Abstract
Aims: Clonality analysis using polymerase chain reaction (PCR) amplification of the immunoglobulin heavy chain (IgH) gene is an important aid to the diagnosis of B cell lymphoproliferative diseases. However, the method has a relatively high false negative rate. In an attempt to improve detection rates simple PCR strategies for clonality analysis of B cell populations using amplification of Ig light chain genes have been developed.
Methods: Novel PCR protocols, designed to amplify Igκ and Igλ light chain genes, were evaluated using high molecular weight DNA samples from 28 selected cases of B cell lymphoma with known light chain expression and 12 reactive lymphoid specimens. Products were run on 10% polyacrylamide minigels using heteroduplex analysis. Conventional IgH PCR analysis was also performed. Twelve randomly selected formalin fixed, paraffin wax processed samples from cases submitted for molecular genetic analysis were also studied.
Results: Polyclonal products were seen in all reactive lymphoid samples. Using Igκ PCR, 24 of 28 lymphomas, including four of five IgH negative cases, displayed monoclonal patterns. Using Igλ PCR, eight of 12 Igλ expressing tumours, including two of five IgH negative cases, showed monoclonal patterns. Standard IgH PCR demonstrated monoclonality in 23 of 28 B cell lymphomas. The detection rate was improved to 27 of 28 lymphomas using heavy and light chain PCR. Efficient amplification was achieved using paraffin wax processed samples, seven of which showed monoclonality compared with eight using IgH PCR.
Conclusions: Ig light chain PCR, used in conjunction with heavy chain analysis, enables improved detection of B cell monoclonality using routine histological specimens and can provide additional clone specific markers for the study of the biology of B cell tumours.
Keywords: immunoglobulin light chain genes, polymerase chain reaction, clonality analysis
Clonality analysis of B cell populations using polymerase chain reaction (PCR) amplification of immunoglobulin (Ig) genes1 has become an important aid to the diagnosis of problematical B cell lymphoproliferative diseases, largely replacing more cumbersome hybridisation techniques.2 The Ig heavy (IgH) chain gene has been the preferred target because most B cell tumours have rearranged IgH genes.3,4 However, a large proportion of B cell tumours (up to 30% depending on lymphoma type and grade) are not amenable to amplification of IgH because of the loss of target sequences or alteration as a result of somatic hypermutation.1,4 Cases that escape amplification using IgH PCR may be amplified using Ig light chain PCR. Strategies for the amplification of Ig light chain genes have been devised,5–7 but have not become well established, especially for the amplification of routinely fixed, paraffin wax embedded samples in which the DNA may be highly degraded. We have developed simple PCR strategies aimed at routine analysis of paraffin wax embedded tissue samples for clonality analysis using primers directed at the CDR3 regions of the rearranged Igκ and Igλ light chain genes.
“A large proportion of B cell tumours are not amenable to amplification of IgH because of the loss of target sequences or alteration as a result of somatic hypermutation”
MATERIALS AND METHODS
Primer design
Igκ and Igλ light chain primers were designed from consensus fragments of the FR3 and J regions after analysis of all known V and J family members (MRC Centre for Protein Engineering; http://www.mrc-cpe.cam.ac.uk/imt-doc/vbase-home-page.html). The selected Igκ primers amplified fragments of approximately 130 to 150 base pairs using two Vκ and two Jκ primers in a single PCR reaction (table 1 ▶).
Table 1.
Details of the primers used
| Primer | Sequence (5`→3`) | Regions targeted |
| VκFR3-1 | TTCAG(C/T)GGCAGTGG(A/G)TCTGG | VκI, III, IV, V, VI |
| VκFR3-2 | TTCAGTGGCAGTGGG(G/T)CAGG | VκII |
| Jκ1 | TTTGAT(A/T/C)TCCACCTTGGTCCC | JκI-IV |
| Jκ2 | TTTAATCTCCAGTCGTGTCCC | JκV |
| Vλ-FR3 | GA(T/C)GAGGCTGA(T/C)TATTACTG | VλI-VII |
| Jλ1 | AGGACGGTGACCTTGGTCCC | JλI |
| Jλ2 | AGGACGGTCAGCT(G/T)GGT(C/G)CC | JλII, III, VII |
The Igλ primers amplified fragments from about 70 to 90 base pairs using a single Vλ and two Jλ primers (table 1 ▶). Each Jλ region primer was used with the Vλ region primer in separate reactions because a single multiplex reaction yielded non-specific products.
Reactions were optimised using high molecular weight DNA from reactive lymphoid samples and from B cell lymphomas with known light chain rearrangements by Southern blot analysis.
Primer evaluation
Primers were evaluated using high molecular weight DNA extracted from 28 B cell lymphomas (20 marginal zone, four follicular, two lymphocytic, one mantle cell, and one lymphoplasmacytic) with known light chain expression and shown to be monoclonal by IgH Southern blot analysis. Twelve reactive lymphoid tissue samples (10 lymph nodes and two tonsils) were used as polyclonal controls. Duplicate aliquots of 100 ng of DNA from each case were amplified in reaction mixtures containing 1× reaction buffer, 1.5mM MgCl2, 200μM of each dNTP, 250 ng of each primer, and 0.5 units of Platinum Taq polymerase (Life Technologies, Paisley, UK). Thirty seven cycles of 93°C for 30 seconds, 58°C (60°C for Igλ) for 30 seconds, and 72°C for one minute were performed using high molecular weight DNA samples and 40 cycles using paraffin wax embedded tissue extracts; a final primer extension at 72°C for five minutes completed the reaction. After amplification, products were heated for five minutes at 95°C and transferred to ice for 60 minutes (heteroduplex analysis) before electrophoresis for 75 (Igκ) or 45 (Igλ) minutes at 200 V on 10% polyacrylamide minigels. Products with clearly visible dominant bands were interpreted as monoclonal, those with a smear of products as polyclonal. All samples were studied using IgH PCR with FR3/JH primers as routinely used in our department.3
Amplification of routine paraffin wax embedded samples
To evaluate the efficiency of amplification using routine samples, 12 extracts3 from randomly selected formalin fixed, paraffin wax embedded samples of cases submitted for IgH clonality analysis were also studied using Igκ and Igλ PCR.
Sensitivity
DNA extracted from a B cell lymphoma was serially diluted with DNA from a reactive tonsil to give estimated tumour proportions of 50%, 25%, 10%, 5%, and 1%. Each dilution was amplified using Igκ and Igλ PCR.
Specificity
To confirm the specificity of amplified Ig light chain products, dominant bands from each of the reactions were sequenced.8 PCR products were purified from 1.5% agarose gels using QIAquick gel extraction kit (Qiagen Ltd, Crawley, West Sussex, UK), ligated into the pGEM™-T vector and transformed into JM109 competent cells (Promega, Southampton, UK). The transformed cells were selected on LB ampicillin agar plates containing X-gal and IPTG. White colonies were screened using PCR with vector primers (Sp6 and T7). The PCR products showing the expected insert sizes were sequenced in both directions using an ABI sequencer with dRhodamine terminators (Perkin-Elmer, Foster City, California, USA). At least six PCR clones from each sample were sequenced.
RESULTS
Primer evaluation
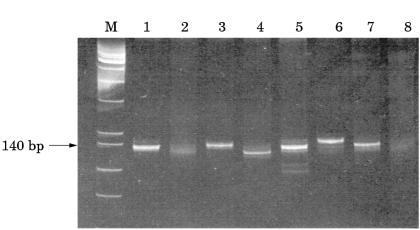
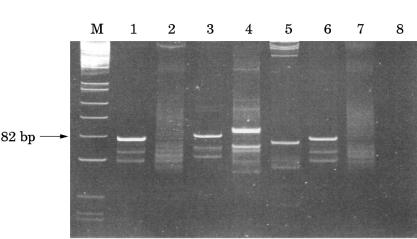
All primer sets yielded products within the predicted size ranges when applied to selected high molecular weight DNA samples. Using Igκ PCR, 24 of 28 B cell lymphomas, including four of five IgH PCR negative cases, showed a monoclonal pattern and four cases yielded a polyclonal smear pattern (fig 1 ▶; table 2 ▶). Using Igλ PCR, eight of 28 lymphomas were monoclonal, including eight of 12 Igλ expressing tumours and two of five IgH PCR negative cases. The remainder showed a polyclonal smear pattern (fig 2 ▶; table 2 ▶). As expected, no Igκ expressing lymphomas were amplified using Igλ primers because Igκ rearrangement precedes Igλ. With both Igκ and Igλ PCR 25 of the 28 lymphomas were monoclonal. Using standard IgH PCR 23 of the 28 B cell lymphomas were monoclonal. The detection rate was improved to 27 of 28 lymphomas using heavy and light chain PCR. Polyclonal products were seen in all reactive lymphoid samples studied. Fourteen of the 16 B cell lymphomas expressing Igκ and 10 of the 12 expressing Igλ were successfully amplified with Igκ primers and eight of the 12 expressing Igλ were amplified with Igλ primers.
Figure 1.
Polyacrylamide gel (10%) of Igκ PCR products. Lane M, PhiXHinfI molecular weight markers; lane 1, positive monoclonal control marginal zone lymphoma (fresh tissue; FT); lane 2, follicular lymphoma (paraffin wax processed tissue; PT); lane 3, lymphoplasmacytic lymphoma (FT); lane 4, marginal zone lymphoma (FT); lane 5, follicular lymphoma (PT); lane 6, follicular lymphoma (FT); lane 7, follicular lymphoma (FT); lane 8, reactive lymph node (FT).
Table 2.
PCR detection of monoclonal, rearranged immunoglobulin genes in DNA samples extracted from fresh/frozen B cell lymphomas and reactive lymphoid legions
| Sample group | Number of cases | Igκ PCR | Igλ PCR | Igκ+λ PCR | IgH PCR |
| B cell lymphomas | 28 | 24 (86%) | 8 (29%) | 25 (89%) | 23 (82%) |
| κ+ B cell lymphomas | 16 | 14 (87%) | 0 | 15 (94%) | 15 (94%) |
| λ+ B cell lymphomas | 12 | 10 (83%) | 8 (67%) | 11 (92%) | 8 (67%) |
| Reactive | 12 | 0 | 0 | 0 | 0 |
| IgH PCR negative | 5 | 4 | 2 | 4 | 0 |
Figure 2.
Polyacrylamide gel (10%) of Igλ PCR products. Lane M, PhiXHinfI molecular weight markers; lane 1, positive control lymphoplasmacytic lymphoma (fresh tissue; FT); lane 2, marginal zone lymphoma (paraffin wax processed tissue; PT); lane 3, follicular lymphoma (PT); lane 4, marginal zone lymphoma (FT); lane 5, follicular lymphoma (FT); lane 6, lymphoplasmacytic lymphoma (PT; same case as lane 1); lane 7, reactive lymph node (PT); lane 8, negative control (no DNA).
Routine paraffin wax embedded samples
All DNA extracts from routine, formalin fixed, paraffin wax embedded samples yielded products within the expected size range, suggesting amplification of specific Ig light chain products (figs 1 and 2 ▶ ▶). Six of 12 showed a monoclonal pattern (five with Igκ and three with Igλ primers) and the remainder a polyclonal pattern (table 3 ▶). Ten samples were B cell lymphomas that were expected to show a monoclonal pattern, and two (one reactive and one nodular lymphocyte predominant Hodgkin's lymphoma) were expected to show a polyclonal pattern.
Table 3.
PCR detection of monoclonal, rearranged immunoglobulin genes in DNA extracted from randomly selected formalin fixed, paraffin wax embedded tissue samples
| Sample | Diagnosis | Igκ PCR | Igλ PCR | IgH PCR |
| 1 | MZL | P | M | M |
| 2 | MZL | M | P | M |
| 3 | MZL | P | M | M |
| 4 | MZL | P | P | M |
| 5 | FL | P | P | M |
| 6 | FL | P | P | P |
| 7 | FL | M | P | P |
| 8 | SMZL | M | P | M |
| 9 | DLBCL | M | P | M |
| 10 | DLBCL | M | M | M |
| 11 | NLPHD | P | P | P |
| 12 | Reactive | P | P | P |
| Total | 5/12 | 3/12 | 8/12 | |
| Igκ/Igλ PCR | 7/12 | |||
| Igκ/Igλ/IgH PCR | 9/12 | |||
DLBCL, diffuse large B cell lymphoma; FL, follicular lymphoma; M, monoclonal; MZL, marginal zone lymphoma; NLPHD, nodular lymphocyte predominant Hodgkin's lymphoma; P, polyclonal; SMZL, splenic marginal zone lymphoma.
Sensitivity
Tumour clones were detected clearly to a dilution of 10% and weakly to 5% of tumour cells using both Igκ and Igλ primers (data not shown).
Specificity
The sequences of amplified products are shown in fig 3 ▶. Comparison of these sequences with those from VBASE confirms amplification of Igκ and Igλ variable regions. The Igκ and Igλ (Vλ-FR3/Jλ2) products were confirmed to be monoclonal because all clones sequenced were identical from each primer set. The Igλ (Vλ-FR3/Jλ1) products yielded three different sequences (two clones each). This may reflect amplification of one or two alleles of the dominant clone and additional background clones. Cloning and sequencing were performed without size selection on total amplificates that would be expected to contain products from reactive B cells in addition to the tumour clone.
Figure 3.
Light chain sequences derived from PCR products (primer sequences are not included). The asterisk denotes the number of identical clones with sequence shown. The nucleotide shown in lower case is the only putative somatic mutation.
DISCUSSION
We have developed PCR methods for the analysis of Igκ and Igλ light chain genes suitable for the detection of expanded B cell clones, including some lymphomas not amplifiable using conventional IgH PCR analysis. Efficient amplification was achieved using DNA extracts from either fresh/frozen or paraffin wax embedded tissue samples. Igκ and Igλ light chain amplification will thus be useful to complement conventional IgH PCR clonality analysis and to study lymphoproliferative diseases with biased expression of light chains, such as human herpesvirus 8 positive multicentric Castleman's disease, which almost invariably expresses Igλ.8
Amplification of products of the expected size range using each primer set suggested that products were specific and this was confirmed by sequencing. Sensitivities of 5–10% tumour cells were lower than figures published for IgH PCR (typically 1–5%9) although there is much variation built into such experiments owing to the rough estimation of tumour cell and competing reactive B cell numbers, and variability in precise fit of primers to individual rearrangements targeted. Nevertheless, the overall assessment of the techniques using a range of lymphomas suggests appropriate sensitivity.
Take home messages.
When used in conjunction with heavy chain analysis, Ig light chain PCR results in improved detection of B cell monoclonality using routine histological specimens
It can provide additional clone specific markers for the study of the biology of B cell tumours
Heteroduplex analysis of PCR products was found to be essential for the efficient resolution of monoclonal bands within polyclonal backgrounds, especially with Igλ primers, probably because of the limited size heterogeneity of products.10
This investigation shows that immunoglobulin light chain PCR techniques permit the detection of monoclonality in a high proportion of B cell lymphomas, including some that are not amplifiable using IgH PCR, using routine tissue samples. The Igκ PCR is particularly useful because B cell lymphomas expressing either κ or λ light chain may be amplified. The protocols described should offer improvements over existing techniques for the analysis of paraffin wax processed samples because they amplify smaller fragments5,6 or target a wider range of germline fragments.7
“The Igκ PCR is particularly useful because B cell lymphomas expressing either κ or λ light chain may be amplified”
Improvements to the primer design, such as the inclusion of κ deleting element primers that target rearrangements with deleted κ regions,11 which are frequent in Igλ expressing cells, and redesign of Igλ primers to permit amplification in a single tube, would improve the overall efficiency of the analysis.
These protocols will expand the possibilities for the detection of monoclonal B cell populations and provide clone specific markers for the study of the distribution and progression of B cell tumours.
Acknowledgments
This work was partly supported by the Cancer Research Campaign.
Abbreviations
IgH, immunoglobulin heavy chain
PCR, polymerase chain reaction
REFERENCES
- 1.Diss TC, Pan L. Polymerase chain reaction in the assessment of lymphomas. Cancer Surv 1997;30:21–44. [PubMed] [Google Scholar]
- 2.Sioutos N, Bagg A, Michaud GY, et al. Polymerase chain reaction versus Southern blot hybridization. Detection of immunoglobulin heavy-chain gene rearrangements. Diagn Mol Pathol 1995;4:8–13. [DOI] [PubMed] [Google Scholar]
- 3.Diss TC, Pan L, Peng H, et al. Sources of DNA for detecting B cell monoclonality using PCR. J Clin Pathol 1994;47:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan JH, Trainor KJ, Brisco MJ, et al. Monoclonality in B cell lymphoma detected in paraffin wax embedded sections using the polymerase chain reaction. J Clin Pathol 1990;43:888–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuppers R, Zhao M, Rajewsky K, et al. Detection of clonal B cell populations in paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1993;143:230–9. [PMC free article] [PubMed] [Google Scholar]
- 6.Kuppers R, Willenbrock K, Rajewsky K, et al. Detection of clonal lambda light chain gene rearrangements in frozen and paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 1995;147:806–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Gong JZ, Zheng S, Chiarle R, et al. Detection of immunoglobulin kappa light chain rearrangements by polymerase chain reaction. An improved method for detecting clonal B-cell lymphoproliferative disorders. Am J Pathol 1999;155:355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du MQ, Liu H, Diss TC, et al. Kaposi sarcoma-associated herpesvirus infects monotypic (IgM lambda) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood 2001;97:2130–6. [DOI] [PubMed] [Google Scholar]
- 9.Trainor KJ, Brisco MJ, Story CJ, et al. Monoclonality in B-lymphoproliferative disorders detected at the DNA level. Blood 1990;75:2220–2. [PubMed] [Google Scholar]
- 10.Garcia-Sanz R, Lopez-Perez R, Langerak AW, et al. Heteroduplex PCR analysis of rearranged immunoglobulin genes for clonality assessment in multiple myeloma. Haematologica 1999;84:328–35. [PubMed] [Google Scholar]
- 11.Bertoni N, Zucca E, Genini D, et al. Immunoglobulin light chain kappa deletion rearrangement as a marker of clonality in mantle cell lymphoma. Leuk Lymphoma 1999;36:147–50. [DOI] [PubMed] [Google Scholar]