore, analysis of the chemical properties of amino acids constituting the identified groove revealed conserved patterns (fig 2 ▶), thereby suggesting a common function.
Figure 1.
Comparison of the structural backbone of (A) papain with those of (B) lipocalin Mus m 1, (C) ragweed Amb a 5, and (D) defensin 1ICA. Papain is shown in blue and the superimposed structures of the other allergens in red. In all four structures the α–β motif forms a groove of similar dimensions.
Figure 2.
Comparison of the structural backbone of (A) papain with those of (B) lipocalin Mus m 1, (C) ragweed Amb a 5, and (D) defensin 1ICA, based on analysis of the chemical properties of the constituent amino acids. Amino acids are shown in red (acidic), blue (basic), yellow (polar) and grey (non-polar). Amino acids of similar nature occupy similar positions within the α–β groove.
MULTIPLE SEQUENCE ALIGNMENTS
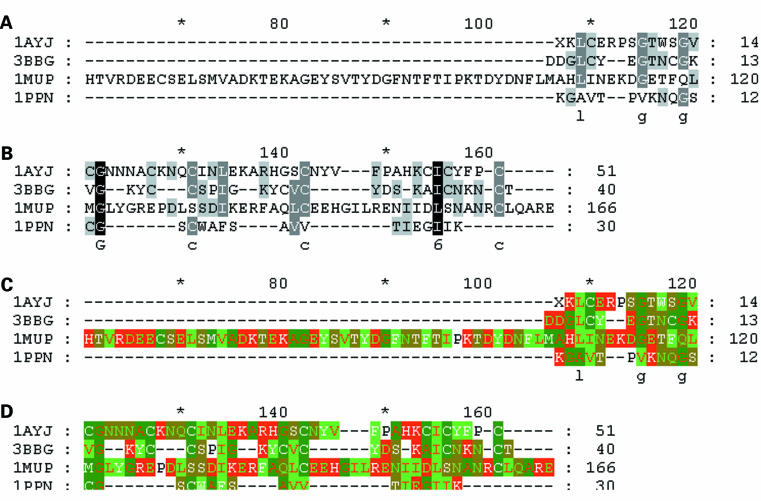
Multiple sequence alignments demonstrated remote sequence homology motifs (fig 3 ▶).
Figure 3.
Sequence alignments of papain (1PPN) with lipocalin (1MUP), ragweed (3BBG), and defensin (1AYJ). Residues that are conserved are shown in white on a black background, negatively charged in green on a red background, positively charged in white on a red background, amide in green on a khaki background, alcohol in white on a khaki background, aliphatic in red on a light green background, aromatic in white on a light green background, small in red on a dark green background, sulphur containing in white on a dark green background, and proline in black on a white background.
Discussion
In keeping with the notion that allergenicity might result from an intrinsic biochemical property of the molecule, allergens have previously been classified into the following broad biochemical groupings: enzymes and enzyme inhibitors, transport proteins, regulatory proteins, and allergens with biochemical activities that do not fall into the previous categories.1 Although it is tempting to suggest that such biochemical properties might contribute to allergenicity, it is difficult to imagine how such relatively diverse biochemical activities could all lead to stimulation of Th2 responses and subsequent bias towards IgE synthesis.
Remote homology among allergens is probably the result of convergent evolution, a process whereby molecules use different building blocks to achieve similar active sites. For instance, the two allergenic serine proteases trypsin and subtilisin differ drastically in their backbone structures, yet share similar catalytic sites.20 Another example is the lipocalin family of transport proteins, members of which only have up to 20% sequence homology, yet perform similar functions.21 With these observations in mind, we conducted a search for remote homologues of Der p 15 with sequences similar to those surrounding the catalytic cysteine residue (Cys34), Met19–Leu48. This 30 amino acid long sequence of Der p 1 forms the interdomain groove, with an α helix on one side and an antiparallel β sheet on the other side, and is the most conserved stretch in Der p 1 and in other cysteine protease allergens, such as papain, chymopapain, and actinidin. A comparison of amino acid sequences and their three dimensional structures revealed that allergens of diverse sources have a common structural motif; namely, a groove located between an α helix and an antiparallel β sheet.
The structural motif identified in our study (the α–β groove) was also found in a large number of toxins and defensins. Defensins are natural antibiotics that contribute to the innate resistance of plants, insects, and mammals to invading bacteria, fungi, and viruses.22 Plant defensins have remarkable structural similarity to scorpion neurotoxins and insect defensins23; they possess a common structural motif (an α helix connected by cysteine bridges to an antiparallel β sheet)14 that is similar to the fold identified in our study. We think that this similarity is not accidental because it complements our current understanding of allergens as toxins24 and substances that are involved in host defence (for example, lysozyme, phospholipase A2, lipocalins, Bet v 1, conalbumin, lactoferrin, class I chitinases, and thaumatin-like proteins).25 Therefore, it is reasonable to assume that similar host defence functions can be performed by allergens using a similar α–β groove, which could potentially enable them to bind to a common ligand. Thus, within the context of the immune system, such a ligand could be expressed on antigen presenting cells, such as dendritic cells, and be involved in antigen recognition and capture. The recent demonstration of the existence of two human dendritic cell subsets, which provide different cytokine microenvironments that determine the differentiation of either Th1 or Th2 cells,26 is particularly relevant in this connection. Clearly, future mutagenesis experiments should help to define the biological importance of the identified groove.
Acknowledgments
RF is supported by a University of Nottingham Postgraduate Scholarship and a Jack and Pat Mallabar Charitable Foundation grant.
References
- 1.Stewart GA, Thompson PJ. The biochemistry of common aeroallergens. Clin Exp Allergy 1996;26:1020–44. [PubMed] [Google Scholar]
- 2.Shakib F, Schulz O, Sewell HF. A mite subversive: cleavage of CD23 and CD25 by Der p 1 enhances allergenicity. Immunol Today 1998;19:313–16. [DOI] [PubMed] [Google Scholar]
- 3.Gough L, Schulz O, Sewell HF, et al. The cysteine protease activity of the major dust mite allergen Der p 1 selectively enhances the immunoglobulin E antibody response. J Exp Med 1999;190:1897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grobe K, Becker WM, Schlaak M, et al. Grass group I allergens (β-expansins) are novel, papain-related proteinases. Eur J Biochem 1999;263:33–40. [DOI] [PubMed] [Google Scholar]
- 5.Chua KY, Stewart GA, Thomas WR, et al. Sequence analysis of cDNA coding for a major house dust mite allergen, Der p 1. Homology with cysteine proteases. J Exp Med 1988;167:175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper NM, Karran EH, Turner AJ. Membrane protein secretases. Biochem J 1997;321:265–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modelling. Electrophoresis 1997;18:2714–23. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander C, Schneider R. Database of homology derived protein structures and the structural meaning of sequence alignment. Proteins 1991;9:56–68. [DOI] [PubMed] [Google Scholar]
- 11.Harris GW, Pickersgill RW, Howlin B, et al. The segmented anisotropic refinement of monoclinic papain by the application of the rigid-body TLS model and comparison to bovine ribonuclease A. Acta Crystallogr B 1992;48:67–75. [DOI] [PubMed] [Google Scholar]
- 12.Bocskei Z, Groom CR, Flower DR, et al. Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography. Nature 1992;360:186–8. [DOI] [PubMed] [Google Scholar]
- 13.Metzler WJ, Valentine K, Roebber M, et al. Proton resonance assignments and three-dimension solution structure of the ragweed allergen Amb a V by nuclear magnetic resonance spectroscopy. Biochemistry 1992;31:8697–705. [DOI] [PubMed] [Google Scholar]
- 14.Cornet B, Bonmatin JM, Hetru C, et al. Refined three-dimensional solution structure of insect defensin A. Structure 1995;3:435–48. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher NLF, Sailer M, Niemczura WP, et al. Three-D structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 1997;36:15062–72. [DOI] [PubMed] [Google Scholar]
- 16.Koiwa H, Kato H, Nakatsu T, et al. Crystal structure of tobacco PR-5d protein at 1.8 A resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J Mol Biol 1999;286:1137–45. [DOI] [PubMed] [Google Scholar]
- 17.Fant F, Vranken W, Broekaert W, et al. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J Mol Biol 1998;279:257–70. [DOI] [PubMed] [Google Scholar]
- 18.Landon C, Cornet B, Bonmatin JM, et al. 1H-NMR-derived secondary structure and the overall fold of the potent anti-mammal and anti-insect toxin III from the scorpion Leiurus Quinquestriatus quinquestriatus. Eur J Biochem 1996;236:395–404. [DOI] [PubMed] [Google Scholar]
- 19.Zhao B, Carson M, Ealick SE, et al. Structure of scorpion toxin variant-3 at 1.2 Å resolution. J Mol Biol 1992;227:239–52. [DOI] [PubMed] [Google Scholar]
- 20.Branden C, Tooze J. Introduction to protein structure, 2nd ed. New York: Garland; 1999.
- 21.Rautiainen J, Auriola S, Rouvinen J, et al. Molecular and crystal properties of Bos d 2, an allergenic protein of the lipocalin family. Biochem Biophys Res Commun 1998;247:746–50. [DOI] [PubMed] [Google Scholar]
- 22.Nidecker A, Writer S. Keratonicytes found to produce natural antibiotic. Skin and Allergy News 1998;29:24–9. [Google Scholar]
- 23.Kushmerick C, de Souza Castro, Santos Cruz J, et al. Functional and structural features of zeathionins, a new class of sodium channel blockers. FEBS Lett 1998;440:302–6. [DOI] [PubMed] [Google Scholar]
- 24.Madan T, Arora N, Sarma PU. Identification and evaluation of a major cytotoxin of A. fumigatus. Mol Cell Biochem 1997;167:89–97. [DOI] [PubMed] [Google Scholar]
- 25.Salcedo G, Diaz-Perales A, Sanchez-Monge R. Fruit allergy: plant defence proteins as novel potential panallergens. Clin Exp Allergy 1999;29:1158–60. [DOI] [PubMed] [Google Scholar]
- 26.Rissoan M-C, Soumelis V, Kadowaki N, et al. Reciprocal control of T helper cell and dendritic cell differentiation. Science 1999;283:1183–6. [DOI] [PubMed] [Google Scholar]