Abstract
Aims—Chromosome 3p allele loss is a frequent event in many common sporadic cancers including lung, breast, kidney, ovarian, and head and neck cancer. To analyse the extent and frequency of 3p allelic losses in T1N0 and T1N1 invasive sporadic breast cancer, 19 microsatellite markers spread along 3p were analysed in 40 such breast carcinomas with known clinicopathological parameters.
Methods—Loss of heterozygosity analysis was carried out using 3p microsatellite markers that were non-randomly distributed and chosen to represent regions that show hemizygous and/or homozygous losses in lung cancer (lung cancer tumour suppressor gene region 1 ( LCTSGR1) at 3p21.3 and LCTSGR2 at 3p12), and regions demonstrating suppression of tumorigenicity in breast, kidney, lung, and ovarian cancer.
Results—Allelic loss was seen at one or more loci in 22 of these clinically early stage sporadic breast tumours, but none had complete 3p allele loss. Several regions with non-overlapping deletions were defined, namely: (1) 18 tumours showed loss at 3p21–22, a physical distance of 12 Mb; (2) 11 tumours showed loss at 3p12 within a physical distance of 1 Mb, this region is contained within LCTSGR2; (3) six tumours showed loss at 3p25–24, including the von Hippel-Lindau (VHL) locus; (4) five tumours showed loss at 3p14.2, including the fragile histidine triad (FHIT) locus.
Conclusions—This is the largest study to date defining the extent and range of 3p allelic losses in early stage invasive breast cancer and the results indicate that region 3p21–22 containing LCTSGR1 and a region at 3p12 within LCTSGR2 are the most frequent sites of 3p allelic loss in these breast carcinomas. This suggests that tumour suppressor genes located in these regions may play important roles in the development of breast cancer. There was an association between increasing 3p allelic loss and increasing tumour grade and loss of progesterone (p = 0.0098) and oestrogen (p = 0.0472) receptor expression, indicating a link between 3p allelic loss and the regulation of differentiation.
Keywords: chromosome 3p, tumour suppressor genes, early invasive breast cancer
In the Western world breast cancer is the most prevalent malignancy in women. The incidence of breast cancer is rising and it is estimated that one in 10 women will develop breast cancer during her life time.1 An understanding of the genetic alterations involved in breast cancer development and progression may aid earlier detection and management.
Certain alterations, such as amplification of the oncogene ERBB2, can be found in a proportion of both in situ and invasive breast cancers, as can mutations of the tumour suppressor gene (TSG) p53 at 17q13, suggesting that they could play a role in the development of these tumours. Alterations to other oncogenes and TSGs, such as MYC and RB1, are associated with more advanced disease.2 Loss of heterozygosity (LOH) on chromosomes 1, 3p, 6q, 7q, 8p, 11p, 13q, 17p, 17q, 18q, and 22q has been reported in breast carcinomas and other tumours (reviewed in Bieche and Lidereau3 and Schwab4), indicating a role for TSGs located in these regions in the development and progression of different cancers. For familial breast cancer, two major genes have been isolated, BRCA1 at 17q21 and BRCA2 at 13q12–13 (reviewed in Buchholz and colleagues5 and Yang and Lippman6), and a third locus (at least) is also thought to exist.7,8 BRCA1 and BRCA2 do not show inactivating mutations in sporadic breast tumours, and their role in sporadic cancers is not known.
By means of hemizygosity and homozygosity mapping, cytogenetic analysis, and functional studies, distinct regions on 3p (3p25–26, 3p 21–22, 3p14.2, and 3p12) have been shown to be important for the development of several common sporadic cancers including lung, breast, kidney, ovarian, cervical, and head and neck cancer (reviewed in Kok and colleagues9).10–13 The region 3p25 contains the von Hippel-Lindau (VHL) TSG,14,15 which is inactivated in patients with von Hippel-Lindau disease and approximately 70% of sporadic clear cell renal carcinomas.16–19 However, mutations of VHL are rare in other common sporadic cancers that show 3p allele loss, such as lung and gonadal tumours.17,20 The fragile histidine triad (FHIT) gene at 3p14.2 undergoes homozygous deletions and alterations in its mRNA in many sporadic cancers (reviewed in Huebner and colleagues21 and Sozzi and colleagues22). High amounts of allele loss at the FHIT locus have been found in low grade ductal carcinoma in situ of the breast and the well differentiated tubular carcinomas,23 suggesting that alterations at the FHIT locus may be important in development of low grade breast cancer.
We have previously reported homozygous 3p deletions in sporadic breast cancers. The region at 3p21.3 (lung cancer tumour suppressor gene region 1; LCTSGR1) is defined by four overlapping homozygous deletions in three small cell lung cancer cell lines and one primary breast tumour.24–27 The 3p12 region (LCTSGR2) contains two overlapping homozygous deletions in small cell lung cancer cell lines and one breast tumour cell line.28 Several genes have been isolated from LCTSGR1 and one from LCTSGR2, but so far none shows frequent inactivating mutations in lung cancer.27,28 Recently, a small study (n = 8) demonstrated 3p allelic loss in benign breast lesions preceding invasive breast cancer.29 Another study demonstrated 3p loss in normal tissue adjacent to breast carcinomas, the most frequent loss being 3p22–25.30 However, only a low frequency of 3p loss has been found in comparative genomic hybridisation studies of ductal carcinoma in situ.31
We have analysed 3p allelic losses in T1N0 and T1N1 sporadic invasive breast carcinomas to determine their extent and frequency. We focused on specific 3p regions, (3p25–26, 3p21–22, 3p14.2, and 3p12) implicated in tumorigenesis in breast and other cancers. We found 3p loss in most of these breast tumours, and the two most frequently lost regions on 3p included LCTSGR1 and LCTSGR2. In addition, a trend was found between 3p allelic loss, higher tumour grade, and loss of oestrogen and progesterone receptors (ER and PR, respectively), suggesting that there are genes on 3p that are associated with differentiation and that their loss results in breast cancers with more aggressive features.
Materials and methods
PATIENTS AND SAMPLES
A total of 40 invasive breast carcinomas (39 infiltrating ductal carcinomas and one infiltrating lobular carcinoma) were studied. Twenty five were detected by mammographic screening and the others had presented symptomatically. Carcinomas were excised at Glenfield Hospital NHS Trust between July 1995 and July 1997. Tumours of 20 mm or less in maximum diameter were examined (range, 10–20 mm; mean, 17), and 15 had nodal metastases (T1N0 or T1N1, no more than three lymph nodes involved in positive cases). None of the tumours was from a woman with a known family history of breast or other cancers.
All tissues were fixed in 4% formaldehyde in saline for 18–36 hours. After slicing, selected blocks were processed through graded alcohols and xylene to paraffin wax.
The carcinomas were reported according to the Royal College of Pathologists' working party guidelines (1990). Infiltrating ductal carcinomas were graded using the modified Bloom and Richardson system.32 All tissue histological assessments were performed by RAW.
ER and PR immunohistochemistry was undertaken as described previously.33
DNA EXTRACTION AND MICRODISSECTION FROM PARAFFIN WAX EMBEDDED SECTIONS
Formalin fixed, paraffin wax embedded tissue from breast tumours and non-involved nodes served as the source of tumour and normal DNA, respectively. For each tumour–normal pair, DNA was extracted from 10 μm thick paraffin wax embedded sections, as described previously.34
MICROSATELLITE REPEAT ANALYSIS
Polymerase chain reaction (PCR) amplification of dinucleotide, trinucleotide, and tetranucleotide microsatellite sequences was carried out. Nineteen markers were selected spanning the regions of interest on 3p. All are available through Genome Database with the exception of new primers for the D3S1621 locus (forward primer, 5′-CCTCACTACTCCTGG AATTG-3′; reverse primer, 5′-CCAAGGAA GGGTTTTA CTTA-3′; PCR product size 140 bp, annealing temperature 55°C). LOH analysis was carried out as described previously.11 Electrophoresis was carried out for two to four hours at 90 W constant power to achieve adequate separation of alleles. After drying the gel was exposed to x ray film (Fuji, Tokyo, Japan).
We defined LOH as a complete absence of, or significantly decreased signal intensity of, one of the constitutional alleles in tumour DNA as determined by visual examination.
STATISTICAL ANALYSIS
Comparisons were made by Fisher's exact test and the χ2 test as appropriate. p Values of < 0.05 were taken as significant.
Results
LOH ANALYSIS USING 3P MICROSATELLITE MARKERS
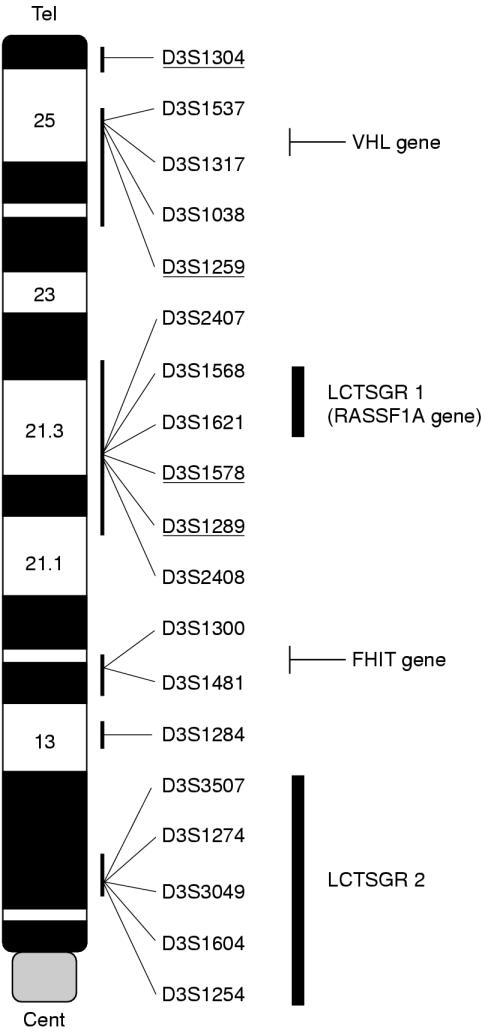
Figure 1 ▶ shows the location of the 19 microsatellite markers from 3p used to screen the 40 tumour–normal DNA pairs of breast carcinomas. The markers are non-randomly distributed and were chosen to represent regions showing hemizygous or homozygous losses and regions that show evidence of suppression of tumorigenicity in several common sporadic cancers including lung, breast, kidney, and ovary (fig 1 ▶).
Figure 1.
Summary of polymorphic markers and regions of interest on chromosome 3p. Genetic markers are listed in descending order from telomere to centromere according to published maps (UDB) and websites (http://apollo.uthscsa.edu/) and approximate cytogenetic (ideogram) positions. The regions LCTSGR1 and LCTSGR2 implicated in lung cancer development at 3p21.3 and 3p12 are also shown. The underlined markers represent loci shown to be important for suppression of tumorigenicity in an ovarian tumour cell line.11,40 The positions of the VHL (von Hippel-Lindau), RASSF1A and FHIT (fragile histidine triad) genes are also shown.
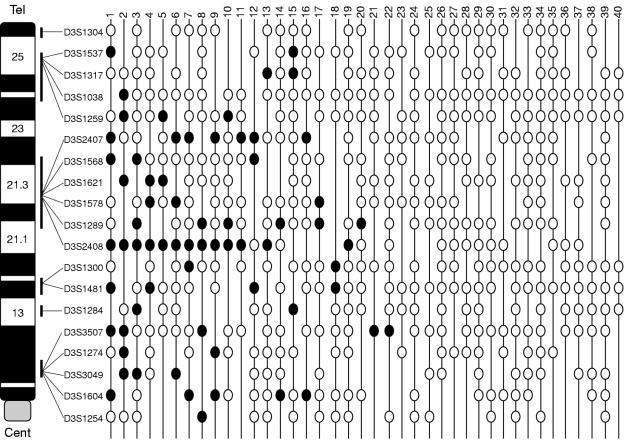
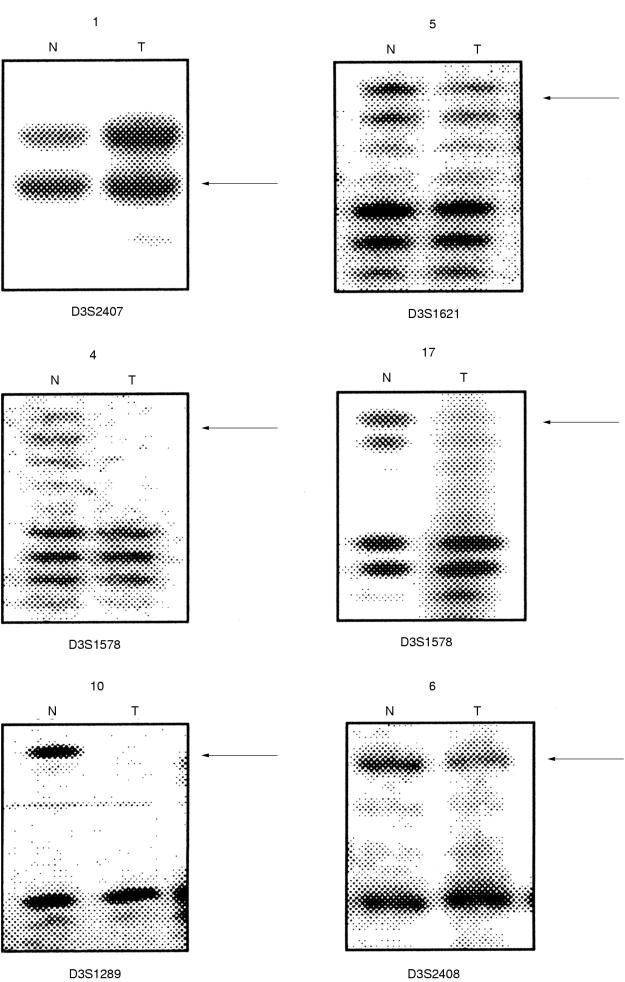
Allelic loss of one or more markers at 3p was seen in 22 tumours. None showed loss of every informative marker and 18 tumours showed no loss of informative markers (fig 2 ▶). The 22 tumours showing partial losses of 3p markers were analysed to identify regions of minimal overlapping deletions (fig 2 ▶). The highest loss (18 of 38 informative tumours) was seen at 3p21–22, between D3S2407 and D3S2408. This interval includes the region LCTSGR1, represented by microsatellite markers D3S1568 (equivalent to D3S4615) and D3S1621 (equivalent to D3S4623) and the region shown to be functionally important in ovarian cancer development. Within the region 3p21–22, the highest loss was observed for D3S2408 (13 of 25 informative tumours). The physical distance of this region bounded by D3S2407 and D3S2408 is 12 Mb (according to the unified database; UDB). Within this large region, smaller regions of overlapping allelic loss identified a region between D3S1289 and D3S2408, with a physical distance of 0.65 Mb, as the candidate region (tumours 20 and 14 both show loss at 1289 and retention at 2408, whereas tumours 9, 11, and 19 show retention at 1289 and loss at 2408) (figs 2 and 3 ▶ ▶). Four tumours showed loss only at 3p21–22 and retention of all informative markers at other 3p regions (tumours 17, 11, 19, and 20; fig 2 ▶). The next most frequently lost region was at 3p12, within the LCTSGR2 region, which was lost in 11 of 39 informative tumours. The highest 3p12 loss was seen for D3S1604 (five of 22 informative tumours); the markers D3S3507, D3S1274, D3S3049, and D3S1604 are located within < 1 Mb of each other. Nine tumours showing LOH at 3p12 also showed loss of distal markers, the remaining two tumours showing loss at 3p12 only were not informative for all distal markers. The region at 3p25–24, which includes the VHL locus, was lost in six tumours; however, all of these tumours also showed LOH at 3p21 and/ or 3p12/3p14. Five tumours showed LOH at the FHIT locus at 3p14.2, but again four of these were accompanied by more distal and or proximal losses, and the one remaining tumour was not informative for all distal and proximal markers.
Figure 2.
Summary of loss of heterozygosity (LOH) analysis. LOH pattern for all 40 early sporadic breast tumours (39 infiltrating ductal carcinomas and one infiltrating lobular carcinoma) for 3p markers. Each column represents a tumour and each row represents a 3p microsatellite marker listed in descending order from telomere (D3S1304) to centromere (D3S1254). The cases are arranged from left to right in decreasing order of chromosome 3p deletions. Status of each 3p locus is indicated: black circles, loss; white circles, retention; no symbols, uninformative loci.
Figure 3.
Representative examples of 3p loss of heterozygosity (LOH). Representative autoradiographs showing allelic losses for the markers at 3p21–22 (D3S2407, D3S1621, D3S1578, D3S1289, and D3S2408) in breast tumours. For each autoradiograph the case number is on the top. N, normal; T, tumour DNA. Arrowheads represent LOH.
ASSOCIATION WITH CLINICOPATHOLOGICAL PARAMETERS
A trend was seen between increasing 3p loss and higher tumour grade. Two of the eight grade 1 tumours, 11 of the 21 grade 2 tumours, and eight of the 11 grade 3 tumours showed 3p loss. Although the above figures are not significant, they do demonstrate a trend between 3p loss and increasing tumour grade.
Next, we investigated whether there was an association between tumour PR and ER expression and 3p allele loss. When the whole 3p arm was analysed we found that PR negative status was significantly more frequent in carcinomas with LOH at any 3p marker (nine of 19) than in those without 3p LOH (one of 16) (p = 0.0098). A significant association was also found for ER negative status and 3p loss (seven of 19) compared with tumours without 3p loss (one of 16) (p = 0.0472) (table 1 ▶). Furthermore, we found that two of the three carcinomas (tumours 11, 17, and 19) that had only 3p21–22 loss and known receptor status had loss of PR and ER expression, whereas the remaining tumour was positive for both receptors. This suggests a correlation between 3p21–22 allele loss and lack of PR and ER expression. The presence or absence of cancer cells in the lymph nodes is an important prognostic parameter; patients with few or no positive nodes have a far better prognosis than those with many. In our series of early stage invasive breast tumours, 14 of 25 lymph node negative tumours had loss of 3p markers, as did seven of 14 lymph node positive tumours. Hence, there was no correlation between lymph node status and 3p loss (p = 0.7496) (table 1 ▶).
Table 1.
3p loss of heterozygosity (LOH), together with grade, lymph node status, progesterone receptor (PR) status, and oestrogen receptor (ER) status
| LOH (number) | Retention (number) | |
| Any locus on 3p | ||
| Grade | ||
| 1 | 2 | 6 |
| 2 | 11 | 10 |
| 3 | 8 | 3 |
| Lymph node status | ||
| – | 14 | 11 |
| + | 7 | 7 |
| p = 0.7496 (NS) | ||
| Hormone receptor PR | ||
| – | 9 | 1 |
| + | 10 | 15 |
| p = 0.0098 | ||
| Hormone receptor ER | ||
| – | 7 | 1 |
| + | 12 | 15 |
| p = 0.0472 | ||
The results show an association between 3p LOH and increasing tumour grade, although this is not significant. There was no correlation between LOH and lymph node status. There was a significant correlation between loss of PR and ER expression (PR, p = 0.0098; ER, p = 0.0472) and loss at any 3p locus.
Discussion
We undertook high resolution deletion mapping on chromosome 3p in T1N0 and T1N1 invasive breast cancers to determine the frequency and importance of 3p loss and to map the precise regions on 3p that may contain TSGs important in breast cancer progression. We found the following: (1) a high incidence of 3p loss in this defined group of sporadic breast cancers; (2) evidence for two major candidate regions for 3p breast TSGs; (3) a trend between 3p allele loss and tumour grade; and (4) a significant correlation between 3p loss and loss of PR and ER expression. Although several other studies have analysed 3p LOH in breast cancer and reported LOH frequencies of 27–51%,35–38 these studies included all tumour stages and did not use a high density of markers for candidate breast cancer TSG regions. We found that just over a half of early stage invasive breast tumours showed loss of one or more 3p markers and defined two minimal regions of loss that may contain TSGs involved in the progression of breast cancer. Within the group studied we saw a trend between 3p LOH and increasing tumour grade. A recent study using comparative genomic hybridisation analysis of unselected breast cancers reported higher 3p loss in grade 3 breast tumours than in grade 1 tumours.39 These findings suggest that there are genes on 3p that are involved in the control of differentiation. Loss of these will result in the development of more aggressive cancers and this is confirmed by our finding of a significant correlation between chromosome 3p allele loss (most notably 3p21–22) and lack of PR and ER.
Multiple TSGs map to chromosome 3p and although we observed loss of one or more 3p markers in just over a half of the tumours, no tumour showed complete 3p LOH. We found that LOH was most frequent at 3p21–22. This region contains the LCTSGR1 (represented by microsatellite markers D3S1568 and D3S1621), which demonstrates overlapping homozygous deletions in lung and breast tumour cell lines.24–27 Although the physical distance between the markers at borders of this region is 12 Mb, the smaller regions of overlapping allelic losses within this interval narrow the candidate region to 0.65 Mb. This region between D3S1289 and D3S2408 overlaps with one of three candidate regions for suppression of tumorigenicity of an ovarian tumour cell line (fig 1 ▶).11,40 Multiple genes have been isolated from LCTSGR1,27 but to date, inactivating mutations in these genes are absent or rare in human cancers. Very recently, we and others have identified a gene (RASSF1A) from LCTSGR1 at 3p21.3 that is epigenetically inactivated in most lung cancers and to a lesser extent in breast tumours.41–43
The second most frequent region of LOH in early breast cancer that we identified was at 3p12. Within this region, the marker D3S1604 gave the highest loss, and the candidate interval corresponded to the LCTSGR2 region, which is defined by the presence of homozygous deletions in two lung cancer cell lines.28,44,45 LCTSGR2 also contains a homozygous deletion in one breast tumour cell line and this region is cloned in a 8 Mb yeast artificial chromosome contig. A candidate TSG, DUTT1, was isolated from this region. DUTT1 is a member of the NCAM family of genes, which includes the DCC (deleted in colorectal cancer) gene; however, DUTT1 mutations have so far not been identified in lung cancer and mutation analysis of DUTT1 in breast cancer has not been reported.28,46 Recently, the 3p12 region implicated in renal cell carcinoma by microcell mediated tumour suppression studies was shown to overlap with LCTSGR2.47,48
We also observed 3p LOH at 3p14.2 and 3p24–25 in our panel of breast tumours. However, loss in these regions was less frequent than at 3p21–22 and 3p12, and no tumour that was informative at 3p12 and 3p21 demonstrated LOH at 3p14.2 and not 3p12 or 3p21. Similarly, 3p24–25 LOH was always seen in tumours with coexisting 3p21–22 or 3p12 LOH. The VHL TSG maps to chromosome 3p25, but VHL gene mutations have not been identified in breast cancer. The 3p24–25 region is one of three candidate intervals for an ovarian cancer TSG; however, although a distal 3p TSG may be involved in some breast cancers, other 3p TSGs appear to be more important. We found five breast tumours that showed allelic losses at the FHIT locus. Abnormalities of FHIT mRNA transcripts and hemizygous and homozygous allelic losses at the FHIT locus have been reported in various cancers including lung, kidney, breast, and digestive tract cancers (reviewed in Huebner and colleagues21 and Sozzi and colleagues22). Although the role of the FHIT gene in tumorigenesis is still controversial, our results suggest that other 3p TSGs are more important for breast tumorigenesis. The pattern of discontinuous regions of 3p LOH with frequent losses at 3p21–22 and 3p12 is similar to that reported in lung cancer and preneoplastic lung lesions.12,49 The similarities between 3p LOH in lung and breast cancer suggest that the relevant 3p TSGs are involved in both tumour types.
In summary, we have demonstrated that regions on 3p (3p21–22 and 3p12) containing overlapping homozygous deletions in lung and breast tumours and tumour cell lines and a region involved in ovarian tumour suppression (3p21) show a high percentage of allelic losses in T1N0 and T1N1 invasive cancer, irrespective of node status, hence providing evidence for the involvement of genes residing at these regions in sporadic breast cancer development and progression. We also demonstrated increasing 3p loss with tumour grade and loss of ER and PR expression, resulting in the formation of more aggressive tumours. Larger studies are required to evaluate whether the 3p allelic losses described in this report can form a useful screening tool for identifying and understanding more aggressive disease.
Acknowledgments
This work was supported in part by a Breast Cancer Campaign grant, the University of Antioquia, Medellin (AM), and a fellowship from COLCIENCIAS (AM).
References
- 1.Harris JR, Lippman ME, Veronesi U, et al. Breast cancer (3). N Engl J Med 1992;327:473–80. [DOI] [PubMed] [Google Scholar]
- 2.Walker RA, Jones JL, Chappell S, et al. Molecular pathology of breast cancer and its application to clinical management. Cancer Metastasis Rev 1997;16:5–27. [DOI] [PubMed] [Google Scholar]
- 3.Bieche I, Lidereau R. Genetic alterations in breast cancer. Genes Chromosomes Cancer 1995;14:227–51. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M. Amplification of oncogenes in human cancer cells. Bioessays 1998;20:473–9. [DOI] [PubMed] [Google Scholar]
- 5.Buchholz TA, Weil MM, Story MD, et al. Tumor suppressor genes and breast cancer. Radiat Oncol Invest 1999;7:55–65. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Lippman ME. BRCA1 and BRCA2 in breast cancer. Breast Cancer Res Treat 1999;54:1–10. [DOI] [PubMed] [Google Scholar]
- 7.Bishop DT. BRCA1, BRCA2, BRCA3 … a myriad of breast cancer genes. Eur J Cancer 1994;30A:1738–9. [DOI] [PubMed] [Google Scholar]
- 8.Seitz S, Rohde K, Bender E, et al. Strong indication for a breast cancer susceptibility gene on chromosome 8p12–p22: linkage analysis in German breast cancer families. Oncogene 1997;14:741–3. [DOI] [PubMed] [Google Scholar]
- 9.Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res 1997;71:27–92. [DOI] [PubMed] [Google Scholar]
- 10.Wistuba II, Montellano FD, Milchgrub S, et al. Deletions of chromosome 3p are frequent and early events in the pathogenesis of uterine cervical carcinoma. Cancer Res 1997;57:3154–8. [PubMed] [Google Scholar]
- 11.Fullwood P, Sergio M, Rader JS, et al. Detailed genetic and physical mapping of tumor suppressor loci on chromosome 3p in ovarian cancer. Cancer Res 1999;59:4662–7. [PubMed] [Google Scholar]
- 12.Wistuba II, Behrens C, Milchgrub S, et al. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene 1999;18:643–50. [DOI] [PubMed] [Google Scholar]
- 13.Wistuba II, Behrens C, Virmani AK, et al. High resolution chromosome 3p allelotyping of lung cancer and preneoplastic bronchial epithelium reveals multiple sites of 3p allele loss and frequent breakpoints in the 600 kb 3p21.3 region. Cancer Res 2000;60:1949–60. [PubMed] [Google Scholar]
- 14.Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 1993;260:1317–20. [DOI] [PubMed] [Google Scholar]
- 15.Kaelin WG, Jr, Maher ER. The VHL tumour-suppressor gene paradigm. Trends Genet 1998;14:423–6. [DOI] [PubMed] [Google Scholar]
- 16.Foster K, Prowse A, van den Berg A, et al. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet 1994;3:2169–73. [DOI] [PubMed] [Google Scholar]
- 17.Gnarra JR, Tory K, Weng Y, et al. Mutation of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85–90. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A 1994;91:9700–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clifford SC, Prowse AH, Affara NA, et al. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumorigenesis. Genes Chromosomes Cancer 1998;22:200–9. [DOI] [PubMed] [Google Scholar]
- 20.Foster K, Osborne RJ, Huddart RA, et al. Molecular genetic analysis of the von Hippel-Lindau disease (VHL) tumour suppressor gene in gonadal tumors. Eur J Cancer 1995;31A:2392–5. [DOI] [PubMed] [Google Scholar]
- 21.Huebner K, Garrison PN, Barnes LD, et al. The role of the FHIT/FRA3B locus in cancer. Annu Rev Genet 1998;32:7–31. [DOI] [PubMed] [Google Scholar]
- 22.Sozzi G, Huebner K, Croce CM. FHIT in human cancer. Adv Cancer Res 1998;74:141–66. [DOI] [PubMed] [Google Scholar]
- 23.Man S, Ellis IO, Sibbering M, et al. High levels of allele loss at the FHIT and ATM genes in non-comedo ductal carcinoma in situ and grade I tubular invasive breast cancers. Cancer Res 1996;56:5484–9. [PubMed] [Google Scholar]
- 24.Daly MC, Xiang RH, Buchhagen D, et al. A homozygous deletion on chromosome 3 in a small cell lung cancer cell line correlates with a region of tumor suppressor activity. Oncogene 1993;8:1721–9. [PubMed] [Google Scholar]
- 25.Wei MH, Latif F, Bader S, et al. Construction of a 600 kb cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumour suppressor gene (TSG) locus on human chromosome 3p21.3. Cancer Res 1996;56:1487–92. [PubMed] [Google Scholar]
- 26.Sekido Y, Ahmadian M, Wistuba II, et al. Cloning of a breast cancer homozygous deletion junction narrows the region of search for a 3p21.3 tumor suppressor gene. Oncogene 1998;16:3151–7. [DOI] [PubMed] [Google Scholar]
- 27.Lerman MI, Minna JD, for The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. The 630 kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res 2000;60:6116–33. [PubMed] [Google Scholar]
- 28.Sundaresan V, Chung G, Heppell-Parton A, et al. Homozygous deletions at 3p12 in breast and lung cancer. Oncogene 1998;17:1723–9. [DOI] [PubMed] [Google Scholar]
- 29.Euhus DM, Maitra A, Wistuba II, et al. Loss of heterozygosity at 3p in benign lesions preceding invasive breast cancer. J Surg Res 1999;83:13–18. [DOI] [PubMed] [Google Scholar]
- 30.Deng G, Lu Y, Zlotnikov G, et al. Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 1996;274:2057–9. [DOI] [PubMed] [Google Scholar]
- 31.Buerger H, Otterbach F, Simon R, et al. Comparative genomic hybidization of ductal carcinoma in situ of the breast—evidence of multiple genetic pathways. J Pathol 1999;187:396–402. [DOI] [PubMed] [Google Scholar]
- 32.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991;19:403–10. [DOI] [PubMed] [Google Scholar]
- 33.Chappell SA, Walsh T, Walker RA, et al. Loss of heterozygosity at chromosome 6q in preinvasive and early invasive breast carcinomas. Br J Cancer 1997;75:1324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw JA, Walsh T, Chappell SA, et al. Microsatellite instability in early sporadic breast cancer. Br J Cancer 1996;73:1393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali IU, Lidereau R, Callahan R. Presence of two members of c-erbA receptor gene family (c-erbA beta and c-erbA2) in smallest region of somatic homozygosity on chromosome 3p21–p25 in human breast carcinoma. J Natl Cancer Inst 1989;81:1815–20. [DOI] [PubMed] [Google Scholar]
- 36.Sato T, Akiyama F, Sakamoto G, et al. Accumulation of genetic alterations and progression of primary breast cancer. Cancer Res 1991;51:5794–9. [PubMed] [Google Scholar]
- 37.Matsumoto S, Kasumi F, Sakamoto G, et al. Detailed deletion mapping of chromosome arm 3p in breast cancers: a 2-cM region on 3p14.3–21.1 and a 5-cM region on 3p24.3–25.1 commonly deleted in tumors. Genes Chromosomes Cancer 1997;20:268–74. [PubMed] [Google Scholar]
- 38.Braga E, Pugacheva E, Bazov I, et al. Comparative allelotyping of the short arm of human chromosome 3 in epithelial tumors of four different types. FEBS Lett 1999;454:215–19. [DOI] [PubMed] [Google Scholar]
- 39.Roylance R, Gorman P, Harris W, et al. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res 1999;59:1433–6. [PubMed] [Google Scholar]
- 40.Rimessi P, Gualandi F, Morelli C, et al. Transfer of human chromosome 3 to an ovarian carcinoma cell line identifies three regions on 3p involved in ovarian cancer. Oncogene 1994;9:3467–74. [PubMed] [Google Scholar]
- 41.Dammann R, Li C, Yoon JH, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet 2000;25:315–19. [DOI] [PubMed] [Google Scholar]
- 42.Burbee DG, Forgacs E, Zochbauer S, et al. The RASSF1A locus in the 3p21.3 homozygous deletion region: epigenetic inactivation in lung and breast cancer and suppression of the malignant phenotype. J Natl Cancer Inst 2001;93:691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agathanggelou A, Honorio S, Macartney DP, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumors. Oncogene 2001;20:1509–18. [DOI] [PubMed] [Google Scholar]
- 44.Rabbitts P, Bergh J, Douglas J, et al. A submicroscopic homozygous deletion at the D3S3 locus in a cell line isolated from a small cell lung carcinoma. Genes Chromosomes Cancer 1990;2:231–8. [DOI] [PubMed] [Google Scholar]
- 45.Latif F, Tory K, Modi W, et al. Molecular characterization of a large homozygous deletion in the small cell lung cancer line U2020: a strategy for cloning the putative tumor suppressor gene. Genes Chromosomes Cancer 1992;5:119–27. [DOI] [PubMed] [Google Scholar]
- 46.Sundaresan V, Roberts I, Bateman A, et al. The DUTT1 gene, a novel NCAM family member is expressed in developing murine neural tissues and has an unusually broad pattern of expression. Mol Cell Neurosci 1998;11:29–35. [DOI] [PubMed] [Google Scholar]
- 47.Lott ST, Lovell M, Naylor SL, et al. Physical and functional mapping of a tumor suppressor locus for renal cell carcinoma within chromosome 3p12. Cancer Res 1998;58:3533–7. [PubMed] [Google Scholar]
- 48.Lovell M, Lott ST, Wong P, et al. The genetic locus NRC-1 within chromosome 3p12 mediates tumor suppression in renal cell carcinoma independently of histological type, tumor microenvironment, and VHL mutation. Cancer Res 1999;59:2182–9. [PubMed] [Google Scholar]
- 49.Sundaresan V, Ganly P, Hasleton P, et al. p53 and chromosome 3 abnormalities, characteristic of malignant lung tumours, are detectable in preinvasive lesions of the bronchus. Oncogene 1992;7:1989–97. [PubMed] [Google Scholar]