Abstract
Fluorescence in situ hybridisation (FISH) is an effective method to detect chromosomal alterations in a variety of tissue types, including archived paraffin wax embedded specimens fixed in B5 or formalin. However, precipitating fixatives such as B5 have been known to produce unsatisfactory results in comparison with formalin when used for FISH. This study describes an effective nuclear isolation and FISH procedure for B5 and formalin fixed tissue, optimising the nuclear isolation step and nuclei pretreatments using tonsil and mantle cell lymphoma specimens. The protocol presented can be used to isolate nuclei and perform FISH on B5 or formalin fixed, paraffin wax embedded samples from a variety of tissue types.
Keywords: fluorescence in situ hybridisation, B5, formalin, paraffin wax
Fluorescence in situ hybridisation (FISH) has greatly facilitated the detection of specific chromosomal abnormalities such as additions, deletions, or translocations in interphase or metaphase nuclei.1 The ready applicability of FISH to fixed tissue specimens embedded in paraffin wax has rendered the vast archives of paraffin wax embedded tissue maintained in hospital pathology departments available for retrospective cytogenetic study. Pathology specimens are usually fixed in 10% neutral buffered formalin before further processing, which includes dehydration, paraffin wax embedding, and the preparation of histological slides. A mercuric chloride based precipitating fixative called B5 has some advantages over formalin, providing greater staining intensity and superior preservation of nuclear morphology.2 Because B5 fixation is often used for specimens that are of potential cytogenetic interest, particularly malignant lymphomas or other haematological neoplasms, it is frequently desirable to apply FISH to B5 fixed specimens. However, precipitating fixatives such as B5 have been associated with unsatisfactory results using conventional FISH protocols.3
Most protocols for FISH analysis of fixed, paraffin wax embedded tissue make use of histological sections. These preserve tissue architecture and morphology but impede the enumeration of signals because of the frequent overlap or slicing of nuclei.4 The use of isolated nuclei prepared from paraffin wax embedded tissue largely avoids these problems and can therefore provide more accurate quantitative results.
“Fluorescence in situ hybridisation has greatly facilitated the detection of specific chromosomal abnormalities such as additions, deletions, or translocations in interphase or metaphase nuclei”
To the best of our knowledge, no procedure describing the application of FISH to isolated interphase nuclei from B5 fixed tissue has been published. In our report, we describe the optimisation of such a method. The technical parameters were optimised using lymphoid tissue from hyperplastic tonsils fixed in either formalin or B5. We then successfully applied the technique to archival samples of mantle cell lymphoma (MCL) that were associated with previously documented cytogenetic abnormalities.
MATERIALS AND METHODS
Tissue sources and preparation
Non-neoplastic tonsil tissue associated with reactive lymphoid hyperplasia was received unfixed in the histopathology laboratory, sliced into sections roughly 3 mm thick, fixed in formalin for six to 12 hours, and then subjected to routine tissue processing and paraffin wax embedding. Alternatively, fresh tissue sections were fixed in freshly prepared B5 for four hours, followed by two to eight hours of fixation in 70% ethanol and then ethanol based, formalin free tissue processing. Paraffin wax blocks containing samples of formalin or B5 fixed tissue from MCL cases documented previously by comparative genomic hybridisation (CGH) to have cytogenetic abnormalities (KJ Harrison, unpublished data, 2000) were retrieved from the archives of the histopathology laboratory at Kingston General Hospital. The MCL cases were chosen for their common X chromosome aneuploidy because this could be easily confirmed using a commercially prepared centromeric FISH probe that is less expensive than locus specific probes. Metaphase preparations of peripheral blood specimens from two normal donors were used as normal controls in each hybridisation experiment. All samples analysed were from male donors.
Tissue preparation
The preparation of single cell suspensions from paraffin wax embedded tissue was performed using Hedley's method of preparing samples for flow cytometric DNA content determination, with modifications.5 Briefly, paraffin wax embedded tissue was cut into pieces measuring about 0.5 mm3 and placed into sterile microcentrifuge tubes. We have found that one to three 10–50 μm tissue sections can also be used to isolate sufficient numbers of intact nuclei that are suitable for FISH analysis.6,7 Approximately 20 mg of tissue was dewaxed by incubation with 1 ml of xylene for 30 minutes at room temperature on a rotating platform, followed by a second xylene incubation lasting 10 minutes. The tissue was rehydrated by sequential, 10 minute incubations in 1 ml of 100%, 90%, 70%, and 50% ethanol and washed twice in deionised water. The tissue was then subjected to digestion in 4 mg/ml pepsin (Sigma, Oakville, Ontario, Canada) in 0.9% NaCl, pH 1.5, at 37°C for 15, 30, 45, 60, or 120 minutes, with vortexing every 10 minutes. After manual removal of undigested tissue fragments, the nuclei were pelleted by centrifugation at 8000 ×g for seven to eight minutes, washed twice in phosphate buffered saline (PBS), resuspended in 350 μl of PBS, and counted using a haemocytometer.8 A 10 μl aliquot of the resulting suspension was applied to histological slides (Fisher superfrost plus; Fisher Scientific, Nepean, Ontario, Canada) that were then dried at 65°C for 10 minutes.8
Fluorescence in situ hybridisation
FISH was carried out according to the method of Hyytinen et al, with modifications.9 Slides were incubated in 50% glycerol/0.1× standard saline citrate (SSC) for three minutes at 90°C, and then cooled in 2× SSC at room temperature for two minutes. Slides were dehydrated by sequential immersion in 70%, 80%, 90%, and 100% ethanol and then digested in 8 μg/ml proteinase K in 20mM Tris, 2mM CaCl2, pH 7.5, at 37°C for 0, 7.5, 10, or 15 minutes. The slides were dehydrated again in the same ethanol series and air dried.
Probe specific to the X chromosome centromere (alpha X CEP Spectrum Green; Vysis, Downers, Illinois, USA) was prepared according to the manufacturer's instructions, with exceptions as indicated below. Probe and target were co-denatured using a HYBrite co-denaturation instrument (Vysis) with a one minute melt at 85°C, followed by an overnight hybridisation at 42°C. Slides were washed according to the CEP rapid assay protocol provided by the manufacturer, except that the wash temperature was lowered to 72(± 1)°C. The slides were counterstained with DAPI II (Vysis) and coverslipped.
Nuclear signals were detected with the aid of a Leitz Diaplan type 307-148.001 fluorescent microscope (Leitz, Wetzlar, Germany) using DAPI, fluorescein isothiocyanate, or rhodamine filter sets. Images were captured digitally using the CytoVision capturing system (Applied Imaging, Santa Clara, California, USA).
RESULTS
Isolation of nuclei and pepsin treatment
Using the protocol described above, intact nuclear morphology was seen after pepsin digestion for periods ranging from 15 to 120 minutes. However, with B5 fixed tissue, increasing the time of digestion from 45 to 60 minutes produced a substantial increase in the number of nuclei isolated—from 602/mg to 2723/mg. Because even longer digestion periods failed to increase the yield significantly, a 60 minute pepsin digestion appeared to be the most suitable for B5 fixed samples.
In formalin fixed tissue, pepsin digestion for 15 minutes produced 206 nuclei/mg, whereas digestion for 120 minutes produced 3571 nuclei/mg, without great loss of structural integrity. Therefore, pepsin digestion for 120 minutes was deemed appropriate for formalin fixed material.
FISH pretreatment
Pretreatments with hot glycerol and proteinase K, performed after nuclei are dried on to slides, are thought to increase the permeability of nuclear membranes and other cell constituents to the probe. However, treatment conditions must be optimised to minimise adverse effects on nuclear morphology. The B5 fixed nuclei were found to be more susceptible to damage during pretreatment.
Nuclei from B5 fixed tonsil tissue exhibited a weak or absent signal without proteinase K digestion and strong, easily scorable signals after 7.5 or 10 minute digestions. Digestion for 7.5 minutes produced the clearest signals. The nuclei from B5 fixed MCL tissue produced the clearest and strongest signals after a 12 minute digestion. In formalin fixed tonsil tissue, weak or absent signals were produced after 0 or 7.5 minute proteinase K treatments, whereas 10, 15, and 17.5 minute pretreatments produced relatively strong signals. Although nuclear adhesion to the slide was compromised by the longer digestion times, the number of cells remaining on the slide was more than sufficient for analysis. Nuclei from formalin fixed MCL tissue produced the best signals after a 15 minute digestion.
Hybridisation and analysis
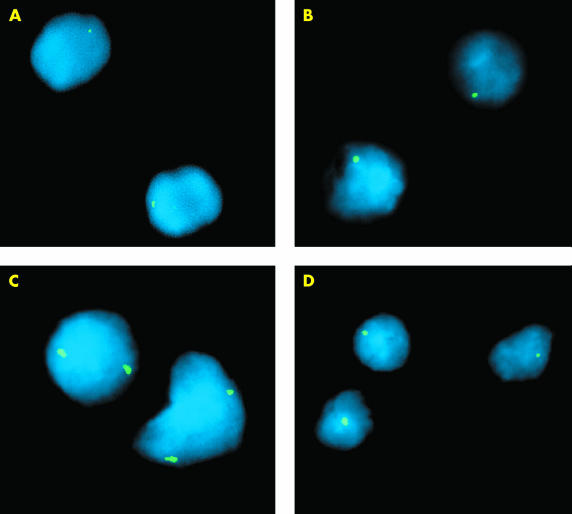
Table 1 ▶ correlates sample type and fixative with the number of signals for each nucleus. In applying the CEP X centromeric probe, nuclei from formalin or B5 fixed tonsil tissue exhibited considerably stronger signals with no background signal when washed in 0.4× SSC/0.3% NP-40 at 72°C rather than at 75°C, the temperature recommended by the probe manufacturer. A reduction in the post hybridisation wash temperature results in a reduction in stringency, thereby producing a stronger hybridisation signal without cross hybridisation to less homologous sequences. The peripheral blood control produced one signal in 93.0% of the nuclei. Hybridisation of the B5 and formalin fixed male tonsil tissue produced comparable results, with one signal in 93.0% and 93.5% of the nuclei, respectively (table 1 ▶; fig 1A, B ▶).
Table 1.
Mean number and percentage of hybridisation signals for each isolated nucleus in blood control, B5 fixed, and formalin fixed nuclei achieved with the X centromere specific probe
| Mean number (%) of cells with the indicated number of hybridisation signals | |||||||
| Specimen | Fixative | Number of nuclei | 0 | 1 | 2 | 3 | 4+ |
| Blood | Carnoy's | 200 | 5 (2.5) | 186 (93.0) | 7 (3.5) | 2 (1.0) | – |
| Tonsil | B5 | 200 | 3 (1.5) | 186 (93.0) | 8 (4.0) | 3 (1.5) | – |
| Tonsil | Formalin | 200 | 6 (3.0) | 187 (93.5) | 7 (3.5) | – | – |
| MCL | B5 | 100 | – | 2 (2.0) | 76 (76.0) | 18 (18.0) | 4 (4.0) |
Figure 1.
Examples of hybridised interphase nuclei isolated from B5 or formalin fixed, paraffin wax embedded tonsil or lymphoma tissue using an X CEP probe. (A) Normal signal distribution of the X CEP probe hybridised to normal male tonsil tissue fixed in formalin. (B) Normal signal distribution of the X CEP probe hybridised to normal male tonsil tissue fixed in B5. (C) X centromere probe exhibiting two X chromosome specific hybridisation signals in nuclei obtained from formalin fixed, male, mantle cell lymphoma (MCL) tissue shown to have X chromosome gain by comparative genomic hybridisation analysis (17.5 minute's proteinase K treatment). (D) Normal signal distribution of the X CEP probe hybridised to nuclei isolated from B5 fixed MCL tissue.
B5 and formalin fixed male MCL tissue with previously identified X chromosome abnormalities showed one signal in only 6% of analysed cells. Gain of X was identified in 76% (two signals) and 18% (three signals) of analysed cells in one MCL (table 1 ▶; fig 1C ▶). This confirmed the X chromosome gain that was initially identified by CGH.
An example of a B5 fixed MCL with normal X chromosome signal distribution, as initially identified by CGH analysis, is shown in fig 1D ▶. All specimens produced consistently bright hybridisation signals that could be easily visualised. We have obtained similar results using locus specific DNA FISH probes (KJ Harrison, data not shown, 2001).
DISCUSSION
Although methodologies describing the application of FISH to formalin fixed tissue sections from a variety of tissue sources have been available for quite some time, FISH analysis of paraffin wax embedded sections has proved to be more difficult than the analysis of conventional cytogenetic preparations. Problems have included unsuccessful hybridisation as a result of poor probe penetration, excessive probe requirement, excessive background, autofluorescence, and sectioned or incomplete nuclei.4,9–11 Indeed, commercial suppliers of FISH probes usually advise that B5 fixed tissue is unsuitable for FISH studies. The isolation of single nuclei and pretreatment to improve probe penetration can overcome some of these limitations, but have not been widely accepted.9,10 Many of the pretreatment protocols are complicated and cannot be easily reproduced. To the best of our knowledge, no procedure describing the application of FISH to isolated interphase nuclei from B5 fixed tissue has been published.
This technical report outlines an efficient and reproducible FISH protocol for the application to nuclei isolated from B5 and formalin fixed, paraffin wax embedded tissue. Optimised conditions for nuclei isolated using pepsin digestion and FISH pretreatments involving hot glycerol and proteinase K are described that can be easily and efficiently applied. The preparation of single cell suspensions from paraffin wax embedded tissue using pepsin digestion was optimised by modifying a method for flow cytometric DNA content determination. 5 A one hour digestion period was optimal for B5 fixed tissue and two hours for formalin fixed tissue. In our own experience, the pepsin treatment time can vary with the type of tissue to be digested, with some tissues requiring a shorter or longer pretreatment time. Longer digestion times for B5 fixed tissue resulted in fragmented nuclei that would be unsuitable for FISH. The more fragile nature of the B5 fixed tissue may be the result of the tissue hardness or brittleness that is produced using fixatives containing mercuric chloride. Prolonged pepsin digestion with frequent vortexing may shear B5 fixed nuclei more easily than the formalin fixed nuclei.
“The more fragile nature of the B5 fixed tissue may be the result of the tissue hardness or brittleness that is produced using fixatives containing mercuric chloride”
FISH pretreatments involving hot glycerol and proteinase K can be applied successfully to both formalin and B5 fixed tissues. These pretreatments were initially described by Hyytinen in 1994 and were applied to formalin fixed, paraffin wax embedded tumours. We have found that this pretreatment can also be successfully applied to B5 fixed tissue using the modified methodology described in our report. To the best of our knowledge, the successful application of FISH to B5 fixed tissues has not been reported previously. In our experience, the B5 fixed tissue did not require proteinase K pretreatment for as long as the formalin fixed tissue, although the same hot glycerol pretreatment time was used. We believe that the major advantages of using the FISH pretreatments described here are that they can be applied to a variety of tissues that have been fixed using either of two commonly used fixatives without major changes in methodology and still yield consistent hybridisation results. The FISH procedure described here is particularly suitable for retrospective studies of archived tumour specimens and for B5 or formalin fixed tissue requiring follow up confirmation of a previously identified chromosomal abnormality.
Take home messages.
This report describes the application of fluorescence in situ hybridisation (FISH) to isolated interphase nuclei from B5 and formalin fixed tissue
The nuclear isolation step and nuclei pretreatments were optimised using tonsil and mantle cell lymphoma specimens: optimal pepsin digestion to isolate nuclei was seen at 60 minutes for B5 fixed samples and 120 minutes for formalin fixed samples; optimal FISH pretreatment to increase permeability was seen at 7.5 minutes for B5 fixed samples and 15 minutes for formalin fixed samples
The protocol presented can be used to isolate nuclei and perform FISH on B5 or formalin fixed, paraffin wax embedded samples from a variety of tissue types
Acknowledgments
The laboratory work was supported by a grant from the Queen's University Department of Pathology Clinical Trust Fund Awards Programme. M J Schurter was supported by a summer studentship from the same grant source. We thank B Thomas for his technical assistance and Dr H Feilotter for helpful discussion.
Abbreviations
CGH, comparative genomic hybridisation
FISH, fluorescence in situ hybridisation
MCL, mantle cell lymphoma
PBS, phosphate buffered saline
SCC, standard saline citrate
REFERENCES
- 1.Blancato JK. Fluorescence in situ hybridization. In: Gersen S, Keagle M, eds. The principles of clinical cytogenetics. Totowa, NJ: Human Press, 1999:443–71.
- 2.Tbakhi A, Totos G, Hauser-Kronberger C, et al. Fixation conditions for DNA and RNA in situ hybridization—a reassessment of molecular morphology dogma. Am J Pathol 1998;152:35–41. [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss LM, Chen YY. Effects of different fixatives on detection of nucleic acids from paraffin-embedded tissues by in situ hybridization using oligonucleotide probes. J Histochem Cytochem 1991;39: 1237–42. [DOI] [PubMed] [Google Scholar]
- 4.Qian J, Bostwick DG, Takahashi S, et al. Comparison of fluorescence in situ hybridization analysis of isolated nuclei and routine histological sections from paraffin embedded prostatic adenocarcinoma specimens. Am J Pathol 1996;149:1193–9. [PMC free article] [PubMed] [Google Scholar]
- 5.Hedley DW, Friedlander ML, Taylor IW, et al. Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J Histochem Cytochem 1983;31:1333–5. [DOI] [PubMed] [Google Scholar]
- 6.Thompson CT, LeBoit PE, Nederlof PM, et al. Thick section fluorescence in situ hybridization on formalin-fixed, paraffin-embedded archival tissue provides a histogenic profile. Am J Pathol 1994;144: 237–43. [PMC free article] [PubMed] [Google Scholar]
- 7.Kuchinka BD, Kalousek DK, Lomax BL, et al. Interphase cytogenetic analysis of single cell suspensions from previously formalin-fixed and paraffin-embedded tissues. Mod Pathol 1995;8: 183–6. [PubMed] [Google Scholar]
- 8.Naaem R. FISH of isolated nuclei from paraffin. www.vysis.com. May 8, 2000.
- 9.Hyytinen E, Visakorpi T, Kallioniemi A, et al. Improved technique for analysis of formalin-fixed, paraffin-embedded tumours by fluorescence in situ hybridization. Cytometry 1994;16:93–9. [DOI] [PubMed] [Google Scholar]
- 10.DiFrancesco LM, Murthy SK, Luider J, et al. Laser-capture microdissection-guided fluorescence in situ hybridization and flow cytometric cell cycle analysis of purified nuclei from paraffin sections. Mod Pathol 2000. 13: 705–11. [DOI] [PubMed] [Google Scholar]
- 11.McKay JA, Murray GI, Keith WN, et al. Amplification of fluorescent in situ hybridisation signals in formalin fixed paraffin wax embedded sections of colon tumour using biotinylated tyramide. J Clin Pathol Mol Pathol 1997;50:322–5. [DOI] [PMC free article] [PubMed] [Google Scholar]