Abstract
Background/Aims: Until recently, the cell line MDA-MB-435 was widely accepted as originating from a breast cancer. However, microarray derived data have suggested that this cell line may in fact originate from an occult melanoma. This study was designed to investigate this hypothesis further.
Methods: Quantitative reverse transcription polymerase chain reaction and immunohistochemistry were used to investigate the tissue of origin of two sublines of MDA-MB-435 (MDA-MB-435 S and MDA-MB-435 HGF). The expression of a panel of genes typical of breast cells or melanocytes was analysed.
Results: The MDA-MD-435 cell lines expressed none of the genes characteristic of breast cancer cells but did express several genes commonly expressed by melanocytes.
Conclusions: These results strongly suggest that MDA-MB-435 is indeed of melanoma origin.
Keywords: MDA-MB-435 cell line, quantitative reverse transcription polymerase chain reaction, melanoma
Our current understanding of the biology underlying health and disease owes much to the development and use of systems that model these situations. In particular, the availability of human cells grown in vitro has increased knowledge in the field of human cell biology. Such in vitro systems are also of value in the early stages of the discovery and development of new medicines, where they are used to investigate putative disease mechanisms and potential treatments. In addition, the technique of growing human tumour cells as xenografts in immunocompromised animals has expanded the range of models available for examining tumour growth characteristics in vivo. Here, we describe an investigation of two MDA-MB-435 derived cell lines. Parental MDA-MB-435 was first reported by Cailleau and colleagues1 in 1978 as a metastatic breast cancer derived cell line and banked with the American Type Culture Collection (ATCC) as MDA-MB-435 S (HTB-129). Our study of MDA-MB-435 S and MDA-MB-435 HGF (MDA-MB-435 transfected with hepatocyte growth factor (HGF))2 was initiated after a recent report indicating that the parental cell line in the National Cancer Institute (NCI60) collection showed more resemblance to melanoma derived cell lines than would have been expected,3 although because other breast cancer derived cells failed to give a simple clustering pattern (as seen for renal cancer and leukaemia) a heterogeneous disease aetiology was proposed as one explanation. Because MDA-MB-435 is a common in vitro and in vivo model for metastatic breast cancer,4–8 we decided to investigate the system further, using both quantitative reverse transcription polymerase chain reaction (RT-PCR) and immunohistochemistry for tissue antigens.
“The technique of growing human tumour cells as xenografts in immunocompromised animals has expanded the range of models available for examining tumour growth characteristics in vivo”
MATERIALS AND METHODS
Cell lines and culture conditions
Mammary gland RNA and G361 RNA were obtained from Clontech (Basingstoke, UK). MDA-MB-453 RNA and T47-D RNA were obtained from Ambion (Huntington, UK). The following human cell lines were obtained from the ATCC: BT474, MCF-7, MDA-MB-435 S, SK MEL2, A2058, and Hs294T. Hs578T was obtained from the ECACC; C8161 was obtained from D Welch, Penn State University, Hershey, Pennsylvania, USA; MDA-MB-435 HGF was obtained from W Birchmeier, Max Delbruck Centre, Berlin, Germany. A2058, MCF-7, and Hs294T were cultured in DMEM with 10% fetal calf serum (FCS) and 2mM glutamine; SK MEL2 in EMEM with 10% FCS, non-essential amino acids (NEAA), 1mM sodium pyruvate, and 2mM glutamine; Hs578T and MDA-MB-435 HGF in DMEM with 10% FCS, 10 μg/ml insulin, and 2mM glutamine; BT474 in DMEM with 10% FCS, NEAA, 1mM sodium pyruvate, 2mM glutamine, and 1mM oxalacetic acid; MDA-MB-435 S in L-15 medium with 10% FCS, 10 μg/ml insulin, and 2mM glutamine; and C8161 in RPMI 1640 medium with 10% FCS, NEAA, 1mM sodium pyruvate, and 2mM glutamine.
RNA isolation
RNA was isolated using Trizol reagent (Life Technologies, Paisley, UK), following the manufacturer’s protocol, and quantified on an RNA 6000 Lab Chip via the Agilent 2100 Bioanalyser (Stockport, UK). A 50 μg aliquot of RNA was treated with DNase 1 (Roche, Lewes, UK), according to the manufacturer’s protocol, and resuspended in RNAsecure resuspension solution (Ambion) to 50 ng/μl.
Selection of gene list
Genes that were expressed differentially in breast epithelium and melanomas were identified via a review of the literature.3,9–15 In addition, electronic cDNA library subtractions of breast and melanoma/melanocyte libraries were performed. Sixty four electronic breast cDNA libraries and five electronic melanoma or melanocyte cDNA libraries were used in the subtractions. Genes were preferentially selected if they were identified in the literature and by library subtraction.
Selected genes, together with respective primer and TaqManTM probe sequences, are listed below.
Quantitative RT-PCR
A 250 ng sample of RNA was amplified using the Platinum Quantitative RT-PCR Thermoscript One Step System (LifeTechnologies), according to the manufacturer’s protocol, using the following primer pairs and TaqMan probes. (1) Mammaglobin: 5′-GCGTTAGAGGGCCTCACATATTTT-3′, 5′-GCTTAGAGGTCAGACTGAGGAAACAA-3′, and 5′ FAM-ATG GTCATGGGTTGAGGACTCTGTGAAGCA-TAMRA 3′; (2) prolactin inducible protein (PIP): 5′-CCCAAGTCAGTACGT CCAAATGAC-3′, 5′-TAAATGCACCTTGTAGAGGGATGC-3′, and 5′-FAM-AAACAGAATTGAAAGAATGCATGGTGGTTAAAACTT-TAMRA-3′; (3) pS2: 5′-GCCCAGACAGAGACGTGTACAGTG-3′, 5′-AACGGTGTCGTCGAAACAGCAG-3′, and 5′ FAM-CCCCC CTGGAAAGACAGAATTGTGGTTTT-TAMRA 3′; (4) retinoid X receptor: 5′-GTCGGCTCCATCTTTGACAGAGTT-3′, 5′-TGGGT TAAAGAGTACAATGGCTCG-3′, and 5′ FAM-CCAAAATG AAAGACATGCAGATGGACAAGTC-TAMRA 3′; (5) tyrosinase: 5′-CACAAGAGAAAGCAGCTTCCTGAA-3′, 5′-GCCCTACTCTAT TGCCTAAGCCTT-3′, and 5′ FAM-TCCTCATGGAGAAAGAG GATTACCACAGCTT-TAMRA 3′; (6) tyrosinase related protein 2 (TYRP2): 5′-TACTGTGGGCTATATTTCTCAATGATAGGG-3′, 5′-AAATCATACATGCAGTTTTTCTAATACTGAAGA-3′, and 5′ FAM-TCAACCTTGGAGAAAATGACCAGATCACTTTACTGCTAMRA 3′; (7) ACP-5: 5′-ATGGGACTGAAGACTCACTGGGTG-3′, 5′-GCAGCCTGGTCTTAAAGAGGGACTT-3′, and 5′ FAM-TTG CCTATGTGGAGATCAGCTCCAAAGAGATGACT-TAMRA 3′; and (8) hypoxanthine ribosyl transferase (HPRT): 5′-AGTGATAGATCCATTCCTATG-3′, 5′-GTCTGAATTGTTTTGCC AGTG-3′, and 5′ FAM-TCAGACTGAAGAGCTATTGTAATGAC-TAMRA 3′. The final concentrations of the primers and TaqMan probes were 0.5μM and 0.2μM, respectively. RT-PCR was performed at 50°C for 20 minutes then 95°C for five minutes, followed by 40 cycles of 94°C for 45 seconds and 60°C for 45 seconds in the Prism 7700 (PE Biosystems, Warrington, UK). Data were collected at the 60°C stage of the reaction. In addition, a dilution series of known amounts of total RNA expressing the gene of interest was amplified in the same machine run as the cell line samples. A standard curve of Ct values against log RNA input was drawn to convert the Ct values from the cell lines into an arbitrary RNA amount. The data were normalised for RNA input by dividing by the HPRT control to generate a relative RNA amount, as described in the ABI PRISM 7700 user bulletin 2.
Immunohistochemistry
MDA-MB-435 HGF cells were implanted (5 × 106 cells) into one of the number 4 (abdominal) mammary fat pads of three female severe combined immunodeficient (SCID) mice. Animals were killed 26 days later when the primary tumours were between 0.46 and 1.53 cm3 in size. Tumours were excised and fixed in 10% neutral buffered formalin for 24 hours. They were processed by standard histological techniques to wax blocks and 3 μm sections were cut for glass slides. Sections were dewaxed in xylene and rehydrated from 95% ethanol. Digestion with 0.1% trypsin was used for anticytokeratin sections. Endogenous peroxidase was blocked with 3% aqueous hydrogen peroxidase. An antigen retrieval method where sections were immersed in 0.01M citrate buffer (pH 6.0) and heated in a microwave oven for up to 15 minutes was used for anti-progesterone receptor (PR), anti-melan A, and anti-oestrogen receptor (ER) stains. Protein block was performed with normal serum (1/20 dilution). The primary antibodies were diluted in Tris buffered saline (pH 7.6) and incubated at room temperature as follows: mouse monoclonal antibody to human epithelial membrane antigen (EMA; MU057-UC; BioGenex, San Ramon, USA; 1/50 dilution; two hour incubation), mouse monoclonal antibody to human PR (MU328-UC; BioGenex; 1/20 dilution; one hour incubation), mouse monoclonal antibody to human melan A (NCL-Melan A; Novocastra, Newcastle upon Tyne, UK; 1/20 dilution; one hour incubation), mouse monoclonal antibody to human cytokeratin (M0821; Dako; Ely, UK; 1/50 dilution; one hour incubation), rabbit polyclonal antibody to bovine S100 (Z311; Dako; 1/500 dilution; one hour incubation), and mouse monoclonal antibody to human ER (M7047; Dako; 1/50 dilution; one hour incubation). Negative controls were included for all procedures by exclusion of the primary antibody. Isotype matched control antibodies were used during method development for these procedures. The positive controls were as follows: EMA, human stomach (Dako control slide T1160); PR, Novocastra PR control slide; ER, mouse uterus (personal source); melan A and S100, human melanoma (personal source); cytokeratin, A431 (human epidermoid carcinoma) tumour cell line as tumour explant (personal source). Secondary (biotinylated rabbit antimouse (E0464; Dako) or sheep antirabbit immunoglobulins (2AB02B; Serotec, Kidlington, UK)) and tertiary antibody layers (K0377; streptABComplex/HRP; Dako) with appropriate buffer washes were applied. Visualisation was by a diaminobenzidine (DAB) chromogen procedure (liquid DAB substrate pack; BioGenex).
RESULTS
Expression analysis by quantitative RT-PCR
In addition to the two MDA-MB-435 derived cell lines described above a selection of breast and melanoma cell lines were chosen as positive controls for gene expression: Hs578T, T47-D, MDA-MB-453, BT474, and MCF-7 as breast lines and SK MEL2, A2058, C8161, Hs294T, and G361 as melanoma lines.
As noted previously, the gene panels were chosen by two complementary methods. First, by searching the scientific literature and second by performing in silico cDNA library subtractions. The breast typical genes—pS2, prolactin inducible protein, and mammaglobin—are well documented,9–11 and were not present in the melanoma/melanocyte electronic cDNA libraries. The melanoma typical genes—tyrosinase, tyrosinase related protein 2, ACP-5, and retinoid X receptor γ—were cited in the literature as being discriminatory between breast and melanoma12–15; however, only tyrosinase and tyrosinase related protein 2, both involved in melanin biosynthesis, were present in the in silico library subtraction results.
Expression patterns of the breast and melanoma genes were analysed by quantitative RT-PCR, using TaqMan to detect the amplification products. Data were normalised to the “housekeeping” gene HPRT to allow comparison of expression between samples.
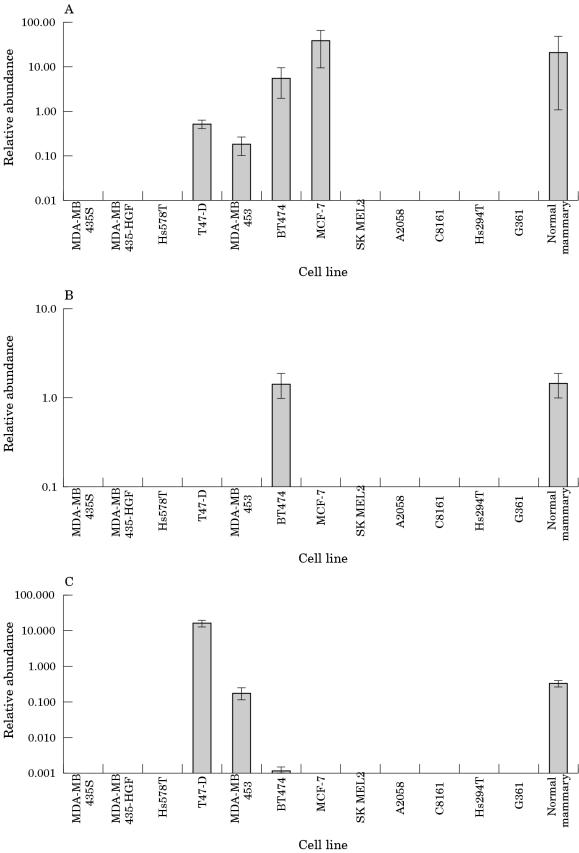
Figure 1 ▶ shows the expression of breast genes in the cell lines analysed. All the breast cancer cell lines except Hs578T, MDA-MB-435 S, and MDA-MB-435 HGF expressed at least one of the characteristic breast genes. These data are not sufficient to suggest that MDA-MB-435 S and MDA-MB-435 HGF are not breast derived because this cell line is considered to be a late stage model of breast cancer and is likely to differ, possibly significantly, from its tissue of origin.
Figure 1.
Relative expression of breast genes in the cell lines and normal mammary gland. A, pS2; B, mammaglobin; C, prolactin inducible protein.
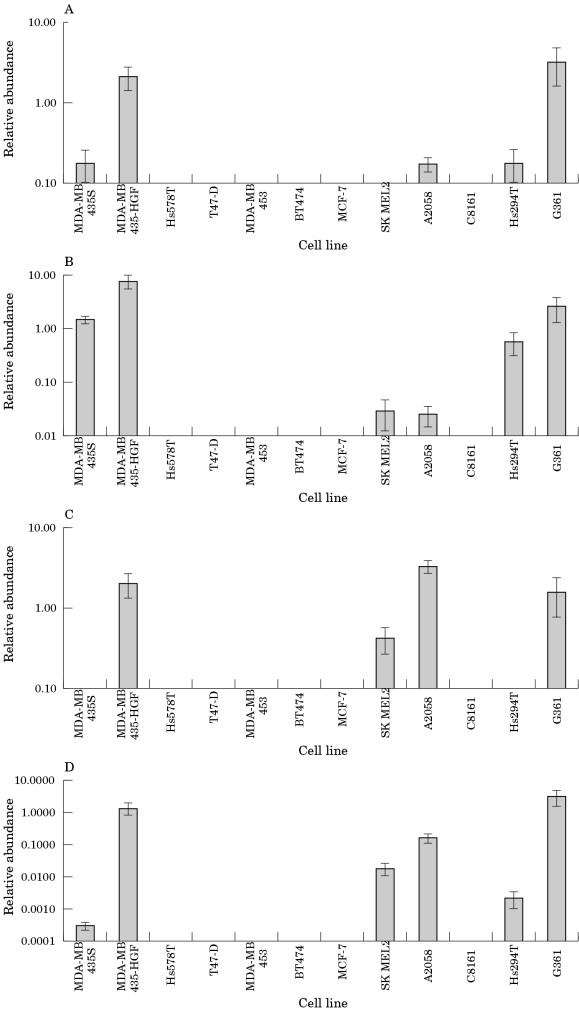
Figure 2 ▶ shows the expression of genes characteristic of melanoma in the cell line samples. All the melanoma cell lines except for C8161 express at least three of the four typical melanoma genes. None of the breast cell lines, excluding MDA-MB-435 S and MDA-MB-435 HGF, expressed these genes. However, MDA-MB-435 HGF expressed all four typical melanoma genes at degrees equivalent to or higher than that seen in the melanoma cell lines. MDA-MB-435 S expressed three of the typical melanoma genes (retinoid X receptor, ACP-5, and tyrosinase related protein 2). The degree of expression was not as high as that seen in the MDA-MB-435 HGF cells but was equivalent to the other melanoma cell lines analysed. The C8161 melanoma cell line expressed none of the typical melanoma genes, but this could be because the cell line has diverged significantly from its original cell type.
Figure 2.
Relative expression of melanocytic genes in the cell lines. A, retinoid X receptor; B, ACP-5; C, tyrosinase; D, tyrosine related protein.
Immunohistochemical analysis
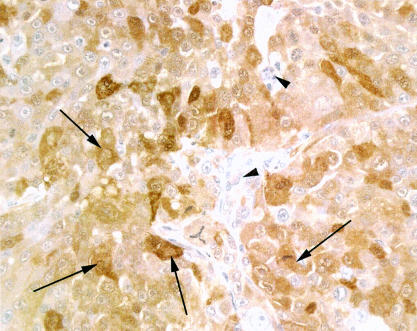
Staining for S100 was positive, strong, and discrete in MDA-MB-435 HGF tumours (fig 3 ▶). Occasional cells adjacent to hair follicles in the skin also stained positive. The positive control sample (human melanoma) showed a similar positive, strong, and discrete staining of tumour cells.
Figure 3.
Section of tumour derived from MDA-MB-435 cells implanted into severe combined immunodeficient mice and stained positively for S100 protein by immunohistochemistry. The cytoplasm of the tumour cells has stained diffusely but discretely brown with a variable intensity (examples are indicated by the arrows). Stromal cells remain unstained (arrowheads). Original magnification, ×400.
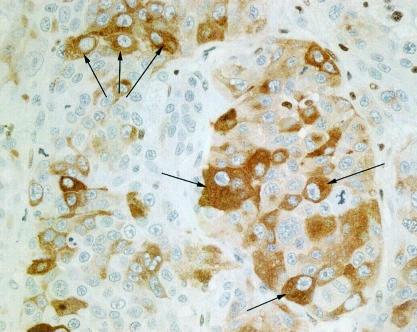
Scattered groups and individual tumour cells showed clear, positive, and discrete cytoplasmic melan A staining (fig 4 ▶). There was some staining of sebaceous gland cells in the skin and mononuclear cells throughout the tissues. Interestingly, mitotic and apoptotic tumour cells did not show positive staining. This may reflect a reduced melanocyte phenotype during cell division and cell degeneration. The positive control sample (human melanoma) showed clearly positive cytoplasmic staining.
Figure 4.
Section of tumour derived from MDA-MB-435 cells implanted into severe combined immunodeficient mice and stained positively for melan A by immunohistochemistry. The cytoplasm of many tumour cells shows a variable but discrete brown staining (as indicated by the arrows) although some cells remain unstained. Original magnification, ×400.
Although MDA-MB-435 HGF tumour cells showed some overall light cytoplasmic staining for PR, this did not appear to be discrete or specific. Many other cell types were also stained, including hair follicles, sebaceous glands, adipocytes, and striated muscle. However, the positive control sample (Novocastra PR control slide) did show strongly positive staining of scattered cells (data not shown). In addition, no specific cytokeratin, ER, or EMA staining of tumour cells was seen. Appropriate positive controls showed strong, discrete, and specific staining (data not shown).
DISCUSSION
Cell lines and tumours can express genes not normally expressed in their tissue of origin. For example, growth in vitro may select for or induce the expression of genes not expressed in vivo.16–18 However, there is no evidence to suggest that the typical melanocyte genes assayed in our study by RT-PCR would confer any growth advantage to a breast cancer cell, particularly because these genes were not expressed in the other breast derived lines investigated. Therefore, it seems most likely that the melanocytic genes are expressed, and the expression of the breast genes is not detected, because MDA-MB-435 S and MDA-MB-435 HGF are of melanocytic origin.
The conclusions drawn from the RT-PCR results are supported by the immunohistochemical stains performed on tumours derived by implantation of MDA-MB-435 HGF cells into SCID mice, which also illustrate that they show protein expression more typical of melanoma than mammary carcinoma. The cells showed a clear positive reaction with antibodies against S100 (protein expressed in neuroectodermal tissue including melanocytes) and melan A (melanocyte differentiation marker). Neither of these markers would be expected to be positive in mammary carcinoma cells. In addition, typical antibody markers for general epithelial or mammary tumours (cytokeratin, EMA, PR, and ER) were either clearly negative or ambiguous.
“We have provided further evidence that cell line MDA-MB-435 is not derived from a breast carcinoma, but is more probably derived from a melanoma”
An interesting ambiguity is evident from the expression data relating to tyrosinase. Because MDA-MB-435 HGF is itself a derivative of MDA-MB-435 S, the detection of tyrosinase overexpression in MDA-MB-435 HGF but not MDA-MB-435 S was surprising. Several explanations for this observation are possible: first, it is feasible that MDA-MB-435 S has itself changed since its isolation, loosing its original higher levels of relative expression; second, it may be that transfection with HGF has in some way induced higher amounts of tyrosinase expression than were originally present in MDA-MB-435 S. However, whatever the true explanation, this example does offer a warning against the use of single markers to differentiate or identify cells derived from abnormal tissues.
Take home messages.
Neither of the MDA-MD-435 cell lines expressed the characteristic breast genes, but they did express several genes commonly expressed by melanocytes
This suggests that the MDA-MB-435 cell line is not derived from a breast carcinoma, but is probably derived from a melanoma
MDA-MB-435 should not be considered for primary use in breast cancer research
These results highlight the extreme caution required when choosing control cell lines for use in cancer research
Another important issue highlighted by our data concerns the selection of control materials. It is apparent from figs 1 and 2 ▶ ▶ that the cell line Hs578T, selected as a control breast cancer derived line, overexpresses none of the selected “breast” tissue genes. Because this cell line has also been categorised as “central nervous system-like” by Ross et al, it would be interesting to investigate this cell line further (it would also be interesting to study the C8161 cell line, in which the expression of melanocytic genes was not detected). This highlights the extreme caution required when choosing control material in these types of biological experiments. Indeed, it would seem that the choice of control cell lines shares many of the same pitfalls as the selection of a single gene transcript or protein as a disease/tissue type marker.
In conclusion, the work described above was initiated following an earlier report that, according to its gene expression profile, MDA-MB-435 did not appear to be typical of breast tissue. Further investigation was therefore warranted because this is a widely used model of cancer cell biology. In addition, Ross and colleagues3 had indicated that the gene expression pattern seen in the NCI60 MDA-MB-435 cells was not identical to that detected in the ATCC MDA-MB-435 S (HTB-129) cells, suggesting potential differences in archived cell lines. We have provided further evidence that cell line MDA-MB-435 is not derived from a breast carcinoma, but is more probably derived from a melanoma. We recommend that MDA-MB-435 should not be considered for primary use in breast cancer research.
Acknowledgments
Thanks to R Somers for immunohistochemical procedures and S Wedge for manuscript review and advice.
Abbreviations
ATCC, American Type Culture Collection
DAB, diaminobenzidine
ECACC, European Collection of Cell Cultures
EMA, epithelial membrane antigen
FCS, fetal calf serum
HGF, hepatocyte growth factor
HPRT, hypoxanthine ribosyl transferase
NEAA, non-essential amino acids
RT-PCR, reverse transcription polymerase chain reaction
SCID, severe combined immunodeficient
REFERENCES
- 1.Cailleau R, Olive M, Cruciger QV. Long-term human breast carcinoma cell lines of metastatic origin: preliminary characterization. In Vitro 1978;14:911–15. [DOI] [PubMed] [Google Scholar]
- 2.Meiners S, Brinkmann V, Naundorf H, et al. Role of morphogenetic factors in metastasis of mammary carcinoma cells. Oncogene 1998;16:9–20. [DOI] [PubMed] [Google Scholar]
- 3.Ross DT, Scherf U, Eisen MB, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 2000;24:227–35. [DOI] [PubMed] [Google Scholar]
- 4.Xin H, Stephans JC, Duan XZ, et al. Identification of a novel aspartic-like protease differentially expressed in human breast cancer cell lines. Biochim Biophys Acta 2000;1501:125–37. [DOI] [PubMed] [Google Scholar]
- 5.Perlino E, Tommasi S, Moro L, et al. TGF-beta 1 and IGF-1 expression are differently regulated by serum in metastatic and non-metastatic human breast cancer cells. Int J Oncol 2000;16:155–60. [DOI] [PubMed] [Google Scholar]
- 6.Hurst DR, Li H, Xu XY, et al. Development and characterization of a new polyclonal antibody specifically against tissue inhibitor of metalloproteinases 4 in human breast cancer. Biochem Biophys Res Commun 2001;281:166–71. [DOI] [PubMed] [Google Scholar]
- 7.Huang LL, Pardee AB. Suberoylanilide hydroxamic acid as a potential therapeutic agent for human breast cancer treatment. Mol Med 2000;6:849–66. [PMC free article] [PubMed] [Google Scholar]
- 8.Bemis LT, Schedin P. Reproductive state of rat mammary gland stroma modulates human breast cancer cell migration and invasion. Cancer Res 2000;60:3414–18. [PubMed] [Google Scholar]
- 9.Hanby AM, Poulsom R, Singh S, et al. Spasmolytic polypeptide and pS2 in the stomach. Gastroenterology 1993;105:1110–16. [DOI] [PubMed] [Google Scholar]
- 10.Watson MA, Fleming TP. Mammaglobin, a mammary specific member of the uteroglobin gene family, is overexpressed in human breast cancer. Cancer Res 1996;56:860–5. [PubMed] [Google Scholar]
- 11.Murphy LC, Tsuyuki D, Myal Y, et al. Isolation and sequencing of a cDNA clone for a prolactin-inducible RT protein (PIP). Regulation of PIP gene expression in the human breast RT cancer cell line, T-47D. J Biol Chem 1987;262:15236–41. [PubMed] [Google Scholar]
- 12.Mangelsdorf DJ, Borgmeyer U, Heyman RA, et al. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev 1992;6:329–44. [DOI] [PubMed] [Google Scholar]
- 13.Lord DK, Cross NCP, Bevilacqua MA, et al. Type 5 acid phosphatase. Sequence, expression and chromosomal localization of a differentiation-associated protein of the human macrophage. Eur J Biochem 1990;189:287–93. [DOI] [PubMed] [Google Scholar]
- 14.Giebel LB, Strunk KM, Spritz RA. Organization and nucleotide sequences of the human tyrosinase gene and a truncated tyrosinase-related segment. Genomics 1991;9:435–45. [DOI] [PubMed] [Google Scholar]
- 15.Bouchard B, del Marmol V, Jackson IJ, et al. Molecular characterization of a human tyrosinase-related-protein-2 cDNA. Patterns of expression in melanocytic cells. Eur J Biochem 1994;219:127–34. [DOI] [PubMed] [Google Scholar]
- 16.Boyce BF, Yoneda T, Guise TA. Factors regulating the growth of metastatic cancer in bone. Endocr Relat Cancer 1999;6:333–47. [DOI] [PubMed] [Google Scholar]
- 17.Mayer B, Lorenz C, Babic R, et al. Expression of leukocyte cell adhesion molecules on gastric carcinomas—possible involvement of LFA-3 expression in the development of distant metastases. Int J Cancer 1995;64:415–23. [DOI] [PubMed] [Google Scholar]
- 18.Fedarko NS, Fohr B, Robey PG, et al. Factor H binding to bone sialoprotein and osteopontin enables tumor cell evasion of complement-mediated attack. J Biol Chem 2000;275:16666–72. [DOI] [PubMed] [Google Scholar]