Summary
Background
Our study aimed to develop a machine learning (ML) model utilizing grayscale ultrasound (US) to distinguish ≤3 cm small hepatocellular carcinoma (sHCC) from non-HCC lesions.
Methods
A total of 1052 patients with 1058 liver lesions ≤3 cm from 55 hospitals were collected between May 2017 and June 2021, and 756 liver lesions were randomly allocated into train and internal validation cohorts at a 8:2 ratio for the development and evaluation of ML models based on multilayer perceptron (MLP) and extreme gradient boosting (XGBoost) methods (ModelU utilizing US imaging features; ModelUR adding US radiomics features; ModelURC employing clinical features further). The diagnostic performance of three models was assessed in external validation cohort (312 liver lesions from 14 hospitals). The diagnostic efficacy of the optimal model was compared to that of radiologists in external validation cohort. The SHapley Additive exPlanations (SHAP) method was employed to interpret the optimal ML model by ranking feature importance. The study was registered at ClinicalTrials.gov (NCT03871140).
Findings
ModelURC based XGBoost showed the best performance (AUC = 0.934; 95% CI: 0.894–0.974) in the internal validation cohort. In the external validation cohort, ModelURC also achieved optimal AUC (AUC = 0.899, 95% CI: 0.861–0.931). Upon conducting a subgroup analysis, no statistically significant differences were observed in the diagnostic performance of the ModelURC neither between tumor sizes of ≤2.0 cm and 2.1–3.0 cm nor across different HCC risk stratifications. ModelURC exhibited superior ability compared to all radiologists and ModelURC assistance significantly improved the diagnostic AUC for all radiologists (all P < 0.0001).
Interpretation
A diagnostic model for sHCC was developed and validated using ML and grayscale US from large cohorts. This model significantly improved the diagnostic performance of grayscale US for sHCC compared with experts.
Funding
This work was supported by National Key Research and Development Program of China (2022YFC2405500), Major Research Program of the National Natural Science Foundation of China (92159305), National Science Fund for Distinguished Young Scholars (82325027), Key project of National Natural Science Foundation of China (82030047), Military Fund for Geriatric Diseases (20BJZ42), National Natural Science Foundation of China Special Program (82441011). National Natural Science Foundation of China (82402280), National Natural Science Foundation of China (32171363), Key Research and Development Program for Social Development of Yunnan Science and Technology Department (202403AC100014).
Keywords: sHCC, Ultrasound, Radiomics, SHAP, Multi-center
Research in context.
Evidence before this study
We searched for the term “(ultrasound OR gray ultrasound) AND (deep learning OR machine learning OR radiomics) AND (small hepatocellur cacinoma OR small liver cancer OR sHCC)” on PubMed and Web of Science, until 1 December 2024 without language restrictions. There are few studies focusing on AI-based diagnosis of HCC smaller than 3 cm, with two studies concentrating on using MRI for diagnosing small HCC, and another focusing on MRI detection of small HCC. These studies utilized a limited number of cases. Currently, there is no research specifically utilizing grayscale ultrasound for AI diagnosis of HCC smaller than 3 cm. As a convenient and cost-effective method for liver nodule detection in clinical practice, grayscale ultrasound has limitations in diagnosing small HCC. Therefore, it is necessary to develop a radiomics-based approach to extract information from grayscale ultrasound images to assist radiologists in diagnosing HCC ≤3 cm.
Added value of this study
This study proposes, for the first time, a diagnostic system for HCC smaller than 3 cm based on liver nodule grayscale ultrasound radiomics combined with ultrasound semantic features and clinical characteristics. It is designed for early diagnosis of small HCC in clinical practice. The system was tested on multi-center data for assisting radiologists in diagnosis and could improve the diagnostic performance of radiologists in diagnosing small HCC using grayscale ultrasound.
Implications of all the available evidence
Our model demonstrated excellent performance in diagnosing HCC smaller than 3 cm, outperforming experienced radiologists. Additionally, it can assist radiologists with varying levels of experience in improving the accuracy of HCC diagnosis, providing clinically useful support for the diagnosis of small HCC.
Introduction
Hepatocellular carcinoma (HCC) constitutes the most prevalent form of primary liver cancer, accounting for 90% of cases, and ranks as the fourth leading cause of cancer-related mortality worldwide.1,2 HCC up to 3 cm is considered as small hepatocellular carcinoma (sHCC) and accurate diagnosis of sHCC may enhance the therapeutic window for HCC, thereby improving overall prognosis.3,4 The diagnosis of hepatic nodule often relies on costly, time-consuming, and invasive examination methods such as enhanced ultrasound (CEUS), computed tomography (CT), and magnetic resonance imaging (MRI).3 These methods not only impose a large burden on medical resources but also involve prolonged waiting times, especially for MRI, during which disease progression may occur.5 In clinical practice, B-mode ultrasound (US) serves as the first line of defense for liver disease screening and is commonly used to detect suspicious liver nodules.6 It is inexpensive, facilitates real-time diagnosis, and does not involve radiation exposure or nephrotoxicity.7, 8, 9 However, ultrasound faces challenges in distinguishing between different hepatic nodules due to its poor specificity for various pathology types and the absence of dynamic perfusion characteristics that are often critical in tumor identification.10 Additionally, US diagnostics are highly dependent on the expertise, experience, and attention to detail of the radiologist.11
According to meta-analysis, the sensitivity of US for diagnosing resectable HCC (defined as a single nodule <5 cm or 2–3 nodules, each <3 cm in diameter) only 53% (95% CI: 38%–67%).6 This low sensitivity leads to a significant number of HCC cases being missed, posing a major clinical challenge. Improving the diagnostic accuracy of US for HCC, particularly for sHCC, represents a significant challenge in advancing early diagnosis.
Recent advances of machine learning (ML) within healthcare have significantly enhanced the diagnostic performance of US in tumor detection. ML approaches employ statistical methods to discern underlying patterns and classifications within medical data, and have been successfully implemented to augment patient care.12 ML have leveraged US features to develop specialized predictive models that assist in tumor diagnosis. Jin et al. developed an ML model utilizing US characteristics and thyroid-function tests,13 demonstrating higher accuracy than the ACR TI-RADS. Ma et al. employed breast cancer US features to create an ML model for differentiating breast cancer molecular subtypes.14 Additionlly, advances in radiomics now enable the extraction of microscopic features that radiologists cannot directly observe, helping to assist differentiate between various tumor types, including those in the breast,15 thyroid,16 gallbladder17 and parotid tumors.18 However, to our knowledge, few studies have explored the use of ultrasound features or radiomics combined with ML to distinguish liver lesions. Previous research attempted to use US-based radiomics to differentiate between primary and metastatic liver cancer in a small cohort (N = 114).19 Beyond the differentiation between primary and metastatic liver cancer, it is even more critical to distinguish HCC from benign nodules, as atypical hyperplasia (a precancerous lesion) and HCC often share similar ultrasound features, making it particularly difficult to differentiate between them, especially in small-sized liver nodules.20,21 The liver's deep position in the abdominal cavity and its lower imaging resolution compared to superficial organs, such as the thyroid and mammary glands, further complicate its imaging. This challenge is exacerbated by interference from other organs and gases in the abdominal cavity.22,23 These factors not only increase the difficulty for radiologists in diagnosing liver cancer but also present challenges in developing ML tools for this purpose.
In this study, we collected grayscale US images of liver nodules ≤3 cm in 55 Chinese hospitals from May 2017 to June 2021 and developed a ML based diagnostic model to distinguish HCC. To our knowledge, this is the first ML model developed using US images for the diagnosis of sHCC, analyzing US images of liver nodules from 55 hospitals across China, acquired with different equipment and covering various pathology types. The model was trained and internally validated using features on 746 US images and externally validated on the cohort comprising 312 images, demonstrating both its accurate characterization and robustness. Furthermore, our model outperformed both junior and senior in the external validation cohort, highlighting its potential to assist radiologists in improving diagnostic accuracy.
Methods
Patient information
The inclusion criteria were as follows:
(1) Participants presented with definitive benign or malignant hepatic lesions, corroborated by histological or cytological evidence; (2) Individuals with hepatic hemangiomas and focal nodular hyperplasia exhibited CT or MRI diagnostic results and underwent a follow-up period exceeding one year; (3) Lesion size was limited to ≤3.0 cm, with a maximum lesion count of ≤3; (4) Accessible clinical data for patients was a requirement; (5) Grayscale ultrasonography was conducted within one month preceding biopsy or surgical intervention.
Exclusion criteria were as follows: (1) Indeterminate pathological diagnoses (excluding hepatic hemangioma and focal nodular hyperplasia); (2) Incomplete clinical data; (3) Missing or poor-quality US images.
All research was conducted in accordance with both the Declarations of Helsinki and Istanbul. Ethical committee approval and written informed consent was obtained from patients at each hospital. The study was registered at ClinicalTrials.gov (NCT03871140).
US feature and clinical feature acquisition
US features were gleaned from US records, comprising tumor size, shape, margin, echo level, echo distribution, blood flow signal, liver background, bile duct dilation, portal hypertension, splenomegaly, and ascites. Clinical features were extracted from medical records, including age, gender, history of hepatitis and history of tumors. To ensure the robustness of our analysis and avoid any potential biases, we excluded these patients with incomplete data from the final analysis cohort.
Tumor delineation and radiomic feature extraction
The criteria for US image selection for target liver lesions were in Supplementary S1. There were different kinds of ultrasound devices were used across the 55 hospitals (Supplementary S2 and Table S1). All grayscale US images in the study were archived in Digital Imaging and Communications in Medicine (DICOM) format. The outlining of the tumor region of interest (ROI) was performed by an experienced radiologist using ITK-SNAP software (version 3.8.0; [http://www.itksnap.org]) without knowledge of the pathology results. The radiologist had more than 5 years of experience in liver ultrasound. Radiomics features within these ROIs were then extracted using the PyRadiomics package (version 2.2.0).24 In order to reduce the impact of ROI selection on model development, we performed an intraclass correlation coefficient test on ROI selection, as detailed in the Supplementary S3.
Machine learning model development and evaluation
Three datasets were used for model development. The patients are consistent across US features dataset, US-Radiomics dataset and US-Radiomics-clinic dataset, each dataset progressively incorporates more meta-data (radiomics features and clinical data) to support the model. US features dataset included gray ultrasound image semantic features, US-Radiomics dataset includes both gray ultrasound image semantic features and radiomics features extracted from the gray ultrasound images. US-Radiomics-clinic dataset includes gray ultrasound image features, radiomics features, and additional clinical data. The models based on these three datasets were categorized as ModelU, ModelUR, and ModelURC. The features of three datasets were normalized by z-score normalization. And feature correlation was assessed using the Spearman correlation coefficient because the relationships between some features may not be strictly linear. Where the correlation coefficient exceeded 0.9, only one relevant feature was retained. The least absolute shrinkage and selection operator (LASSO) regression model was employed on the discovery dataset for signature construction. Contingent on the regularization weight λ, LASSO diminishes all regression coefficients towards zero and assigns the coefficients of numerous irrelevant features precisely to zero. To ascertain an optimal λ, 10-fold cross-validation with minimum criteria was implemented, wherein the final value of λ produced the minimum cross-validation error. The retained features with nonzero coefficients were utilized for regression model fitting and amalgamated into a signature group. For each dataset, eight common ML models25 were initially developed, and the ML model with the best performance in terms of AUC was identified as the final model for that dataset. The eight ML models (Supplementary S4) include logistic regression (LR), random forest (RF), support vector machine (SVM), and extreme gradient boosting (XGBoost), K-nearest neighbor (KNN), ExtraTrees, Light gradient boosting machine (LightGBM), and multilayer perceptron (MLP). ML models were subsequently validated in the internal validation and external validation cohort. We also compared the performance of the optimal model in different tumor size subgroups, HCC-risk subgroups and liver ground subgroups.
To assess the calibration of the model, calibration curves were plotted. In addition, decision curve analysis (DCA) was performed to determine the clinical utility of the predictive model. Net reclassification index (NRI) and integrated discrimination improvement (IDI) were also analyzed to investigate whether there was a significant improvement in model performance as model parameters were increased.
Diagnostic performance comparison of ML model and radiologists
Three junior radiologists (with less than 5 years of experience) and three senior radiologists (with more than 5 years experience) participated in the evaluation. All radiologists first assessed the grayscale ultrasound images and provided an initial diagnosis with access to additional clinical information, which included age, gender, history of hepatitis and history of tumor. After a one-month washout period, all radiologists participated in the second evaluation. In this assessment, radiologists were also informed of the predictions from the best-performing ML model, in addition to having access to the clinical information. We then compared the diagnostic performance of the ML model, radiologists without ML model assistance, and radiologists with ML model assistance.
Interpretability of the optimal performing machine learning model
Machine learning models often pose interpretability challenges, as the rationale behind accurate predictions for specific patient cohorts may be unclear, leading to a “black box” perception. In our study, we elucidated the optimal ML model using the SHAP algorithm. This unified approach facilitates the explanation of outcomes for any machine learning model. We employed a SHAP tree explainer to assess feature attributions, assuming that each patient feature was a player in a game where the prediction constituted the payout. The contribution of each player indicated the feature's importance to the prediction. Specifically, we utilized SHAP feature importance to rank features by decreasing their importance as a measure of the average absolute Shapley values. The summary plot can reflect both feature importance and feature effects. Each point on the summary plot represents a SHAP value per feature and instance. The summary plot demonstrates the relationship between feature values and their impact on diagnosis. SHAP interpretation waterfall plot can be employed to visualize feature attributions.
Statistical analysis
ML models were developed and validated using Python (version 3.8) and scikit-learn (version 1.1.2). Their diagnostic performance was assessed using six metrics: area under the curve (AUC), sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV). Delong's test was employed to compare AUCs. The AUC, Delong's test, sensitivity, specificity, accuracy, PPV, and NPV were calculated using R (version 4.2.2). The McNemar test was utilized to examine differences in diagnostic accuracy, sensitivity, and specificity between radiologists and the ML model by employing the SPSS software (version 22.0). Differences with P < 0.05 were considered statistically significant.
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. Zhicheng Du and Ping Liang had access to the dataset. All authors decided to publish the study findings.
Results
Clinical characteristics
Between May 2017 and June 2021, a total of 1423 consecutive patients with 1447 hepatic lesions (≤3 cm in maximum diameter) who underwent biopsy and/or hepatectomy or followed up by imaging in 55 hospitals were screened. Ultimately, 1058 hepatic lesions from 1052 patients in 55 hospitals (Table S2) were included as the primary cohort (as shown in Fig. 1). Seven hundred forty-six patients with 746 lesions from 41 hospitals of the primary cohort were randomly divided into train (597 lesions from 597 patients) and internal validation (149 lesions from 149 patients) cohorts with a 8:2 ratio. An additional 306 patients with 312 hepatic lesions from 14 hospitals were included as external validation cohort. In our model development and validation process, we divided the data at the patient level to ensure that all lesions from the same patient were included in the same dataset. Regarding multiple lesions per patient, each lesion was treated as an independent sample for feature extraction and model training. The characteristics of patients in the three cohorts are shown in Table 1. The development cohort was derived from a diverse group of patients, with cases collected across multiple hospitals. This cohort includes patients with various ultrasound features and clinical characteristics, ensuring that it reflects the heterogeneity typically encountered in clinical practice. The patients captured a broad spectrum of liver lesions, which enhances the generalizability of the model. In this study, the total sample size from external validation cohort is considered adequate for assessing the diagnostic performance of the models. External validation cohort is collected from 14 independent hospitals and these patients are independent of each other from the patients in the train cohort and the internal validation cohort, fulfilling our need for external validation.
Fig. 1.
A flowchart of patient and nodule inclusion and exclusion.
Table 1.
Characteristics of patients in three cohorts.
| Characteristics | Train cohort |
Internal validation cohort |
External validation cohort |
|---|---|---|---|
| (N = 597) | (N = 149) | (N = 306) | |
| Age | |||
| Mean (SD) | 55.2 (11.8) | 54.7 (11.1) | 57.8 (11.1) |
| Gender | |||
| Female | 176 (29.5%) | 47 (31.5%) | 98 (32.0%) |
| Male | 421 (70.5%) | 102 (68.5%) | 208 (68.0%) |
| History of hepatitis | |||
| No | 262 (43.9%) | 69 (46.3%) | 104 (34.0%) |
| HBV | 325 (54.4%) | 70 (47.0%) | 180 (58.8%) |
| HCV | 9 (1.5%) | 8 (5.4%) | 15 (4.9%) |
| HBV + HCV | 1 (0.2%) | 2 (1.3%) | 7 (2.3%) |
| History of tumors | |||
| No | 335 (56.1%) | 91 (61.1%) | 206 (67.3%) |
| Extrahepatic tumor | 193 (32.3%) | 45 (30.2%) | 75 (24.5%) |
| Intrahepatic tumor | 69 (11.6%) | 13 (8.7%) | 25 (8.2%) |
| Vascular invasion | |||
| No | 590 (98.8%) | 147 (98.7%) | 301 (98.4%) |
| Yes | 7 (1.2%) | 2 (1.3%) | 5 (1.6%) |
| Lymph node metastasis | |||
| No | 567 (95.0%) | 144 (96.6%) | 302 (98.7%) |
| Yes | 30 (5.0%) | 5 (3.4%) | 4 (1.3%) |
| Intratumoral vascularitya | |||
| No | 395 (66.2%) | 95 (63.8%) | 207 (67.6%) |
| Yes | 202 (33.8%) | 54 (36.2%) | 99 (32.4%) |
| Bile duct dilation | |||
| No | 583 (97.7%) | 145 (97.3%) | 305 (99.7%) |
| Yes | 14 (2.3%) | 4 (2.7%) | 1 (0.3%) |
| Portal hypertension | |||
| No | 521 (87.3%) | 137 (91.9%) | 279 (91.2%) |
| Yes | 76 (12.7%) | 12 (8.1%) | 27 (8.8%) |
| Splenomegaly | |||
| No | 432 (72.4%) | 114 (76.5%) | 236 (77.1%) |
| Yes | 165 (27.6%) | 35 (23.5%) | 70 (22.9%) |
| Ascites | |||
| No | 560 (93.8%) | 140 (94.0%) | 287 (93.8%) |
| Yes | 37 (6.2%) | 9 (6.0%) | 19 (6.2%) |
| Liver bacground | |||
| Cirrhosis | 226 (37.9%) | 63 (42.3%) | 125 (40.8%) |
| Hepatic steatosis | 41 (6.9%) | 12 (8.1%) | 23 (7.5%) |
| Increased echo | 138 (23.1%) | 37 (24.8%) | 27 (8.8%) |
| Normal | 192 (32.2%) | 37 (24.8%) | 131 (42.8%) |
| Tumor sizea | |||
| Mean (SD) | 2.2 (0.5) | 2.2 (0.5) | 2.1 (0.66) |
| Tumor shapea | |||
| Circle | 38 (6.4%) | 10 (6.7%) | 29 (9.3%) |
| Elliptic | 468 (78.4%) | 119 (79.9%) | 224 (71.8%) |
| Irregular | 91 (15.2%) | 20 (13.4%) | 59 (18.9%) |
| Tumor margina | |||
| Clear | 452 (75.7%) | 121 (81.2%) | 225 (72.1%) |
| Unclear | 145 (24.3%) | 28 (18.8%) | 87 (27.9%) |
| Tumor echogenicitya | |||
| Heterogeneous | 18 (3.0%) | 3 (2.0%) | 19 (6.1%) |
| Hyper- | 106 (17.8%) | 27 (18.1%) | 64 (20.5%) |
| Hypo- | 432 (72.4%) | 116 (77.9%) | 209 (67.0%) |
| Iso- | 41 (6.9%) | 3 (2.0%) | 20 (6.4%) |
| Tumor echo distributiona | |||
| Heterogeneous | 286 (47.9%) | 74 (49.7%) | 140 (44.9%) |
| Homogeneous | 311 (52.1%) | 75 (50.3%) | 172 (55.1%) |
| Intratumoral vascularitya | |||
| No | 395 (66.2%) | 95 (63.8%) | 213 (68.3%) |
| Yes | 202 (33.8%) | 54 (36.2%) | 99 (31.7%) |
| Diagnostic methoda | |||
| Biopsy | 436 (73.0%) | 109 (73.2%) | 238 (76.3%) |
| Resection | 126 (21.1%) | 30 (20.1%) | 66 (21.1%) |
| Clinical diagnosticb | 35 (5.9%) | 10 (6.7%) | 8 (2.6%) |
| Pathological typea | |||
| Adenoma | 2 (0.3%) | 0 (0%) | 2 (0.6%) |
| Liver hemangiomas | 32 (5.4%) | 11 (7.4%) | 8 (2.6%) |
| Cholangiocarcinoma | 12 (2.0%) | 2 (1.3%) | 6 (1.9%) |
| Dysplastic nodule | 25 (4.2%) | 6 (4.0%) | 10 (3.2%) |
| Focal nodular hyperplasia | 21 (3.5%) | 4 (2.7%) | 8 (2.6%) |
| Hepatocellur cacinoma | 307 (51.4%) | 77 (51.7%) | 168 (53.8%) |
| Inflammatory pseudotumor | 14 (2.3%) | 3 (2.0%) | 15 (4.8%) |
| Lipoma | 9 (1.5%) | 0 (0%) | 7 (2.2%) |
| Liver abscess | 2 (0.3%) | 2 (1.3%) | 9 (2.9%) |
| Metastatic liver cancer | 161 (27.0%) | 42 (28.2%) | 68 (21.8%) |
| Neuroendocrine tumors | 3 (0.5%) | 1 (0.7%) | 2 (0.6%) |
| Regenerative nodules | 8 (1.3%) | 1 (0.7%) | 8 (2.6%) |
| Sarcoma | 1 (0.2%) | 0 (0%) | 1 (0.3%) |
Note. Qualitative variables are expressed as n (%), and quantitative variables are expressed as Mean ± SD.
Data calculated based on tumor number.
Clinical diagnosis is only for liver hemangiomas and focal nodular hyperplasia.
Development of ML models
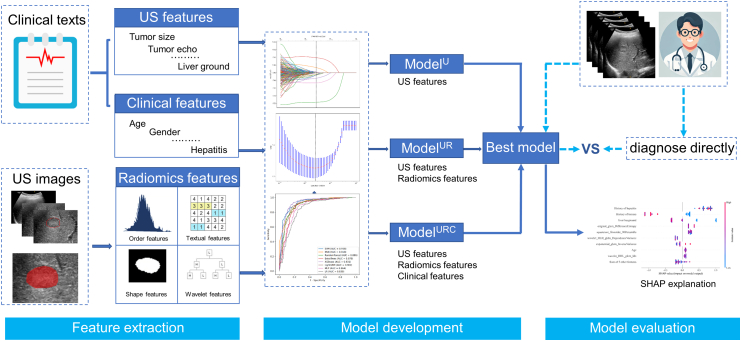
We selected features for the ML models using LASSO regression. The coefficients and mean standard error (MSE) of 10-fold validation in three groups are displayed in Figure S1. Ten features with nonzero coefficients were selected for ModelU, 11 features for ModelUR, and 14 features for ModelURC, respectively (as shown in Figure S2). Further details on the ML model development and validation workflow as shown in Fig. 2.
Fig. 2.
Workflow depicting the development and validation of the machine learning model. (US: ultrasound; SHAP: SHapley Additive exPlanations).
Comparison of diagnostic performance of the ML models
In the internal validation cohort, MLP model exhibited the highest AUC in US features dataset (AUC = 0.743, 95% CI: 0.662–0.819) than SVM model (AUC = 0.730, 95% CI: 0.638–0.813), KNN model (AUC = 0.684, 95% CI: 0.601–0.758), RF model (AUC = 0.702, 95% CI: 0.615–0.782), Extra Trees model (AUC = 0.722, 95% CI: 0.642–0.798), XGBoost model (AUC = 0.721, 95% CI: 0.641–0.802), LigthGBM model (AUC = 0.712, 95% CI: 0.628–0.790) and LR model (AUC = 0.718, 95% CI: 0.635–0.792). XGBoost model exhibited the highest AUC in US-Radiomics dataset (AUC = 0.913, 95% CI: 0.868–0.958) than SVM model (AUC = 0.899, 95% CI: 0.848–0.943), KNN model (AUC = 0.900, 95% CI: 0.844–0.949), RF model (AUC = 0.905, 95% CI: 0.853–0.947), Extra Trees model (AUC = 0.886, 95% CI: 0.829–0.935), LigthGBM model (AUC = 0.905, 95% CI: 0.852–0.950), MLP model (AUC = 0.853, 95% CI: 0.788–0.910) and LR model (AUC = 0.845, 95% CI: 0.781–0.905). XGBoost model exhibited the highest AUC in US-Radiomics-clinic dataset (AUC = 0.934, 95% CI: 0.894–0.974) than SVM model (AUC = 0.931, 95% CI: 0.885–0.967), KNN model (AUC = 0.905, 95% CI: 0.853–0.952), RF model (AUC = 0.918, 95% CI: 0.865–0.959), Extra Trees model (AUC = 0.920, 95% CI: 0.871–0.967), LigthGBM model (AUC = 0.921, 95% CI: 0.874–0.965), MLP model (AUC = 0.922, 95% CI: 0.867–0.964) and LR model (AUC = 0.912, 95% CI: 0.858–0.958) (as shown in Figure S3).
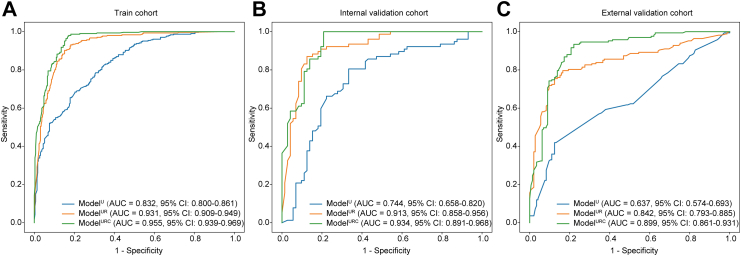
In the training cohort, ModelUR and ModelURC demonstrated superior performance with AUCs of 0.931 (95% CI: 0.911–0.952) and 0.955 (95% CI: 0.938–0.971) compared to ModelU, which had an AUC of 0.832 (95% CI: 0.802–0.862), (P < 0.0001, respectively) (as shown in Table 2, Fig. 3A). The AUC of ModelURC was better than that of ModelUR (P < 0.0001). ModelUR and ModelURC achieved sensitivities of 90.2% and 97.7%, which were superior to ModelU's sensitivity of 68.4% (P < 0.0001 for both comparisons).
Table 2.
Performance of ModelU, ModelUR, and ModelURC in different cohorts.
| Cohort | Model | AUC (95% CI) | SEN (%) | SPE (%) | ACC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| Train | ModelU | 0.832 (0.802–0.862)a, b | 68.4a, b | 79.7 | 73.9a, b | 78.1 | 70.4 |
| ModelUR | 0.931 (0.911–0.952)c | 90.2c | 84.8 | 87.6c | 86.3 | 89.1 | |
| ModelURC | 0.955 (0.938–0.971) | 97.7 | 83.1 | 90.6 | 86.0 | 97.2 | |
| Internal validation | ModelU | 0.744 (0.673–0.814)a, b | 66.2a, b | 77.8 | 71.8a, b | 76.1 | 68.3 |
| ModelUR | 0.913 (0.868–0.958)c | 89.6 | 81.9 | 85.9 | 84.1 | 88.1 | |
| ModelURC | 0.934 (0.894–0.974) | 96.1 | 79.2 | 87.9 | 83.1 | 95.0 | |
| External validation | ModelU | 0.637 (0.574–0.693)a, b | 59.3a, b | 62.1a, b | 60.6b | 64.3 | 57.0 |
| ModelUR | 0.842 (0.793–0.885)c | 73.7c | 87.6c | 80.1c | 87.2 | 74.3 | |
| ModelURC | 0.899 (0.861–0.931) | 92.8 | 77.9 | 85.9 | 82.9 | 90.4 |
AUC: area under curve; CI: confidence interval; ACC: accuracy; SEN: sensitivity; SPE: specificity; PPV: positive predictive value; NPV: negative predictive value.
Compared the performance of ModelU with the performance of ModelUR, P < 0.05.
Compared the performance of ModelU with the performance of ModelURC, P < 0.05.
Compared the performance of ModelUR with the performance of ModelURC, P < 0.05.
Fig. 3.
Performance of ModelU, ModelUR, and ModelURCin different cohorts. (A) Performance of three models in train cohort; (B) Performance of three models in internal validation cohort; (C) Performance of three models in external validation cohort. (AUC: area under curve; CI: confidence interval).
In the internal validation cohort, ModelUR and ModelURC displayed improved performance with AUCs of 0.913 (95% CI: 0.868–0.958) and 0.934 (95% CI: 0.894–0.974) compared to ModelU, which had an AUC of 0.744 (95% CI: 0.673–0.814), (P < 0.0001 for both comparisons) (as shown in Table 2, Fig. 3B). The sensitivities of ModelUR (89.6%) and ModelURC (96.1%) were superior to that of ModelU (66.2%), (P = 0.0003, P < 0.0001). The AUC of ModelURC was better than that of ModelUR (P = 0.0031).
In the external validation cohort, ModelUR and ModelURC displayed improved performance with AUCs of 0.842 (95% CI: 0.793–0.885) and 0.899 (95% CI: 0.861–0.931) compared to ModelU, which had an AUC of 0.637 (95% CI: 0.574–0.693), (P < 0.0001 for both comparisons) (as shown in Fig. 3C). The sensitivities of ModelUR (73.7%) and ModelURC (92.8%) were superior to that of ModelU (59.3%), (P < 0.0001 for both comparisons). To validate the robustness of the model, we further split the external validation cohort into two cohorts (cohort 1: 154 lesions from 8 hospitals, cohort 2: 158 lesions from 6 hospitals). Similarly, ModelURC demonstrated the best diagnostic performance in both external validation cohorts (Table S3 and Figure S4).
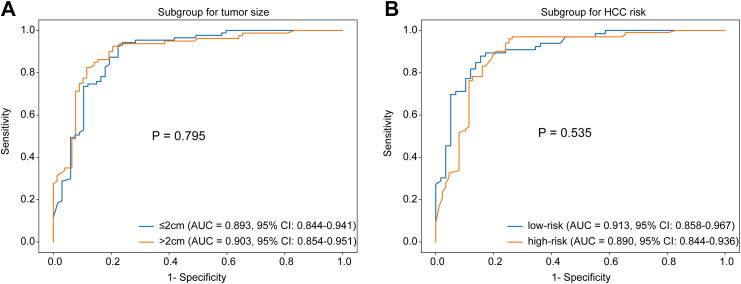
The ModelURC showed the best diagnostic performance and was chosen as the final diagnostic strategy. The performance of ModelURC was also assessed across different tumor size groups (≤2.0 cm and 2.1–3.0 cm) and different HCC risk groups (high-risk and low-risk) (refer to Table 3 for details). The differences in AUC were not statistically significant both in tumor size subgroups and different HCC risk subgroups, indicating that ModelURC exhibited consistent diagnostic performance (Fig. 4). Moreover, we conducted an additional subgroup analysis based on liver background. The results showed that, although ModelURC exhibited lower diagnostic AUC in the Cirrhosis group, there was no statistically significant difference between the Cirrhosis group and Non-cirrhosis group (Table S4 and Figure S5).
Table 3.
Stratification analysis of ModelURC in external validation cohort.
| Cohort | Subgroup | AUC (95% CI) | SEN (%) | SPE (%) | ACC (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| Tumor size | >2 cm | 0.903 (0.854–0.951) | 92.5 | 79.5 | 86.1 | 82.2 | 91.2 |
| ≤2 cm | 0.893 (0.844–0.941) | 93.1 | 76.1 | 85.7 | 83.5 | 89.5 | |
| HCC risk | high-risk | 0.890 (0.844–0.936) | 89.4 | 82.8 | 86.3 | 85.5 | 87.3 |
| low-risk | 0.913 (0.858–0.967) | 95.0 | 74.7 | 85.6 | 81.4 | 92.9 |
AUC: area under curve; CI: confidence interval; ACC: accuracy; SEN: sensitivity; SPE: specificity; PPV: positive predictive value; NPV: negative predictive value.
There were no statistical differences in sensitivity, specificity, accuracy and AUC neither between tumor sizes of ≤2 cm and >2 cm nor across different HCC risk stratifications in the three cohorts.
Fig. 4.
Stratification analysis of ModelURCaccording to tumor size and HCC risk level. (A) The stratification analysis of ModelURC according to tumor size; (B) The stratification analysis of ModelURC according to HCC risk level. (AUC: area under curve; CI: confidence interval; HCC: hepatocellur carcinoma).
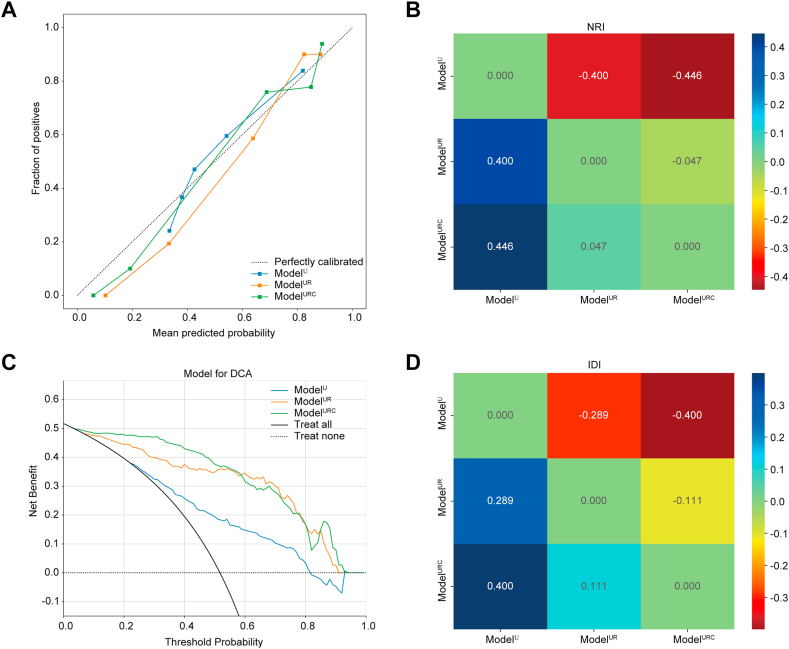
In this study, we also assessed the three models using calibration curve and DCA. The calibration curve of the ModelURC showed good calibration in the internal validation cohort (Fig. 5A). The DCA for ModelU, ModelUR, and ModelURC is presented in Fig. 5C. Compared to scenarios where no prediction model would be employed (i.e., treat-all or treat-none schemes), ModelURC demonstrated a significant benefit for intervention in patients with a prediction probability relative to ModelU and ModelUR. In addition, the model performance progressively improves with the increase of model features, and NRI (Fig. 5B) and IDI (Fig. 5D) showed significant improvement in benefit for ModelUR vs ModelU and for ModelURC vs ModelUR. Preoperative HCC prediction using ModelURC has been shown to provide better clinical benefits.
Fig. 5.
Clinical evaluation of models in the internal validation cohort. (A) The calibration curves of three models, the closer curve to the center line, the more stable the model is; (B) Net reclassification index; (C) The decision curves of three models, ModelURC showed the best benefit; and (D) Integrated discrimination improvement of three models. The difference between the new model and the old model represents the degree of improvement between the models, greater than zero indicates the effectiveness of the new model. (NRI: net reclassification index; DCA: decision curve analysis; IDI: integrated discrimination improvement).
Comparison of diagnostic performance of ModelURC and radiologists
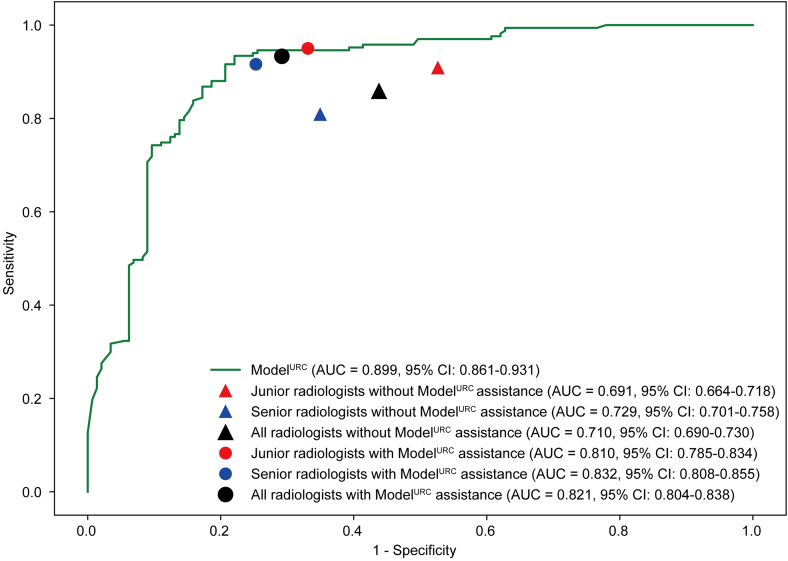
The comparison of ModelURC with radiologists in external validation cohort was shown in Table 4 and Fig. 6. ModelURC displayed better AUC, diagnostic accuracy and specificity than those of junior radiologists (0.899, 95% CI: 0.861–0.931 vs 0.691, 95% CI: 0.664–0.718; 85.9% vs 70.6%; 77.9% vs 47.4%, respectively), (all P < 0.0001). Compared with senior radiologists, ModelURC exhibited better AUC, diagnostic accuracy, sensitivity and specificity (0.899, 95% CI: 0.861–0.931 vs 0.729, 0.701–0.758, 85.9% vs 73.5%, 92.8% vs 80.8%, 77.9% vs 65.1%) (all P < 0.0001). In addition, the AUC, diagnostic accuracy, sensitivity and specificity were improved in junior radiologists with ModelURC assistance (0.810, 95% CI: 0.785–0.834, 81.9%, 95.0% and 66.9%) (P < 0.0001, P < 0.0001, P = 0.0023 and P < 0.0001). And the AUC, diagnostic accuracy, sensitivity and specificity were also improved in senior radiologists with ModelURC assistance (0.832, 95% CI: 0.808–0.855, 83.8%, 91.6% and 74.7%), (all P < 0.0001). We also compared the diagnostic performance of junior and senior radiologists, both with and without ModelURC assistance. Without ModelURC assistance, the AUC of senior radiologists was significantly higher than that of junior radiologists (P = 0.019). And with ModelURC assistance, there was no statistically significant difference in the AUC between senior and junior radiologists (P = 0.088). The evaluations of all radiologists were detailed in Table S5 and Figure S6. In addition, we also analyzed the model-assisted results for the radiologists in two subsets of the external validation cohort. The results showed that ModelURC could also significantly improve the diagnostic performance of radiologists in both subsets of the external validation cohort (Tables S6 and S7, Figure S7).
Table 4.
Average performance comparison of ModelURC and radiologists in external validation cohort.
| AUC (95% CI) | SEN (%) | SPE (%) | ACC (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|
| ModelURC | 0.899 (0.861–0.931) | 92.8 | 77.9 | 85.9 | 82.9 | 90.4 |
| All radiologists | ||||||
| Without ModelURC assistance | 0.710 (0.690–0.730)a, b | 85.8a, b | 56.2a, b | 72.1a, b | 69.3 | 77.5 |
| With ModelURC assistance | 0.821 (0.804–0.838) | 93.3 | 70.8 | 82.9 | 78.6 | 90.2 |
| Junior radiologists | ||||||
| Without ModelURC assistance | 0.691 (0.664–0.718)a, b, c | 90.8a, c | 47.4a, b, c | 70.6a, b | 66.5 | 81.7 |
| With ModelURC assistance | 0.810 (0.785–0.834) | 95.0 | 66.9d | 81.9 | 76.8 | 92.1 |
| Senior radiologists | ||||||
| Without ModelURC assistance | 0.729 (0.701–0.758)a, b | 80.8a, b | 65.1a, b | 73.5a, b | 72.7 | 74.7 |
| With ModelURC assistance | 0.832 (0.808–0.855) | 91.6 | 74.7 | 83.8 | 80.7 | 88.6 |
AUC: area under curve; CI: confidence interval; ACC: accuracy; SEN: sensitivity; SPE: specificity; PPV: positive predictive value; NPV: negative predictive value.
Compared the average performance of radiologists without ModelURC assistance to the average performance of radiologists with ModelURC assistance, P < 0.05.
Compared the average performance of radiologists without ModelURC assistance to the performance of ModelURC, P < 0.05.
Compared the average performance of junior radiologists without ModelURC assistance to the performance of senior radiologists without ModelURC, P < 0.05.
Compared the average performance of junior radiologists with ModelURC assistance to the performance of senior radiologists with ModelURC, P < 0.05.
Fig. 6.
Average performance comparison of ModelURCand radiologists in overall external validation cohort. (AUC: area under curve).
Interpretability of ModelURC
SHAP values indicate the contribution of each feature to the final prediction and can effectively clarify and explain model predictions for individual patients. The SHAP feature importance plots revealed that history of hepatitis, history of tumors, liver background, orginal_glcm_DifferenceEntropy, wavelet_HHL_glcm_Idn and squareroot_firstorder_90Percentile were the six most crucial features in ModelURC (as shown in Fig. 7A). The SHAP explanation waterfall plot for a representative case demonstrate how the features influence the model's output from the baseline (as shown in Fig. 7B–C). The arrows indicate the effect of each factor on prediction. The blue and red arrows denote whether the factor reduced (blue) or increased (red) the risk of HCC. The combined effects of all factors provided the final SHAP value, which corresponded to the prediction score. We also develop an ML model based the six most crucial features and it also showed a well diagnostic performance but the AUC is decreasing compared with ModelURC (based 14 features, Table S8).
Fig. 7.
SHAP interpretation of the ModelURC. (A) The importance ranking of the risk factors with stability and interpretation using ModelURC. The higher SHAP value of a feature is given, the higher risk of HCC patient would be. The red part in feature value represents higher value. A case of HCC in the ModelURC: (B) Grayscale ultrasonic image of HCC; (C) The SHAP value of ModelURC in this case. (SHAP: SHapley Additive exPlanations; HCC: hepatocellur carcinoma).
Discussion
One of the challenges of using ultrasound as a preferred screening tool for HCC is the high demand for physician experience and equipment resolution.5 Another challenge is the heterogeneity of liver grayscale ultrasound images, which can exhibit varying levels of brightness, noise, and differences in liver shape, size, and location.26 These issues lead to a substantial number of liver nodules that cannot be properly identified, resulting in missed diagnoses or necessitating more invasive imaging techniques, which increase both the financial cost and the risk of tumor progression. ML offers a solution by identifying image features with high heterogeneity. The ModelURC we developed has improved the diagnostic performance for HCC by integrating radiomics features, ultrasound features, and clinical data from a large cohort.
Radiomics has garnered significant attention in hepatology, with early studies demonstrating its great potential in the diagnosis of HCC. Several studies have evaluated the effectiveness of radiomics in classifying indeterminate liver nodules. For instance, a multicenter study involving 2143 patients with focal liver lesions developed a deep convolutional neural network ultrasound (DCNN-US) model to classify indeterminate lesions as either HCC or benign nodules.5 The model showed high sensitivity and specificity, outperforming the visual assessment by radiologists. While CT and MRI are commonly used for the accurate diagnosis of HCC.27,28 Kim et al. developed a deep learning model for HCC prediction using multiphase CT images, achieving a sensitivity of 84.8%.29 Hamm et al. constructed a convolutional neural network system based on multiphase MRI imaging data, capable of differentiating six common types of liver nodules, including HCC.30 However, there remains a lack of classification models specifically targeting sHCC, representing an important gap in current research.
Upon review of the existing literature, our retrospective multi-center investigation represents the first endeavor to develop a comprehensible ML tool utilizing US characteristics and radiomics for the classification of sHCC. In our research, the ML model, based solely on US features and radiomics, demonstrated robust performance in diagnosing sHCC within both the internal validation cohort and external validation cohort. The AUC of ModelUR for classifying sHCC from non-HCC for the internal validation cohort achieved 0.913 and for the two subsets of the external validation cohort reached 0.772–0.868. The sensitivities of ModelUR reached 89.6%, 64.7% and 79.8% in the internal validation and two subsets of the external validation cohort, respectively, which surpass previously reported sensitivities in the literature for HCC diagnosis (53.0%).6 Although specificity was limited, enhanced sensitivity provides critical assurance for the screening efficacy of an initial HCC imaging tool.
In clinical practice, radiologists frequently have access to clinical information including age, gender, history of hepatitis, and history of tumors, which can influence their US diagnostic conclusions. Therefore, we added clinical features to the US-Radiomics features to study whether they could improve the diagnostic performance. As a result, the addition of clinical factors likewise significantly improved the model's performance in diagnosing HCC. Even for lesions up to 2.0 cm, ModelURC achieved comparable diagnostic performance to 2–3 cm lesions. This demonstrates the generalization ability of our constructed model for the diagnosis of small HCC of different sizes. In a cirrhotic liver, the presence of nodular regeneration, fibrosis, and altered liver architecture can make it more challenging to distinguish lesions, especially small ones. Although the diagnostic performance of ModelURC decreased in the cirrhosis subgroup, no statistically significant difference was observed, which also reflects the relative robustness of the model.
To validate whether the model we developed could be applied by radiologists at different levels. We reevaluated the US images on the external validation cohort. We found that both junior and senior radiologists were able to achieve better diagnostic performance with the ModelURC assistance than without the ModelURC assistance. Our study has merged quantitative parameters to generate a model that can markedly enhance radiologists' performance in diagnosing small HCC using grayscale ultrasound features. ModelURC can help minimize the diagnostic gap in sHCC between radiologists varying experience levels, reducing the dependence on the radiologist's expertise. Additionally, it allows some HCC patients to decrease the need for subsequent enhancement examinations, enabling them to enter the treatment phase earlier while simultaneously reducing the likelihood of missed malignant tumors.
In our research, we employed the SHAP tree explainer to visually elucidate the output of the XGBoost model, which demonstrated the greatest diagnostic power. XGBoost, an open-source machine learning classifier and a variant of the Gradient Boosting Machine (GBM), has been extensively applied for classification tasks and frequently demonstrates superior classification capabilities compared to other ML algorithms.31,32 The SHAP summary plot provides a visually concise representation of the range and distribution of each feature's impact on the model's output.14 Our analysis revealed that history of hepatitis, history of tumors and liver background were the three most critical features, as shown by the SHAP summary plot. Patients with cirrhosis of any etiology are at a heightened risk for HCC, with annual incidence rates ranging from 1 to 4%.33 A history of hepatitis B infection emerged as the second most significant characteristic. As one of the most prevalent infections worldwide, HBV is the primary cause of HCC, especially in Asian countries.34 Patients infected with HBV are also more likely to arouse suspicion of HCC in clinical practice. A SHAP waterfall plot can be employed to explain a single patient's assessment. Radiologists can directly compare a patient's output SHAP value with the base value. If the output SHAP value exceeds the base value, the patient will be classified as having small HCC. Radiologists can also discern how features impact an individual patient's assessment by observing the arrow's color and length, which indicate the contribution of a specific feature.
Our study has several limitations. First, there is a scarcity of benign liver nodules, as most benign nodules do not undergo biopsy or surgery for pathological confirmation. Second, an inevitable selection bias may be present because a large proportion of non-HCC cases involve non-cirrhotic liver backgrounds. Third, it is essential to expand the international external testing population to more accurately assess the model's performance and its auxiliary effects on radiologists.
In summary, we developed a ML model (ModelURC) utilizing US features, radiomic features and clinical data to diagnose small HCC based on US images from a large cohort. ModelURC demonstrated higher diagnostic accuracy compared to radiologists and significantly improved their performance. Additionally, we applied the SHAP method to interpret ModelURC's output, which may facilitate the integration of interpretable models into clinical practice.
Contributors
All authors read and approved the final version of the manuscript, and ensure it is the case. Ping Liang, Jie Yu and Guojun Zhang had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Zhicheng Du and Fangying Fan are considered co–first authors.
Project administration: Ping Liang and Jie Yu.
Methodology: Zhicheng Du and Fangying Fan; Data curation: Fangying Fan; Formal analysis: Jun Ma and Xing Yan; Validation: Xuexue Chen, Yangfang Dong and Yuling Wang; Investigation: Jiapeng Wu; Resources: Wenzhen Ding and Qinxian Zhao; Software: Zhicheng Du; Writing-original draft: Zhicheng Du; Writing-review&editing: Guojun Zhang, Jie Yu and Ping Liang; Supervision: Guojun Zhang and Ping Liang.
Data sharing statement
We are unable to provide access to our dataset for privacy reasons. The protocol and statistical analysis methods used in the study can be requested directly from the corresponding author after approval.
Declaration of interests
All authors declare no competing interests.
Acknowledgements
This work was supported by National Key Research and Development Program of China (2022YFC2405500), Major Research Program of the National Natural Science Foundation of China (92159305), National Science Fund for Distinguished Young Scholars (82325027), Key project of National Natural Science Foundation of China (82030047), Military Fund for Geriatric Diseases (20BJZ42), National Natural Science Foundation of China Special Program (82441011). National Natural Science Foundation of China (82402280), National Natural Science Foundation of China (32171363), Key Research and Development Program for Social Development of Yunnan Science and Technology Department (202403AC100014).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2025.103098.
Contributor Information
Guojun Zhang, Email: zhangguojun@kmmu.edu.cn.
Jie Yu, Email: Jiemi301@163.com.
Ping Liang, Email: Liangping301@hotmail.com.
Appendix A. Supplementary data
References
- 1.Hepatocellular carcinoma. Nat Rev Dis Prim. 2021;7(1):7. doi: 10.1038/s41572-021-00245-6. [DOI] [PubMed] [Google Scholar]
- 2.Llovet J.M., Kelley R.K., Villanueva A., et al. Hepatocellular carcinoma. Nat Rev Dis Prim. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q.F., Chen S., Yi J.Z., et al. Recommended 10-year follow-up strategy for small hepatocellular carcinoma after radiofrequency ablation: a cost-effectiveness evaluation. Am J Gastroenterol. 2024;119(10):2052–2060. doi: 10.14309/ajg.0000000000002774. [DOI] [PubMed] [Google Scholar]
- 5.Yang Q., Wei J., Hao X., et al. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: a multicentre study. eBioMedicine. 2020;56 doi: 10.1016/j.ebiom.2020.102777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colli A., Nadarevic T., Miletic D., et al. Abdominal ultrasound and alpha-foetoprotein for the diagnosis of hepatocellular carcinoma in adults with chronic liver disease. Cochrane Database Syst Rev. 2021;4(4) doi: 10.1002/14651858.CD013346.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heimbach J.K., Kulik L.M., Finn R.S., et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 8.Tchelepi H., Ralls P.W. Ultrasound of focal liver masses. Ultrasound Q. 2004;20(4):155–169. doi: 10.1097/00013644-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Yu N.C., Chaudhari V., Raman S.S., et al. CT and MRI improve detection of hepatocellular carcinoma, compared with ultrasound alone, in patients with cirrhosis. Clin Gastroenterol Hepatol. 2011;9(2):161–167. doi: 10.1016/j.cgh.2010.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Xu M., Pan F.S., Wang W., et al. The value of clinical and ultrasound features for the diagnosis of infantile hepatic hemangioma: comparison with contrast-enhanced CT/MRI. Clin Imag. 2018;51:311–317. doi: 10.1016/j.clinimag.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 11.Akinyemiju T., Abera S., Ahmed M., et al. The burden of primary liver cancer and underlying Etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ngiam K.Y., Khor I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–e273. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 13.Jin Z., Pei S., Ouyang L., et al. Thy-Wise: an interpretable machine learning model for the evaluation of thyroid nodules. Int J Cancer. 2022;151(12):2229–2243. doi: 10.1002/ijc.34248. [DOI] [PubMed] [Google Scholar]
- 14.Ma M., Liu R., Wen C., et al. Predicting the molecular subtype of breast cancer and identifying interpretable imaging features using machine learning algorithms. Eur Radiol. 2022;32(3):1652–1662. doi: 10.1007/s00330-021-08271-4. [DOI] [PubMed] [Google Scholar]
- 15.Magnuska Z.A., Roy R., Palmowski M., et al. Combining radiomics and autoencoders to distinguish benign and malignant breast tumors on US images. Radiology. 2024;312(3) doi: 10.1148/radiol.232554. [DOI] [PubMed] [Google Scholar]
- 16.Ren J.Y., Lin J.J., Lv W.Z., et al. A comparative study of two radiomics-based blood flow modes with thyroid imaging reporting and data system in predicting malignancy of thyroid nodules and reducing unnecessary fine-needle aspiration rate. Acad Radiol. 2024;31(7):2739–2752. doi: 10.1016/j.acra.2024.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Wang L.F., Wang Q., Mao F., et al. Risk stratification of gallbladder masses by machine learning-based ultrasound radiomics models: a prospective and multi-institutional study. Eur Radiol. 2023;33(12):8899–8911. doi: 10.1007/s00330-023-09891-8. [DOI] [PubMed] [Google Scholar]
- 18.He Y., Zheng B., Peng W., et al. An ultrasound-based ensemble machine learning model for the preoperative classification of pleomorphic adenoma and Warthin tumor in the parotid gland. Eur Radiol. 2024;34(10):6862–6876. doi: 10.1007/s00330-024-10719-2. [DOI] [PubMed] [Google Scholar]
- 19.Mao B., Ma J., Duan S., Xia Y., Tao Y., Zhang L. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur Radiol. 2021;31(7):4576–4586. doi: 10.1007/s00330-020-07562-6. [DOI] [PubMed] [Google Scholar]
- 20.Ling W., Wang M., Ma X., et al. The preliminary application of liver imaging reporting and data system (LI-RADS) with contrast-enhanced ultrasound (CEUS) on small hepatic nodules (≤ 2cm) J Cancer. 2018;9(16):2946–2952. doi: 10.7150/jca.25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forner A., Vilana R., Ayuso C., et al. Diagnosis of hepatic nodules 20 mm or smaller in cirrhosis: prospective validation of the noninvasive diagnostic criteria for hepatocellular carcinoma. Hepatology. 2008;47(1):97–104. doi: 10.1002/hep.21966. [DOI] [PubMed] [Google Scholar]
- 22.Gerstenmaier J.F., Gibson R.N. Ultrasound in chronic liver disease. Insights Imaging. 2014;5(4):441–455. doi: 10.1007/s13244-014-0336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rafailidis V., Sidhu P.S. In: Imaging of the Liver and Intra-Hepatic Biliary Tract: Volume 1: Imaging Techniques and Non-Tumoral Pathologies. Quaia E., editor. Springer International Publishing; Cham: 2021. Ultrasound of the liver; pp. 51–76. [Google Scholar]
- 24.van Griethuysen J.J.M., Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedregosa F., Varoquaux G., Gramfort A., et al. 2011. Scikit-learn: machine learning in Python; pp. 2825–2830. 12(null %J J. Mach. Learn. Res.) [Google Scholar]
- 26.Xu Y., Zheng B., Liu X., et al. Improving artificial intelligence pipeline for liver malignancy diagnosis using ultrasound images and video frames. Briefings Bioinf. 2023;24(1) doi: 10.1093/bib/bbac569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nie P., Yang G., Guo J., et al. A CT-based radiomics nomogram for differentiation of focal nodular hyperplasia from hepatocellular carcinoma in the non-cirrhotic liver. Cancer Imag. 2020;20(1):20. doi: 10.1186/s40644-020-00297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Z., Lin K., Fu J., et al. An MR-based radiomics model for differentiation between hepatocellular carcinoma and focal nodular hyperplasia in non-cirrhotic liver. World J Surg Oncol. 2021;19(1):181. doi: 10.1186/s12957-021-02266-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D.W., Lee G., Kim S.Y., et al. Deep learning-based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur Radiol. 2021;31(9):7047–7057. doi: 10.1007/s00330-021-07803-2. [DOI] [PubMed] [Google Scholar]
- 30.Wang C.J., Hamm C.A., Savic L.J., et al. Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol. 2019;29(7):3348–3357. doi: 10.1007/s00330-019-06214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Deng C.Y., Wang S.Z. Imbalance-XGBoost: leveraging weighted and focal losses for binary label-imbalanced classification with XGBoost. Pattern Recognit Lett. 2020;136:190–197. [Google Scholar]
- 32.Li Q., Yang H., Wang P., Liu X., Lv K., Ye M. XGBoost-based and tumor-immune characterized gene signature for the prediction of metastatic status in breast cancer. J Transl Med. 2022;20(1):177. doi: 10.1186/s12967-022-03369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Serag H.B., Mason A.C. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340(10):745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 34.Chan S.L., Wong V.W., Qin S., Chan H.L. Infection and cancer: the case of hepatitis B. J Clin Oncol. 2016;34(1):83–90. doi: 10.1200/JCO.2015.61.5724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.