Abstract
Aims: To examine the expression of ADAM12 (meltrin α), a member of the disintegrin and metalloprotease (ADAM) family, in human giant cell tumours of the bone, skeletal muscle tissue from human embryos, and human adult skeletal muscle tissue.
Methods: ADAM12 mRNA was detected by reverse transcription polymerase chain reaction and in situ hybridisation.
Results: ADAM12 mRNA was detected in 14 of the 20 giant cell tumours of bone and in three of the six tumour cell cultures. The expression of ADAM12 in cells cultured from the tumour was linked to the presence of multinucleated giant cells. ADAM12 mRNA could not be detected in the five adult skeletal muscle tissue samples, although it was found in the two embryonic skeletal muscle tissue samples. ADAM12 mRNA was localised to the cytoplasm of multinucleated giant cells and some mononuclear stromal cells.
Conclusions: These results indicate that multinucleated giant cells are formed by the cell fusion of mononuclear stromal cells in giant cell tumours of bone and that ADAM12 is involved in the cell fusion process.
Keywords: giant cell tumour of bone, ADAM12 (meltrin-α), cell fusion
A DAM (a disintegrin and metalloprotease) molecules are a recently discovered family of membrane anchored cell surface glycoproteins. They have a unique domain organisation, comprising (from the N-terminus to the C-terminus) a prodomain, metalloprotease domain, disintegrin domain, cysteine rich domain, transmembrane domain, and cytoplasmic domain.1–4 Structurally, ADAMs are different from other membrane anchored adhesion proteins or proteases, although they have a similar structure to and share about 30% sequence identity with snake venom metalloproteases (SVMPs).5 Full length SVMPs are processed to generate a metalloprotease, which is able to degrade proteins of the basement membrane, such as type IV collagen and laminin,6 and a disintegrin domain, which can inhibit the function of platelets by interacting with platelet integrin GPIIα-IIIβ.7 Because of their structural features, ADAMs have been implicated in proteolysis, cell adhesion, cell fusion, and signalling. Recently, new ADAMs have been discovered; to date, 29 types of ADAMs have been reported.8 They are involved in many life processes. For example, ADAM10 is responsible for the proteolytic cleavage and activation of Delta, a ligand of the Notch receptor.9 ADAM11 is a candidate tumour suppressor gene.10 ADAM13 may be involved in neural crest cell adhesion and migration in Xenopus laevis.11 ADAM17 is required for the cleavage of tumour necrosis α from its membrane bound precursor.12–14 In addition, ADAM20 and ADAM21 are expressed in human testis and may be functional equivalents of human sperm fertilin α.15 The first described ADAMs were fertilin α and fertilin β (ADAM1 and ADAM2), which are expressed as heterodimers on the posterior head of mammalian spermatocytes. They are thought to mediate sperm–egg binding and fusion by interacting with integrin α6β1 on the egg surface,2,16–18 although human fertilin α is thought to be non-functional.19,20
In addition to sperm–egg binding and fusion in fertilisation, the process of cell fusion occurs in myotube formation, multinucleated giant cell formation, and placenta syncytiotrophoblast formation. All these processes of cell fusion appear to be similar. Yagami-Hiromasa et al identified meltrin α (ADAM12) when they searched for homologues of fertilin in a mouse myogenic cell line in 1995.21 Gilpin et al identified human ADAM12 in 1998.22 The human ADAM12 gene is located at 10q26.3 and has typical ADAM family structure sequences—from the N-terminus to the C-terminus, the ADAM12 gene encodes a prodomain, metalloprotease domain, disintegrin domain, cysteine rich domain, transmembrane domain, and cytoplasmic domain. There is a zinc binding active site in the metalloprotease domain, and a fusion peptide-like sequence in the cysteine rich domain.22
“Because of their structural features, ADAMs have been implicated in proteolysis, cell adhesion, cell fusion, and signalling”
Using northern blotting to analyse ADAM12 (meltrin α) in adult mouse tissues—including bone, brain, liver, heart, and muscle—only bone, which contains multinucleated osteoclasts, expressed ADAM12.21 Strong expression of mouse ADAM12 was also shown in neonatal skeletal muscle and bone and in cultured mouse C2 myoblast cells after differentiation. ADAM12 has fusogenic properties and plays a role in the formation of myotubes by the fusion myoblasts.21 Furthermore, high expression of ADAM12 in adult bone also suggests that ADAM12 might be involved in the formation of multinucleated osteoclasts by the fusion of mononuclear stromal cells.
Human giant cell tumours of bone are a distinctive neoplasm of the bone, which are characterised clinically by locally expansive osteolytic lesions. Histopathologically, giant cell tumours consists of abundant multinucleated giant cells (MGCs) and short spindle shaped stromal cells. MGCs in the tumour tissue are assumed to be potentially active bone resorbers.23 Increasing evidence also strongly supports the view that the MGCs of the tumour are osteoclastic. However, the process of MGC formation is still unknown.
Here, we report the expression of ADAM12 in human giant cell tumours of bone. Human embryo and adult skeletal muscle tissues were also examined and served as controls.
MATERIALS AND METHODS
Tissue specimens
Twenty samples of giant cell tumour of bone were obtained from surgical excision and five samples of adult skeletal muscle tissue were obtained at the same time as controls. Two samples of human embryo skeletal muscle tissue were obtained from 4 month naturally aborted fetuses. All tissues of giant cell tumour of the bone were divided into three parts: one for ADAM12 mRNA reverse transcription polymerase chain reaction (RT-PCR), one for RNA in situ hybridisation, and one for cell culture.
Cell culture
Six giant cell tumour of bone samples were cultured successfully. The tissues were cut into 1 mm3, seeded into plastic flasks with 0.5 ml of fresh RPMI 1640 medium (Gibco BRL, Gaitherburg, Massachusetts, USA), and supplemented with antibiotics (100 IU/ml of penicillin and 100 μg/ml streptomycin) and 20% fetal calf serum. The cultures were incubated for 24–48 hours at 37°C in a humidified atmosphere of 95% air and 5% CO2. The medium was changed and 4 ml of fresh medium was added to the cultures when cells began to grow out from the margin of the seeded tissues. The cells were subcultured once a week when they reached confluence. The cells were harvested at the first to 12th passages for mRNA extraction.
RT-PCR
Total RNA was extracted from both the tissues and the cultured cells using Total RNA Extracting Liquid (Da An Gene Co, Guangzhou, China). Specific primers for detecting ADAM12 mRNA were designed from a known sequence of the human ADAM12 gene.22 The oligonucleotide primers for the RT-PCR were 5′-ACT TCC GGA GGC AAA GTC TCG AAG AGT TC-3′ and 5′-ACT TCC GGA GCA AGA AGA CAC AGG ATG GT-3′. The amplified products are 498 bp long and contain the major cysteine rich domain and a part of the transmembrane domain of ADAM12. For RT-PCR, 1 μg of total RNA was added to the RT-PCR reaction reagent (Titan™ One Tube RT-PCR Kit; Roche, Germany) in a total volume of 50 μl. The amplification protocol consisted of 30 seconds of denaturation at 94°C, 30 seconds of annealing at 60°C, and 45 seconds of extension at 68°C. At the 11th cycle, the extension time was increased by five seconds after each cycle and a total of 30 cycles were performed. β Actin mRNA was also amplified from both the tumours and the cultured cells as an internal control using the same RT-PCR protocol as above. The amplified products were detected using 1.5% agarose gel electrophoresis.
RNA in situ hybridisation
The plasmid containing the full length ADAM12 sequence was kindly provided by Dr UM Wewer (University of Copenhagen, Denmark).22 The plasmid was cut with BamHI and separated on a 1% agarose gel. A 3.3 kb fragment was obtained and used as a template for RNA probe synthesising. For probe labelling, 1.8 μg of template was added to 20 μl of reaction reagent (DIG RNA Probe Labelling Kit; Roche, Germany). The digoxigenin (DIG) labelled sense probe synthesised by T7 RNA polymerase is 524 bp long and was used to detect the 469 bp sequence of ADAM12 mRNA, which contains a 5′-untranslated region, a signal peptide region, and part of the prodomain. A DIG labelled nonsense probe synthesised by T3 RNA polymerase was used as a negative control.
Frozen tissue sections (5 μm) were cut on a cryomicrotome at −20°C and placed on diethyl pyrocarbonate treated siliconised glass slides. After fixation in 4% paraformaldehyde in DEPC treated phosphate buffered saline (PBS) for 20 minutes, the sections was digested with 25 μg/ml protease K for 25 minutes, and rinsed twice in PBS for 15 minutes each. The DIG labelled probe of ADAM12 RNA was diluted with the hybridisation solution to a final concentration of 20 μg/ml. The mixtures were heated at 65°C for 10 minutes and then 30 μl was placed on each slide, which was coverslipped and incubated at 50°C for 20 hours in a humidified chamber. The coverslips were removed and the slides were washed twice in 2× saline sodium citrate (SSC) at 37°C, twice with 1× SSC at room temperature, and finally in buffer I for 10 minutes and buffer II for 30 minutes at room temperature. The hybridisation probes were detected with anti-DIG-POD, set according to the manufacturer’s instructions (Roche). Briefly, the slides were incubated sequentially in anti-DIG-POD for 60 minutes at 37°C in a humidified chamber. The slides were then washed briefly in a washing buffer at 37°C, stained with diaminobenzidine, and the colour allowed to develop. For negative controls, slides were hybridised with 20 μg/ml of nonsense probe.
RESULTS
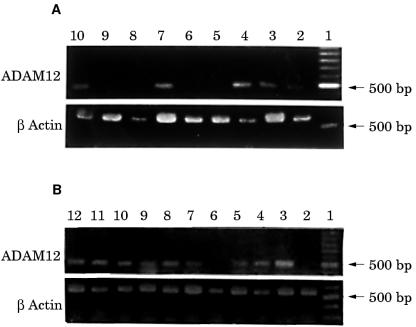
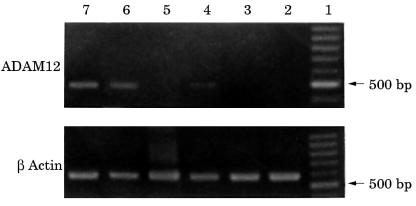
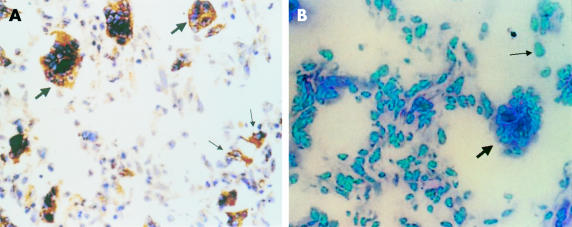
The positive RT-PCR amplified product of ADAM12 was 498 bp long, as expected. Fourteen of the 20 giant cell tumour of the bone samples were ADAM12 mRNA positive, whereas six samples were negative (fig 1 ▶). Three of the six cultures grown from these tumours were positive for ADAM12 mRNA and three were negative (fig 2 ▶). The expression of ADAM12 mRNA in cultured cells was dependent on the cell passage. At the first, fourth, and sixth passages, the cells were positive for ADAM12 mRNA. The expression of ADAM12 decreased as the cell passage number increased. After the eighth passage, the cells became ADAM12 mRNA negative. Some MGCs survived in these cultures, but their numbers decreased with increased passaging and they eventually disappeared. After the fourth passage, there were no MGCs—only mononuclear stromal cells were present and they continued to grow. Therefore, the expression of ADAM12 in cultured cells of the tumour appeared to be dependent on the existence of MGCs. ADAM12 mRNA could not be detected in the five adult skeletal muscle tissue samples, although it was positive in the two embryonic skeletal muscle tissue samples. The DIG labelled RNA probe hybridised with the ADAM12 mRNA and positive signals were seen as brown granules in the cytoplasm. Twelve of the 20 giant cell tumours of the bone were positive; almost all MGCs and some mononuclear stromal cells were positive (fig 3 ▶). The five adult skeletal muscle tissue samples were negative, whereas the two embryonic skeletal muscle tissue samples were positive. The negative control probe detected no signals in either the tumour or the muscle tissues. Tables 1 and 2 ▶ ▶ summarise the expression of ADAM12 mRNA in giant cell tumours of bone tissues and in cultured cells.
Figure 1.
Expression of ADAM12 in 20 cases of giant cell tumour. The ADAM12 reverse transcription polymerase chain reaction (RT-PCR) product is 498 bp long. (A) Lane 1, 100 bp DNA ladder; lanes 2–10, giant cell tumour samples. Five of nine cases show positive expression of ADAM12 (lanes 2, 3, 4, 7, and 10). (B) Lane 1, 100 bp DNA ladder; lanes 2–12, giant cell tumour tissues. Nine of 11 cases show positive expression of ADAM12 (lanes 3–5, 7–12). The β actin RT-PCR product from the same tumour tissues was used as an internal control (lower panels).
Figure 2.
The expression of ADAM12 in six cell cultures from cases of giant cell tumour. The ADAM12 reverse transcription polymerase chain reaction (RT-PCR) product is 498 bp long. Lane 1, 100 bp DNA ladder; lane 2, eighth passage cells; lane 3, 10th passage cells; lane 4, sixth passage cells; lane 5, 12th passage cells; lane 6, fourth passage cells; lane 7, first passage cells. The β actin RT-PCR product from the same culture cells was used as an internal control (lower panel).
Figure 3.
The expression of ADAM12 mRNA in giant cell tumour tissues. (A) Positive signal for ADAM12 mRNA detected by in situ hybridisation; the brown granules are seen in the cytoplasm of multinucleated giant cells (thick arrow) and in some mononuclear stromal cells (thin arrow). (B) Negative ADAM12 mRNA control using a nonsense probe. Original magnification, ×200.
Table 1.
The expression of ADAM12 mRNA in 20 giant cell tumours of bone
| Tumour tissues | RT-PCR | RNA in situ hybridisation |
| 10 cases | + | + |
| 2 cases | – | + |
| 4 cases | + | – |
| 4 cases | – | – |
RT-PCR, reverse transcriptase polymerase chain reaction.
Table 2.
The expression of ADAM12 mRNA in different passages of cells cultured from giant cell tumours of bone
| Passage | RT-PCR | RNA in situ hybridisation |
| 1st passage | + | + |
| 4th passage | + | + |
| 6th passage | + | + |
| 8th passage | – | – |
| 10th passage | – | – |
| 12th passage | – | – |
RT-PCR, reverse transcriptase polymerase chain reaction.
DISCUSSION
ADAM12 is a member of the recently identified ADAM family of membrane proteins, which contain both a disintegrin and metalloprotease domain. Its full length gene has a sequence similar to that of the gene encoding the fusion peptide fertilin. Among the ADAM family, only fertilin and ADAM12 have this fusion peptide gene sequence, implicating ADAM12 in cell fusion.
Using northern blotting, Yagami-Hiromasa et al detected the specific expression of ADAM12 in mouse muscle at embryonic and neonatal stages, in mouse bone from embryo to adult, and in the mouse myoblast cell line C2.21 When the expression of ADAM12 was blocked in C2 cells, myotube formation was suppressed,21 indicating that ADAM12 is necessary for myotube formation. This also suggests that the role of ADAM12 in cell fusion might involve cell–cell and/or cell–matrix interactions. Gilpin et al identified a secreted form of ADAM12 that could induce myogenesis in vivo.22 However, among bone, brain, liver, heart, and muscle of the adult mouse, only bone expressed ADAM12 mRNA.21 It is possible that this is because bone has many multinucleated osteoclasts, which are formed by mononuclear progenitors, and the fusion mechanism is similar to that seen in myotube formation. Thus, in view of the above studies, we believe ADAM12 might play a crucial role in osteoclast formation.
“Using RNA in situ hybridisation, we found that ADAM12 mRNA was localised to the cytoplasm of multinucleated giant cells and was also seen in mononuclear stromal cells”
Histopathologically, giant cell tumour of the bone consists of abundant MGCs and many short spindle shaped stromal cells. The MGCs are scattered among a mass of mononuclear stromal cells. However, the mechanism of MGC formation is still unknown. Our previous studies have shown that MGCs and mononuclear stromal cells in giant cell tumours of bone have the capability to resorb bone matrix and that the stromal cells may secret macrophage colony stimulating factor, which could facilitate the formation of MGCs.23 If ADAM12 is involved in the formation of MGCs, it should be overexpressed in this tumour. Using RT-PCR, we detected, for the first time, the expression of ADAM12 in almost three quarters of the giant cell tumours of the bone examined. Furthermore, using RNA in situ hybridisation, we found that ADAM12 mRNA was localised to the cytoplasm of MGCs and was also seen in mononuclear stromal cells. This suggests that MGCs may be formed by the fusion of mononuclear stromal cells, which express ADAM12.
Take home messages.
The multinucleated giant cells found in giant cell tumours of the bone appear to be formed by the cell fusion of mononuclear stromal cells
ADAM12, a member of the disintegrin and metalloprotease family, appears to be involved in the cell fusion process, although its precise role is unclear
In addition, a passage dependent expression of ADAM12 mRNA was detected in the cells cultured from giant cell tumours of the bone. With increasing cell passages ADAM12 mRNA gradually disappeared, along with the MGCs. The studies of Abe et al indicate that after treatment with 1,25(OH)2D3 (a potent inducer of osteoclast and giant cell formation), murine mononuclear alveolar macrophages expressed ADAM12 mRNA before cell fusion. Moreover, the addition of ADAM12 antisense oligonucleotides to the cultures caused a 50% inhibition of giant cell formation. Similarly, the addition of ADAM12 antisense oligonucleotides to co-cultures of bone marrow and osteoblastic cells caused a 70% inhibition of MGCs expressing tartrate resistant acid phosphatase.24
What is the mechanism by which ADAM12 could facilitate cell fusion? Some SVMP disintegrin domains contain an RGD (arginine–glycine–aspartic acid) integrin ligand sequence in a β loop structure, which binds with high affinity to αVβ3 and αIIβ3 integrins; such a sequence could play an important role in cell–cell and/or cell–matrix interactions.25,26 However, ADAM12 does not contain a RGD sequence; it is the cysteine rich domain of ADAM12 that supports cell adhesion, in a dose dependent manner.27 The disintegrin domain and cysteine rich domain of mouse ADAM12 could form an active cell adhesion domain, supporting myoblast cell adhesion in a divalent cation dependent manner, thus facilitating myoblast fusion.28
In addition, we think that it is highly likely that MGCs are formed by the cell fusion of mononuclear stromal cells in giant cell tumours of bone and that ADAM12 plays an important role in this cell fusion process. However, ADAM12 may only have a role as a membrane anchored receptor—its ligand for cell fusion is, at present, unknown.
Acknowledgments
This work was supported by a grant from the Chinese Medical Board (CMB) and a PhD student training programme grant of the Ministry of National Education, P R China (No. 200046). We thank Dr UM Wewer (Institute of Molecular Pathology, University of Copenhagen, Copenhagen, Denmark) for providing the plasmid used to produce the mRNA in situ hybridisation probe.
Abbreviations
ADAM, disintegrin and metalloprotease
DIG, digoxigenin
MGC, multinucleated giant cell
PBS, phosphate buffered saline
RT-PCR, reverse transcription polymerase chain reaction
SSC, saline sodium citrate
SVMP, snake venom metalloprotease
REFERENCES
- 1.Huovila APJ, Almeida EA, White JM. ADAMs and cell fusion. Curr Opin Cell Biol 1996;8:692–9. [DOI] [PubMed] [Google Scholar]
- 2.Wolfsberg TG, White JM. ADAMs in fertilization and development. Dev Biol 1996;180:389–401. [DOI] [PubMed] [Google Scholar]
- 3.Blobel CP. Metalloprotease-disintegrins: links to cell adhesion and cleavage of TNF-α and Notch. Cell 1997;90:589–92. [DOI] [PubMed] [Google Scholar]
- 4.Wolfsberg TG, Primakoff P, Myles DG, et al. ADAM, a novel family of membrane proteins containing a disintegrin and metalloprotease domain: multipotential functions in cell–cell and cell–matrix interactions. J Cell Biol 1995;131:275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox JW, Bjarnason JB. The reprolysins: a family of metalloproteases defined by snake venom and mammalian metalloproteases. In: Hooper NM. Zinc metalloproteases in health and disease. London: Taylor and Francis, 1996:47–81.
- 6.Hite LA, Jia LG, Bjarnason JB, et al. cDNA sequences for four snake venom metalloproteinases: structure, classification, and their relationship to mammalian reproductive proteins. Arch Biochem Biophys 1994;308:182–91. [DOI] [PubMed] [Google Scholar]
- 7.Paine MJI, Desmond HP, Theakstont RDG, et al. Purification, cloning, and molecular characterization of a high molecular weight hemorrhagic metalloprotease, jararhagin, from bothropis jararaca venom. J Biol Chem 1992;267:22869–76. [PubMed] [Google Scholar]
- 8.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet 2000;16:83–7. [DOI] [PubMed] [Google Scholar]
- 9.Qi H, Rand MD, Wu X, et al. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science 1999;283:91–4. [DOI] [PubMed] [Google Scholar]
- 10.Emi M, Katagiri T, Harada Y, et al. A novel metalloprotease disintegrin-like gene at 17q12.3 is somatically rearranged in two primary breast cancers. Nat Genet 1993;5:151–7. [DOI] [PubMed] [Google Scholar]
- 11.Alfandari D, Wolfsberg TG, White JM, et al. ADAM13: a novel ADAM expressed in somatic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol 1997;182:314–30. [DOI] [PubMed] [Google Scholar]
- 12.Black RA, Rauch CT, Kozlosky CJ, et al. A metalloproteinase disintegrin that releases tumor necrosis factor-alpha from cells. Nature 1997;385:729–33. [DOI] [PubMed] [Google Scholar]
- 13.Moss ML, Jin SL, Milla ME, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumor necrosis factor-α. Nature 1997;385:733–6. [DOI] [PubMed] [Google Scholar]
- 14.Moss ML, Jin SL, Becherer JD, et al. Structural features and biochemical properties of TNF-α converting enzyme (TACE). J Neuroimmunol 1997;72:127–9. [DOI] [PubMed] [Google Scholar]
- 15.Hooft van Huijsduijnen-R. ADAM20 and ADAM21: two novel human testis-specific membrane metalloproteases with similarity to fertilin-α. Gene 1998;206:273–82. [DOI] [PubMed] [Google Scholar]
- 16.Almeida EA, Huovila AP, Sutherland AE, et al. Mouse egg integrin α6β1 functions as a sperm receptor. Cell 1995;81:1095–104. [DOI] [PubMed] [Google Scholar]
- 17.Myles DG, Primakoff P. Why did the sperm cross the cumulus? To get to the oocyte. Functions of the sperm surface proteins PH-20 and fertilin in arriving at, and fusing with the egg. Biol Reprod 1997;56:320–7. [DOI] [PubMed] [Google Scholar]
- 18.Carroll DJ, Dikegoros E, Koppel DE, et al. Surface expression of the pre-beta subunit of fertilin is regulated at a post-translational level in guinea pig spermatids. Dev Biol 1995;168:429–37. [DOI] [PubMed] [Google Scholar]
- 19.Jury JA, Frayne J, Hall L. The human fertilin α gene is non-functional: implications for its proposed role in fertilization. Biochem J 1997;321:577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jury JA, Frayne J, Hall L. Sequence analysis of a variety of primate fertilin-α genes: evidence for non-functional genes in the gorilla and man. Mol Reprod Dev 1998;51:92–7. [DOI] [PubMed] [Google Scholar]
- 21.Yagami-Hiromasa T, Sato T, Kurisaki T, et al. A metalloprotease-disintegrin participating in myoblast fusion. Nature 1995;377:652–6. [DOI] [PubMed] [Google Scholar]
- 22.Gilpin BJ, Loechel F, Mattei MG, et al. A novel, secreted form of human ADAM12 (meltrin-α) provokes myogenesis in vivo. J Biol Chem 1998;273:157–66. [DOI] [PubMed] [Google Scholar]
- 23.Wen J, Dan X, Yao J, Zhang M, et al. Effect of cytokines on in vitro bone resorption by cells isolated from giant cell tumor of bone. Chin Med J (Engl) 1999;112:443–7. [PubMed] [Google Scholar]
- 24.Abe E, Mocharla H, Yamate T, et al. Meltrin-alpha, a fusion protein involved in multinucleated giant cell and osteoclast formation. Calcif Tissue Int 1999;64:508–15. [DOI] [PubMed] [Google Scholar]
- 25.Marcinkiewicz C, Vijay-Kumar S, McLane MA, et al. Significance of RGD loop and C-terminal domain of echistatin for recognition of αIIbβ3 and αVβ3 integrins and expression of ligand-induced binding site. Blood 1997;90:1565–75. [PubMed] [Google Scholar]
- 26.Tselepis VH, Green LJ, Humphries MJ. An RGD to LDV motif conversion within the disintegrin kistrin generates an integrin antagonist that retains potency but exhibits altered receptor specificity: evidence for a functional equivalence of acidic integrin-binding motifs. J Biol Chem 1997;272:21341–8. [DOI] [PubMed] [Google Scholar]
- 27.Iba K, Albrechtsen R, Gilpin BJ, et al. Cysteine-rich domain of human ADAM12 (meltrin-α) supports tumor cell adhesion. Am J Pathol 1999;154:1489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zolkiewska A. Disintegrin-like/cysteine-rich region of ADAM12 is an active cell adhesion domain. Exp Cell Res 1999;252:423–31. [DOI] [PubMed] [Google Scholar]