Abstract
Classic cytogenetic and comparative genomic hybridisation (CGH) data on osteosarcomas have been reported extensively in the literature. However, the number of paediatric osteosarcoma cases studied below the age of 14 years remains relatively small. This study reports four new cases of paediatric osteosarcoma in patients aged 3 to 13 years, evaluated by classic cytogenetics and CGH analyses. Clonal chromosomal alterations were detected in all the cases and included structural rearrangements at 1p11–13, 1q11, 4q27–33, 6p23–25, 6q16–25, 7p13–22, 7q11–36, 11p10–15, 11q23, 17p11.2–13, 21p11, and 21q11–22. The CGH analysis revealed recurrent gains at 1p, 4q, 17p, and 21q and losses at 3q and 16p. Five amplification sites were observed at 1q11–23, 6p21, 8q13, 8q21.3–24.2, and 17p. The data are discussed and compared with other cytogenetic reports in the literature.
Keywords: paediatric osteosarcoma, chromosome alterations, comparative genomic hybridisation,
Osteosarcoma is the most common primary bone malignancy, occurring mainly in children and young adults.1 Classic cytogenetic studies have shown that most of these tumours present with a complex karyotype, characterised by a high degree of heterogeneous chromosomal aberrations, which include both structural and numerical abnormalities.2–4 More recently, comparative genomic hybridisation (CGH) studies have confirmed the complexity of the karyotypic changes in osteosarcomas, and have identified several areas of DNA copy number gains and losses in these tumours.5–9 Although the combined use of both classic cytogenetic and CGH studies has allowed the accurate identification of most of the chromosomal abnormalities associated with these tumours, the specific genetic changes underlying the onset and progression of osteosarcomas remain poorly understood.
“Studies of early onset (paediatric) tumours may provide better insight into the genetic mechanisms of tumorigenesis”
Cytogenetic studies have provided relevant and detailed information on the chromosomal alterations that occur in osteosarcomas; however, most of this information was gathered from adult patients, and the mechanism of tumorigenesis may be different in children. Although osteosarcomas commonly occur in children, fewer studies have looked at the cytogenetic changes in this group.2–4 Studies of early onset (paediatric) tumours may provide better insight into the genetic mechanisms of tumorigenesis. We report the chromosomal changes seen in four paediatric osteosarcoma cases, in children between the ages of 3 and 13 years, which we evaluated by classic cytogenetics (four cases) and by CGH (three cases).
MATERIALS AND METHODS
Tumour samples
Tumour samples were obtained after surgery and primary excision at Saint Louis University School of Medicine, St Louis, Missouri, between 1995 and 2000. Classic cytogenetic evaluation was performed on all four tumours; in addition, three of the four tumours were evaluated by CGH. CGH was not performed on the fourth case because the DNA in the archival tissues was degraded. Table 1 ▶ details the clinicopathological data on all four patients.
Table 1.
Clinicopathological data on the four patients
| Case | Age/sex | Location | Histology results | Recurrence |
| 1 | 9/M | Left proximal humerus | Cellular lesion with focal areas of calcified osteoid and other areas of spindle cells with a high mitotic rate and focal anaplasia | Lung nodules lesion lumbar body |
| 2 | 3/F | Right proximal tibia | Malignant stroma producing osteoid hyperchromatic pleomorphic cells. Several multinucleated giant cells resembling osteoclosts are present | No |
| 3 | 13/M | Right distal femur | Cellular neoplasm with hyperchromotic cells within a mixoid stoma. Cytological atypia is seen. Focal ostoid is seen | No |
| 4 | 9/F | Right distal femur | Not provided | Lung nodules |
F, female; M, male.
Tissue processing for cytogenetic analysis
Fresh tumour fragments were collected in tissue culture medium shortly after tumour excision, and were routinely processed for chromosomal studies. In brief, tumour fragments were minced and treated with collagenase (0.125 mg/ml) for four hours. Cell cultures were set up on coverslips with Amniomax and Chang media, and chromosomes were prepared 10–14 days later, using conventional in situ culture and harvest techniques. Chromosomal analysis was performed by examining four different coverslips for each case. Chromosomes were GTG banded (G banding using trypsin and Giemsa)10 and classified according to the International System for Human Cytogenetic Nomenclature (ISCN 1995).11
Comparative genomic hybridisation
CGH was performed as described previously.12 Tumour DNA and control DNA were labelled by nick translation and hybridised to karyotypically normal male slides. Quantitative evaluation of hybridisation was performed using commercially available software (Applied Imaging, Pittsburgh, Pennsylvania, USA). Average ratio profiles were computed as the mean value of at least eight ratio images, and were used to identify changes in chromosome copy number.
RESULTS
Table 2 ▶ shows both the classic cytogenetic and CGH results of the four paediatric osteosarcoma cases analysed in our study.
Table 2.
The classic cytogenetic and comparative genomic hybridisation (CGH) results of the four paediatric osteosarcoma cases studied
| Case | Classic cytogenetics | CGH |
| Case 1 (M) | 39∼41,XY, −X, del(1)(p11)×2, add(3)(p21), add(4)(q28), del(4)(q28), −5,tas(5;22;14)(p15.3;q13p13;p13), add(6)(p25), add(7)(p15)×2, add(8)(p21), −9, −10, add(11)(p15), del(11)(p11.1), del(12)(p12), der(13)t(1;13)(p11;q11), −15, −15, add(16)(p11.2), add(17)(p13)×2, add(19)q13.1), der(20)t(1;20)(p22;p13), der(21)t(1;21)(p11;q11), −22, +2−3mar[cp12]/46, XY[4] | +1pter–q24, ++1q11–q23, −3q, +4q11–q28, +5p, −5q, +6q, −7p14–q11.2, −8p, ++ 8q13, ++ 8q21.3–q24.2, +9, −10q23–qter, −13, −15q11–q24, −16pter–q21,++17p, +21q11–q21 |
| Case 2 (F) | 80∼160<7n>XXXXXXX, del(1)(q11), +2, del(3)(q26.1)×2, der(4)t(1;4)(q44;q33)inv dup(1)(q44q11)×2, der(5)dup(5)(q11.2q13)dup(5)(q31q35)×2, add(6)(p23)×2, add(6)(q16), hsr(7)(q11.2)×2, −9(×5), −10−10, inv(11) (q13q23), der(12)t(1;12)(q21;p12)×2, del(13)(q14q14)×2, add(14)(p11.2)×2, −17–17, der(17)t(3;17) (?q12;p11.2)×2, +3−4mar [cp10]/46, XX[20] | +X, −3pter–q22, +4q26, ++6p21, +17p11.2, +18p |
| Case 3 (M) | 43, XY, del(1)(p13), −1, −3, add(4)(q33), −6, add(6)(q25), der(7)t(7;12) (p22;q13), −8, add(8)(q24.3), −10, add(11)(p13), −13, der(14)(q24), −17, add(18)(q12), −19, −19, −21, add(21)(q22), −22, +8mar[9]/86, idem×2[2]/46, XY[9] | NR |
| Case 4 (F) | 50∼56, XX, +del(1)(q10)×2, I(1)(q10), +del(2)(p11.2), +3, +del(3)(?q21), +del(4)(q27q31.1), +6, +del(6)(q21), add(7)(q36)×2, +8, +8, −11, −11, −15, −15, −17, −17, +18, −19, +add(21)(p11.2), + 3–4mars[cp6]/46, XX[7] | +1p31–q22, +2q24–qter, +4, +6p, +7p, +8, +11q14–qter, +13q21–qter, −16p, +17p, −19q, +21 |
F, female; M, male; NR, no results; ++, amplification or increase copy number; +, gain; −, loss.
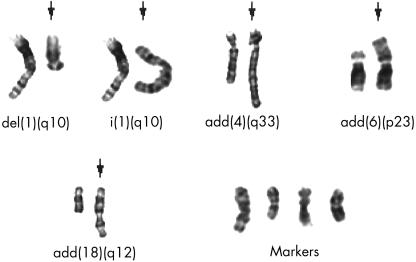
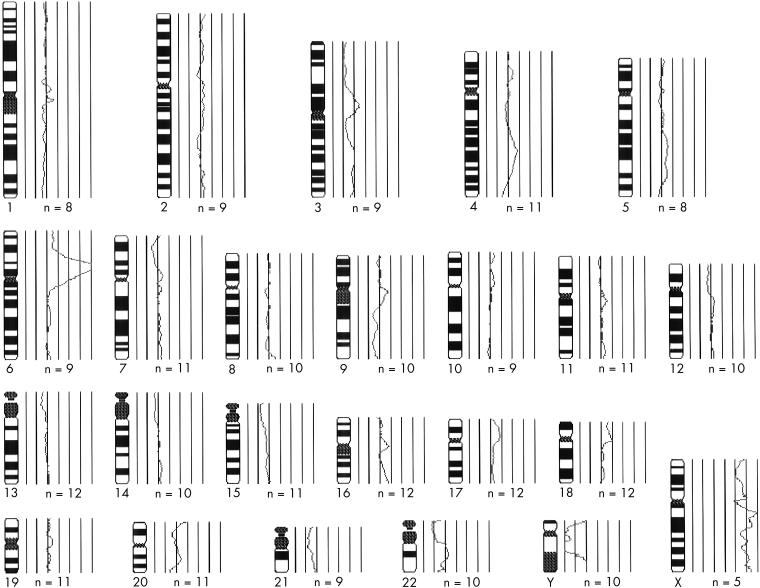
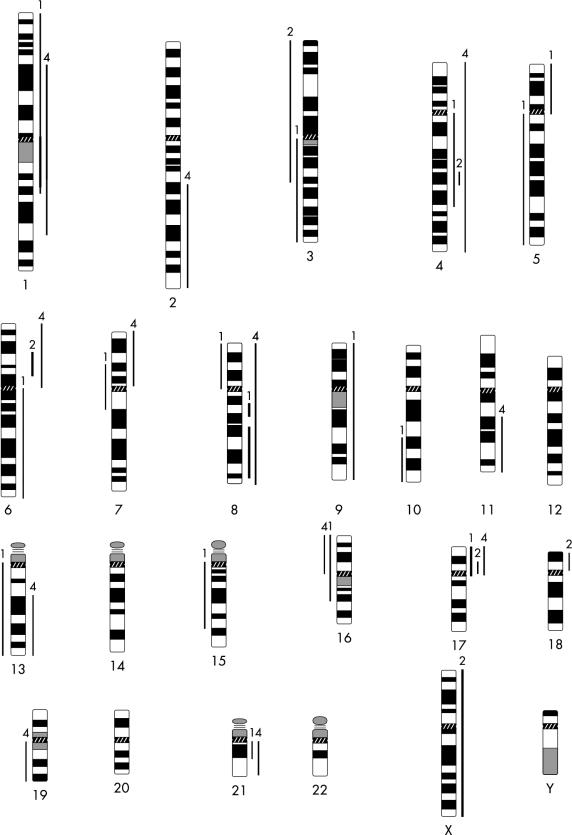
In case 1, most cells were hypodiploid. In case 2, most of the cells were diploid, with some hypertriploid and hypoheptaploid cells. In case 3, a mixed population of cells was observed: hypodiploid, hypotetraploid, and diploid; and in case 4, both hyperdiploid and diploid cells were seen. After GTG banding, clonal chromosomal alterations were seen in all the cases analysed. In general, structural chromosomal abnormalities were more frequent than numerical abnormalities, and preferentially involved the chromosomal regions 1p11–13, 1q11, 4q27–33, 6p23–25, 6q16–25, 7p13–22, 7q11–36, 11p10–15, 11q23, 17p11.2–13, 21p11, and 21q11–22. Marker chromosomes were observed in all the cases analysed. Figure 1 ▶ shows examples of structurally rearranged chromosomes and chromosomal markers detected by GTG banding. The most frequent numerical chromosomal abnormalities included losses of chromosomes 9, 10, 17, 19, and 22. Normal diploid metaphases were present in all four cases, with frequencies ranging from 25% (case 1) to 66% (case 2). CGH analysis revealed changes in the DNA copy number in all three cases analysed. The mean number of chromosomal alterations in each case was 11.5 (range, 5–18) and, in general, gains of chromosomal material were more common than losses (66% and 33%, respectively). The most common gains occurred at the chromosomal arms 1p (two cases), 4q (three cases), 17p (two cases), and 21q (two cases). Losses were more commonly identified at 3q (two cases) and 16p (two cases). Amplification sites were seen at 1q11–23, 8q13, 8q21.3–24.2, and 17p in case 1, and at 6p21 in case 2. In case 4, no high level amplification was observed. Figure 2 ▶ shows the CGH profile of case 2 and a summary of the chromosomal gains and losses detected by CGH in all cases analysed is shown in fig 3 ▶.
Figure 1.
Examples of structurally rearranged chromosomes and chromosomal markers detected by GTG banding. The arrows indicate the rearranged chromosomes.
Figure 2.
Results of comparative genomic hybridisation evaluation of case 2. The vertical lines on the right side of the chromosome ideograms reflect different values of the fluorescence ratio between the test and the normal DNA. The values are 0.5, 0.75, 1.0, 1.25, 1.5, 1.75, and 2.0 from left to right. Ratios of 1.25 or higher reflect gains whereas rations of 0.75 or lower reflect losses. N is the number of chromosomes used to generate each ratio profile. The profile shows an amplification at 6p21, gains at 4q26, 17p11.2, and 18p, an increase in the X chromosome copy number, and loss of 3pter–q22.
Figure 3.
Karyogram of the DNA copy number changes seen in the three osteosarcoma cases. Bars to the right side of the chromosome ideogram indicate a gain whereas bars to the left side indicate a loss of genetic material. Bold lines indicate amplifications. The numbers on the top of each bar refer to the case number.
DISCUSSION
Although osteosarcoma is the most common malignant bone tumour in childhood, most reports have only examined adult cases; few studies have looked at chromosomal changes in the paediatric age group. Our present study evaluated four paediatric osteosarcoma cases, from children between the ages of 3 and 13 years, and characterised their chromosomal aberrations using both classic cytogenetics and CGH analysis. Studies of genomic alterations in early onset (paediatric) tumours may reveal a different pattern of changes than that seen in adult cases, and may be more informative about the molecular mechanisms of tumour onset.
Classic cytogenetic analysis detected numerous and complex clonal chromosome abnormalities in the four paediatric osteosarcoma cases. The structural alterations included rearrangements at 1p11–13, 1q11, 4q27–33, 6p23–25, 6q16–25, 7p13–22, 7q11–36, 11p10–15, 11q23, 12p12, 17p11.2–13, 21p11, and 21q11–22, in addition to the presence of several marker chromosomes. The most common numerical changes were losses of chromosomes 9, 10, 17, 19, and 22. Although cytogenetic data on paediatric osteosarcomas are limited and the results are heterogeneous, reported chromosome abnormalities include structural alterations at 1p32–36, 1q32–44, 3q11–27, 5p14, 11p15, 12p13, and 17p11–13; in addition, numerical alterations involving monosomies of chromosomes 6, 8, 10, 16, 18, and 19 and trisomies of chromosomes 20 and 22 have also been reported.2–4 Some of the alterations described in our study, such as rearrangements at 11p15, 12p12, and 17p11.2–13 and losses of chromosomes 10 and 19, are in agreement with the published data.2–4 However, these changes were not specific to the paediatric cases and have been reported in adults. These aberrant regions, detected in both adult and paediatric osteosarcoma cases, are assumed to harbour genes involved in the onset and progression of these tumours.
“Interestingly, in all four paediatric cases that we studied, we found rearrangements at the 4q region by classic cytogenetics and in three cases, 4q gain was detected by comparative genomic hybridisation”
In our study, CGH showed several sites of chromosomal amplification, including 1q11–23, 8q13, 8q21.3–24.2 (case 1), and 6p21 (case 2). These chromosomal regions appear to contain amplicons that could be specific to osteosarcoma, as suggested independently by Tarkkanen and colleagues5 and Forus et al.6 We detected an amplification at 8q21.3–24.2 in case 1, which has been reported to be a prognostic marker associated with high grade osteosarcomas.7 Other chromosomal alterations detected by CGH in our study included gains at 1p (two cases), 4q (three cases), and 21q (two cases), and losses at 3q (two cases) and 16p (two cases). Zielenska et al reported 1p as the most frequent region of gain detected by CGH; this study included paediatric osteosarcoma cases.9 The 1p region is also commonly rearranged in adult osteosarcomas.7,8 Gains of 6p have also been reported in retinoblastomas13 and in gastric cancers.14 Other upregulated genes were also described in osteosarcomas. The genes encoding the fibromodulin protein (FMDO), located at 1q32, and the heat shock protein (HSP90β), located at 6p12, were found to be upregulated in three different osteosarcoma cell lines analysed using cDNA microarray by Wolf et al.15 Overexpression of such proteins may be of importance in the pathogenesis of osteosarcomas.
Interestingly, in all four paediatric cases that we studied, we found rearrangements at the 4q region by classic cytogenetics and in three cases, 4q gain was detected by CGH. Stock and colleagues8 detected 4q abnormalities by CGH in eight patients, five of whom were children. Using representational difference analysis, an allelic loss at 4q32–34 was recently identified in 63% of osteosarcomas analysed.16 This particular region on 4q harbours several candidate genes, including those encoding interleukins 2 and 21 on 4q26–27, cyclin A on 4q27, and ATP binding cassette on 4q31. Alterations of 4q may be a common aberration peculiar to the paediatric tumours. Larger studies are needed to confirm this finding in this age group.
Our data showed some differences between the CGH findings and the classic cytogenetic results. However, such observations are not uncommon and have been reported previously when both approaches have been used to evaluate the same specimens.17–20 CGH analysis is capable of detecting chromosomal aberrations when present in a large proportion of cells. Conventional metaphase evaluation shows a cell by cell analysis of the tumour genome, whereas CGH analysis shows the average genomic aberrations in the tumour. These two methods are therefore complementary and offer a comprehensive cytogenetic evaluation of the tumours studied. The finding of cells with a normal karyotype is not uncommon in human solid tumours.21 The methodology and the technical conditions that are used for culturing the tumour cells, including the time of growth in vitro, can result in the growth of cells with normal karyotypes or with no clonal chromosomal abnormalities.22–24 The meaning of such findings remains controversial, and it is not known whether these cells with normal karyotypes are of neoplastic origin or whether selection for non-neoplastic cells occurs during in vitro culture, as suggested by Truong et al.21 Recently, it was shown by Kleivi and colleagues18 that cases of breast cancer that presented normal cells by classic cytogenetic analysis were not really karyotypically normal, but had genetic alterations when CGH analysis was carried out. In our study, CGH showed the presence of genetic imbalances in all three cases studied, reflecting the presence of these aberrations in a high proportion of the tumour cells (usually over 50%), but not in every cell.17 Cells with a normal karyotype may still be present, as demonstrated by classic cytogenetic analysis.
In conclusion, the cytogenetic abnormalities, detected either by classic cytogenetic studies or by CGH, were highly diverse and variable, confirming the complex pattern of chromosomal alterations present in osteosarcomas, as reported previously. It was not possible to identify chromosomal alterations specific to the paediatric group in our study. Even when our paediatric cases were taken together with those previously reported in the literature, the small number of total paediatric osteosarcoma cases, and the similarity to adult osteosarcoma, make it difficult at this time to separate specific paediatric chromosomal changes from those in the adult. It is apparent that additional genetic studies of paediatric osteosarcomas are needed to provide a better “profile” of these tumours and to provide further insight into the mechanisms of tumour onset.
Take home messages
We studied four cases of osteosarcoma in patients aged 3 to 13 years by classic cytogenetics and comparative genomic hybridisation (CGH) analyses because such data are scant in children below the age of 14
Clonal chromosomal alterations were detected in all the cases and included structural rearrangements at 1p11–13, 1q11, 4q27–33, 6p23–25, 6q16–25, 7p13–22, 7q11–36, 11p10–15, 11q23, 17p11.2–13, 21p11, and 21q11–22
The CGH analysis revealed recurrent gains at 1p, 4q, 17p, and 21q and losses at 3q and 16p
Five amplification sites were observed at 1q11–23, 6p21, 8q13, 8q21.3–24.2, and 17p
These abnormalities are highly diverse and variable, confirming the complex pattern of chromosomal alterations present in osteosarcomas and we could not identify chromosomal alterations specific to the paediatric age group
Additional genetic studies of paediatric osteosarcomas are needed to produce a better “profile” of these tumours and to provide further insight into the mechanisms of tumour onset
Abbreviations
CGH, comparative genomic hybridisation
GTG, G banding using trypsin and Giemsa
REFERENCES
- 1.Himelstein BP. Osteosarcoma and other bone cancers. Curr Opin Oncol 1998;10:326–33. [DOI] [PubMed] [Google Scholar]
- 2.Mertens F, Mandahl N, Mitelman F, et al. Cytogenetic analysis in the examination of solid tumors in children. Pediatr Hematol Oncol 1994;11:361–77. [DOI] [PubMed] [Google Scholar]
- 3.Biegel JA, Womer RB, Emanuel BS. Complex karyotypes in a series of pediatric osteosarcomas. Cancer Genet Cytogenet 1989;38:89–100. [DOI] [PubMed] [Google Scholar]
- 4.Bridge JA, Nelson M, McComb E, et al. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet 1997;95:74–87. [DOI] [PubMed] [Google Scholar]
- 5.Tarkkanen M, Karhu R, Kallioniemi A, et al. Gains and losses of DNA sequences in osteosarcomas by comparative genomic hybridization. Cancer Res 1995;55:1334–8. [PubMed] [Google Scholar]
- 6.Forus A, Weghuis DO, Smeets D, et al. Comparative genomic hybridization analysis of human sarcomas: II. Identification of novel amplicons at 6p and 17p in osteosarcomas. Genes Chromosomes Cancer 1995;14:15–21. [DOI] [PubMed] [Google Scholar]
- 7.Tarkkanen M, Elomaa I, Blomqvist C, et al. DNA sequence copy number increase at 8q: a potential new prognostic marker in high-grade osteosarcoma. Int J Cancer 1999;84:114–21. [DOI] [PubMed] [Google Scholar]
- 8.Stock C, Kager L, Fink F-M, et al. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer 2000;28:329–36. [DOI] [PubMed] [Google Scholar]
- 9.Zielenska M, Bayani J, Pandita A, et al. Comparative genomic hybridization analysis identifies gains of 1p35 approximately p36 at chromosome 19 in osteosarcoma. Cancer Genet Cytogenet 2001;130:14–21. [DOI] [PubMed] [Google Scholar]
- 10.Scheres VMJC. Identification of two Robertsonian translocations with a Giemsa banding technique. Hum Genet 1972;15:253–6. [DOI] [PubMed] [Google Scholar]
- 11.Mitelman F, ed. ISCN (1995). An international system for human nomenclature (1995). Basel: Karger.
- 12.Figueiredo BC, Stratakis CA, Sandrini R, et al. Comparative genomic hybridization (CGH) analysis of adrenocortical tumors of childhood. J Clin Endocrinol Metab 1999;84:1116–21. [DOI] [PubMed] [Google Scholar]
- 13.Mairal A, Pinglier E, Gilbert E, et al. Detection of chromosome imbalances in retinoblastoma by parallel karyotype and CGH analyses. Genes Chromosomes Cancer 2000;28:370–9. [PubMed] [Google Scholar]
- 14.Sakakura C, Mori T, Sakabe T, et al. Gains, losses and amplifications of genomic materials in primary gastric cancers analysed by comparative genomic hybridization. Genes Chromosomes Cancer 1999;24:299–305. [DOI] [PubMed] [Google Scholar]
- 15.Wolf M, El-Rifai W, Tarkkanen M, et al. Novel findings in gene expression detected in human osteosarcoma by cDNA microarray. Cancer Genet Cytogenet 2000;123:128–32. [DOI] [PubMed] [Google Scholar]
- 16.Simons A, Schepens M, Forus A, et al. A novel chromosomal region of allelic loss, 4q32–q34, in human osteosarcomas revealed by representational difference analysis. Genes Chromosomes Cancer 1999;6:115–24. [PubMed] [Google Scholar]
- 17.Teixeira MR, Tsarouha H, Kraggerud SM, et al. Evaluation of breast cancer polyclonality by combined chromosome banding and comparative genomic hybridization analysis. Neoplasia 2001;3:204–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleivi K, Lothe RA, Heim S, et al. Genome profiling of breast cancer cells selected against in vitro shows copy number changes. Genes Chromosomes Cancer 2002;33:304–9. [DOI] [PubMed] [Google Scholar]
- 19.Schmid-Braz AT, Cavalli LR, Cornelio DA, et al. Comprehensive cytogenetic evaluation of a mature ovarian teratoma case. Cancer Genet Cytogenet 2002;132:165–8. [DOI] [PubMed] [Google Scholar]
- 20.Persson K, Pandis N, Mertens F, et al. Chromosomal aberrations in breast cancer: a comparison between cytogenetics and comparative genomic hybridization. Genes Chromosomes Cancer 1999;25:115–22. [PubMed] [Google Scholar]
- 21.Truong K, Guilly MN, Gerbault-Seureau M, et al. Evidence for in vitro selection during cell culturing of breast cancer: detection by flow and image cytometry. Cancer Genet Cytogenet 1999;114:154–5. [DOI] [PubMed] [Google Scholar]
- 22.Pandis N, Bardi G, Heim S. Interrelationship between methodological choices and conceptual models in solid tumor cytogenetics. Cancer Genet Cytogenet 1994;76:77–84. [DOI] [PubMed] [Google Scholar]
- 23.Steinarsdottir M, Petursdottir I, Snorradottir S, et al. Cytogenetic studies of breast carcinomas: different karyotypic profiles detected by direct harvesting and short-term culture. Genes Chromosomes Cancer 1995;13:239–48. [DOI] [PubMed] [Google Scholar]
- 24.Cavalli LR, Cavalieri LM, Ribeiro LA, et al. Cytogenetic evaluation of 20 primary breast carcinomas. Hereditas 1997;126:261–8. [DOI] [PubMed] [Google Scholar]