Abstract
Human endogenous retroviruses (HERVs) are a family of viruses within our genome with similarities to present day exogenous retroviruses. HERVs have been inherited by successive generations and it is possible that some have conferred biological benefits. However, several HERVs have been implicated in certain cancers and autoimmune diseases. This article demystifies these retroviruses by providing an insight into HERVs, their means of classification, and a synopsis of HERVs implicated in cancer and autoimmunity. Furthermore, the biological roles of HERVs are explored.
Keywords: human endogenous retroviruses, cancer, autoimmunity
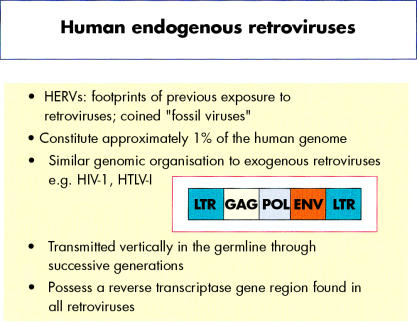
Human endogenous retroviruses (HERVs) represent footprints of previous retroviral infection and have been termed “fossil viruses”. They are transmitted vertically through the germline and are thus inherited by successive generations in a Mendelian manner. Over time, HERVs have been subjected to repeated amplification and transposition events giving rise to multicopy and single copy proviruses that are distributed within the DNA of all cells. Overall, HERVs constitute about 1% of the human genome. HERVs possess a similar genomic organisation to present day exogenous retroviruses such as human immunodeficiency virus (HIV) and human T cell leukaemia virus (HTLV), and are composed of gag, pol, and env regions sandwiched between two long terminal repeats (LTRs) (fig 1 ▶). The LTRs possess nucleotide sequence motifs that are fundamental to the regulation of retroviral gene expression. In brief, the gag and env genes encode retroviral capsid and envelope proteins, respectively, whereas the pol gene encodes enzymes for viral replication, integration, and protein cleavage. Retroviruses in effect are retrograde, because the flow of genetic information is reversed compared with the normal pathway of molecular biosynthesis—DNA → RNA → protein. Indeed, all retroviruses necessitate the conversion of viral RNA into a cDNA intermediary, which is catalysed by the enzyme reverse transcriptase.
Figure 1.
Salient features of human endogenous retroviruses (HERVs) and similarity to exogenous retroviruses.
“Overall, human endogenous retroviruses constitute about 1% of the human genome”
Over 20 HERV families have been identified during the past two decades.1–3 Although many are defective through the accumulation of mutations, deletions, and termination signals within coding sequences, a limited number of HERVs have the potential to produce viral products and, indeed, to produce viral-like particles. Furthermore, some HERVs have been implicated in certain autoimmune diseases and cancers4–7 and might have a role in the aetiology and pathology of disease. However, many HERVs have been present in our genome for a considerable period of time so that their presence may also be of benefit to the human host.
MOLECULAR BIOLOGY AND HERVS
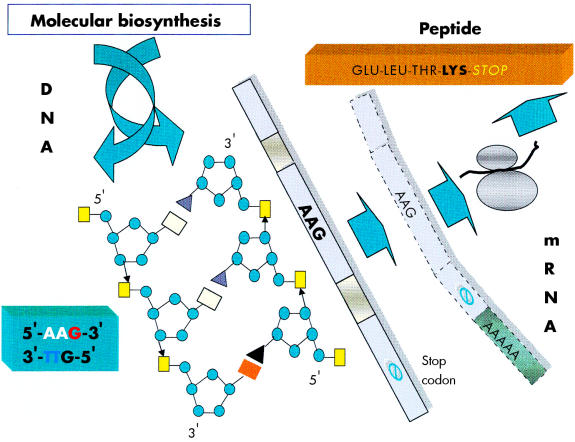
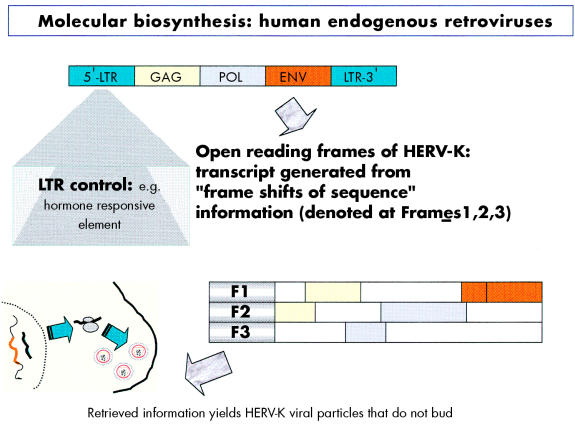
The focus of the human genome project is to sequence the 3 billion DNA bases that compose our human genome.8 Interestingly, only a small proportion (about 3%) appears to constitute the estimated 24 179 to 87 720 genes that are translated into the proteins necessary to life. The function of non-coding DNA (maligned as “junk DNA”) is not readily obvious and it may have important roles in packaging and influencing gene expression. DNA is composed of two helical strands orientated in an antiparallel fashion (fig 2 ▶), where each strand consists of a deoxyribose phosphate backbone and a series of purine and pyrimidine bases denoted A (adenine), G (guanine), T (thymine), and C (cytosine). A unit of sugar, phosphate, and base is strictly termed a nucleotide, although casually represented by its base. The two DNA strands associate through the complementary pairing of bases—for example, A–T and C–G—which is stabilised through hydrogen bonding. Stretches of DNA, measured in base pairs, constitute the numerous genes that are found on the 23 pairs of human chromosomes. Genes vary in size, with some in the order of 20–40 kilobase pairs (kbp), whereas others extend to millions of base pairs. In addition, human genes are composed of exons, which are transcribed and translated into amino acids, and introns, which are interspersed between exons and represent non-translated regions that contribute to the large size of some genes. Retroviruses are remarkably small (in the order of 9–10 kbp) compared with other pathogenic viruses, such as cytomegalovirus, which is 200 kbp. Nevertheless, retroviruses capitalise on their limited size by using frame shifts to retrieve genetic information from multiple overlapping open reading frames (fig 3 ▶). Interestingly, the open reading frames of many defunct HERVs are interrupted by premature stop codons that interrupt the “read through” of genetic information. A previous demystified article9 highlights the use of the polymerase chain reaction (PCR) to detect certain nucleotide regions of HERVs that are common to all retroviruses.7,9
Figure 2.
A schematic diagram of a segment of DNA that is transcribed into mRNA and finally translated into a peptide product. In this example the DNA triplet code AAG is translated into the amino acid lysine.
Figure 3.
Diagram illustrating the influence of the long terminal repeat (LTR) on the production of endogenous viral peptides. Of particular importance is that a human endogenous retrovirus (HERV) product may be generated using different open reading frames.
“Retroviruses capitalise on their limited size by using frame shifts to retrieve genetic information from multiple overlapping open reading frames”
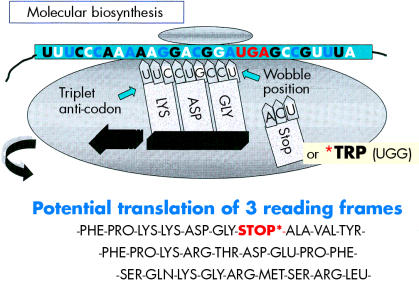
Before protein synthesis, mRNA is transcribed from relevant stretches of DNA (reliant on a triplet base code or codon) that specifies a start signal (for example, ATG), an amino acid, or a termination signal (TAA, TAG, or TGA). The primary mRNA transcript is refined by splicing out unwanted introns and then by adding a series of adenine bases to its 3′ end. The process of polyadenylation is considered crucial to the stability of the mRNA that leaves the nucleus to attach to cytoplasmic ribosomes. Here, individual tRNA molecules attached to a specific amino acid are aligned with mRNA that dictates the assembly of polypeptide chains (fig 4 ▶). Strictly speaking, tRNAs use anticodons for complimentary binding, although a codon, such as AAU for lysine (denoted by K in the single amino acid code), is often used with reference to the tRNA “primer binding site” of a HERV—for example, HERV-K. Interestingly, tRNAs may use the “wobble” phenomenon in that the first two bases are essential for binding but the third base is less crucial. Therefore, it is possible that tRNAs may over-ride stop codons and allow the growth of a polypeptide chain. Ultimately, both exogenous and endogenous retroviruses exploit the cell’s genetic machinery and comply with the same generic principles of protein biosynthesis to produce further viral progeny. However HERVs stop short of viral budding (fig 3 ▶) and consequently are non-infectious.10
Figure 4.
A schematic diagram illustrating that a premature stop codon on mRNA may be over-ridden at the ribosome, thus allowing a potential to produce full length or truncated viral products.
CLASSIFICATION OF HERVS
Historically, retroviruses were shown to cause malignant tumours and leukaemias in chickens and mammals—for example, the Rous sarcoma virus and mouse mammary tumour virus (MMTV), respectively—and were subdivided into three subfamilies: oncovirinae (oncoviruses), lentivirinae (slow viruses), and spumavirinae (foamy viruses).2 Retroviruses were also classified according to morphological and biological criteria: in particular by the observance of retroviral particles in infected cells.11,12 In brief, type A retroviruses were only visible inside cells, devoid of envelope, and referred to as intracisternal particles (A type particles), whereas types B, C, and D were enveloped and produced extracellular particles that varied in size and appearance. Subsequently, in the 1980s, the retroviruses HTLV-I and HIV-1 were reported and a new classification formulated by the International Committee for Taxonomy of Viruses.13 This taxonomy reflected the similarity of a virus to an established retroviral genus, such as murine leukaemia related virus. Set against this background, the first HERV was reported in 1981,14 and many others have subsequently been identified (table 1 ▶) using several different methodologies. These have included the screening of human genomic libraries under low stringency conditions with DNA probes from animal retroviruses (for example, HTDV/HERV-K, HERV-E, and HERV-R), the use of synthetic probes homologous to the primer binding site of known retroviruses (such as HERV-P), and the analysis of human gene loci (HERV-H, ERV-9, and HERV-I), plus information from the human genome project.1,3,15 Consequently, the classification of HERVs has been complex, with arbitrary nomenclatures arising from independent investigators, coupled with a raft of classification criteria including morphological type, copy number, and specificity of the tRNA primer binding site. In this last case, problems have arisen when distantly related families have had similar binding sites or cloned sequences are deficient in this region.2,10,16
Table 1.
Classification of human endogenous retroviruses (HERVs)
| HERV family | Representative accession number |
| Class I HERVs (type C related HERVs) | |
| Group 1, HERV-HF | |
| HERV-H (RTVL-H, RGH) | AF108842 |
| HERV-F | AF070684 |
| Group 2, HERV-RW | |
| HERV-W | AF072506 |
| HERV-R (ERV9) | X57147 |
| HERV-P (HuERS-P, HuRRS-P) | X06279 |
| Group 3, HERV-ERI | |
| HERV-E (4-1, ERVA, NP-2*) | S46403 |
| 51-1 | J00273 |
| HERV-R (ERV3) | M12140 |
| RRHERV-I | M64936 |
| Group 4, HERV-T | |
| HERV T (S71, CRTK1, CRTK6) | M32788 |
| Group 5, HERV-IP | |
| HERV-I (RTVL-I) | X14953 |
| HERV-IP-T47D (ERV-FTD) | U27241 |
| Group 6, ERV-FRD | |
| ERV-FRD | U27240 |
| Class II HERVs (type A, B, and D related HERVs) | |
| Group 1, HERV-K (HML-1) | |
| HERV-K (HML-1.1) | U35102 |
| Group 2, HERV-K (HML-2) | |
| HERV-K10 | M14123 |
| HERV-K-HTDV | X8227 |
| Group 3, HERV-K (HML-3) | |
| HERV K (HML3.1) | U35153 |
| Group 4, HERV-K (HML-4) | |
| HERV-K-T47D | AF020092 |
| Group 5, HERV-K (HML-5) | |
| HERV -K-NMWV2 | AF015995 |
| Group 6, HERV-K (HML-6) | |
| HERV K (HML-6p) | U86698 |
| Group 7, HERV-K (HML-7) | |
| HERV-K-NMWV7 | AF016000 |
| Group 8, HERV-K (HML-8) | |
| HERV-K-NMWV3 | AF015996 |
| Group 9, HERV-K (HML-9) | |
| HERV-K-NMWV9 | AF016001 |
| Group 10, HERV-K (HML-10) | |
| HERV-KC4 | U07856 |
| Class III Foamy virus related HERVs | |
| HERV-L | X89211 |
Recently, HERVs have been classified into three broad classes (table 1 ▶) based on sequence comparison with animal retroviruses.17 Class I HERVs are subdivided into six groups that share homology with infectious mammalian type C viruses. Three families within this class show homology with murine leukaemia virus (MuLV) and baboon endogenous virus (BaEV) in the highly conserved pol region and the gag and env regions. Members include HERV-H, HERV-I, and HERV-R (ERV-9). Class II HERVs show homology to mammalian type B (for example, MMTV) and type D retroviruses and are subdivided into 10 groups. These groups reflect the sequence homology of reverse transcriptase regions of PCR derived clones from normal leucocytes, and clones derived by low stringency hybridisation of an MMTV gag–pol probe to a breast cancer cell genomic library.18–20 Members include HERV-K (fig 5 ▶) and HERV-K (C4).21,22 Interestingly, all class II HERVs possess a lysine tRNA reflecting their derivation from B and D type viruses.10 HERV-K has been termed the biologically most active human endogenous retrovirus family,23 and has been further subdivided into type 1 or type 2, based on the presence or absence of a 292 bp segment at the pol–env boundary.24 Finally, foamy virus related HERVs are classified as class III HERVs and include a solitary member, HERV-L. This system provides a uniformity to the classification of HERVs but does not include the human T cell leukaemia related endogenous retrovirus HRES-1,25 which shows only limited homology to HTLV-I in the LTR region.
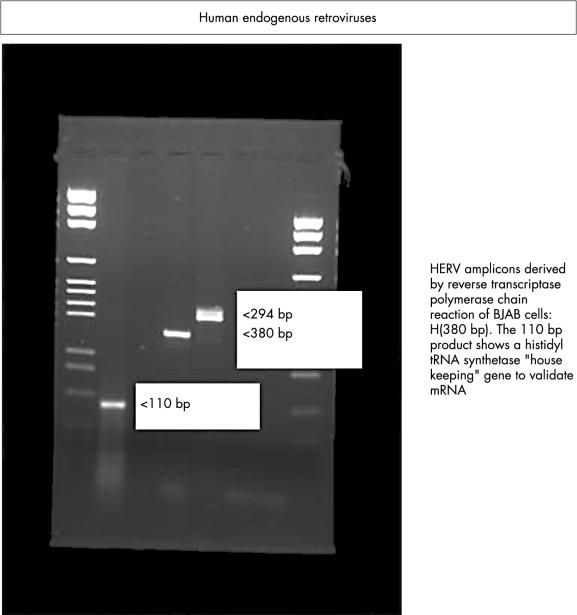
Figure 5.
Expression of endogenous viruses HERV-K and RTVL-H in human B cells as demonstrated by reverse transcriptase polymerase chain reaction.
HERVS AND CANCER
Although the precise role(s) of HERVs in the carcinogenic process has not been fully elucidated there are several studies that, if taken together, put forward a convincing argument for the possible involvement of HERVs in malignancy. HERVs may be involved in carcinogenesis by virtue of the expression of HERV mRNA,26 functional proteins,27 or retroviral-like particles.28 They may also be associated with the generation of new promoters29 or the activation of proto-oncogenes.30 The expression of HERV-R mRNA is increased in some cases of small cell lung carcinoma.26 In addition, a teratocarcinoma cell line has been shown to possess a HERV-K sequence and to secrete retroviral-like particles.28 Testicular germ cell tumours (TGCTs) have been shown to contain proteins of the HERV-K family and patients with TGCT often exhibit a specific immune response to gag and env proteins.27,31 It has been suggested that HERV-K may be important in the progression of TGCT through inhibition of an effective immune response,31 and the HERV env genes have been shown to encode immunosuppressive proteins.32,33 It is clear that overexpressed HERV proteins can elicit high titre IgG responses in some settings (for example, HERV-K10 in patients with renal cancer), as detected by the SEREX method (serological identification of expressed genes),34 suggesting that HERV proteins may in the future provide targets for antitumour immunotherapy.
“It has been suggested that HERV-K may be important in the progression of testicular germ cell tumours through inhibition of an effective immune response”
HERV-K might be important in the pathogenesis of human breast cancer. It has been shown that the T47D human mammary carcinoma cell line produces retroviral particles35 with reverse transcriptase activity.36 Both the HERV-K10 related sequences of T47D cells37 and the reverse transcriptase activity36 are increased by steroid hormone treatment, which is thought to be the result of transcriptional activation via binding of the progesterone receptor to regions on the HERV-K genome that correspond to progesterone and glucocorticoid response elements.
In choriocarcinoma, it has been shown that a HERV type C is inserted into the human growth factor gene, pleiotrophin (PTN). This results in the generation of a novel tissue specific promoter, which results in the expression of HERV–PTN fusion transcripts, leading to the production of biologically active PTN protein. Expression of the PTN protein (which is normally expressed only at very low amounts in a few normal adult tissues38) appears to be responsible for the aggressive and invasive growth of human choriocarcinoma.29
Overexpression is a common mechanism by which proto-oncogenes become activated, leading to subsequent neoplastic transformation.39 In particular, activation of proto-oncogenes of the ras family is common in many tumour types, and some studies have suggested a potential role for HERVs in ras activation. It was shown a methylnitrosourea induced rat mammary carcinoma that insertion of a defective endogenous retrovirus into the intron of c-Ha-ras was responsible for its more than 10 fold overexpression.30
Recently, it has been shown that lymphotropic herpesvirus, Epstein-Barr virus (EBV), itself a potent transforming agent, can transcriptionally activate the env gene of HERV-K18, which possesses superantigen activity (as demonstrated by major histocompatibility complex (MHC) class II dependent preferential activation of TCRVB13 T cells in response to murine B cells transfected with the HERV-K18 env gene).40 The authors suggested that this phenomenon accounts for the previously described EBV associated superantigen activity, which might in turn be crucial to T cell activation by EBV. Although the exact function of HERVs in the carcinogenic process is still under investigation, the evidence implicating HERVs in the carcinogenic process is substantial and further investigation will be required to elucidate the contribution of HERVs to the development of malignancy.
HERVS AND AUTOIMMUNITY
In 1990, an article appeared in the Times newspaper (24 November) with the title “AIDS-like virus may cause arthritis”. The report focused on Robert Garry’s research that identified retroviral particles in lip biopsies taken from patients with primary Sjogren’s syndrome (SS).41 Similarly, in other autoimmune rheumatic diseases, such as rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE), a plethora of articles added to this intriguing observation by providing evidence of retroviral antigens at the site of disease, or the presence of antiretroviral antibodies in the sera of patients.6,42–44 A novel report in 1994 used both PCR (using consensus primers) and serological tests to investigate the presence of retroviruses in a cross section of patients with rheumatoid diseases, including RA, SS, and SLE.45 Interestingly, PCR failed to amplify products relating to HTLV-I or HIV-1, although antibodies to retroviral antigens were detected in the sera of patients. Consequently, there appeared to be a conundrum: antibodies to retroviral products were present but no evidence to implicate exogenous retroviruses could be found. Between 1996 and 1999, some research groups used so called “degenerate” retroviral primers in their PCR reactions.7,46,47 These primers cater for modest variations within two segments common to all retroviruses within the reverse transcriptase encoding pol region and provide an intervening “fingerprint region”, which permits DNA sequencing. In brief, these studies7,46,47 revealed nucleotide homologies to endogenous retroviral families, including viruses with similarity to known exogenous retroviruses. Thus, it was plausible that the presence of HERVs could provide an explanation for the presence of antiretroviral antibodies in certain rheumatoid diseases.6,7,48 HERVs have also been implicated in other autoimmune diseases, such as multiple sclerosis (HERV-W, HERV-H) and insulin dependent diabetes mellitus (IDDM) (HERV-K, IDDM22), in addition to inflammatory vascular diseases.49–54 However, in the case of IDDM, subsequent studies55,56 have not been able to confirm this association. Mechanisms whereby HERVs could influence autoimmunity include molecular mimicry (HERVs sharing amino acids common to host proteins), superantigen motifs that bypass the normal MHC restrictive process of T cell stimulation, aberrant expression of antigens, and the presence of neo-antigens, perhaps as a result of HERV and/or exogenous viral combinations.7,32,57–59 The use of animal models has also served to enhance our understanding of endogenous retroviruses. In a lupus model, an 8.4 kbp endogenous retroviral transcript is expressed in affected mice.60 Furthermore, a retroviral element in one of the introns of the fas apoptosis gene appears to alter the splicing of fas transcripts, resulting in a lupus-like autoimmune disease in MRL-lpr/lpr mice.61 Further investigations using animal models and multicentre patient studies are needed to establish links between specific HERVs and autoimmune diseases because many HERVs are also expressed in varying amounts, or in a coordinated fashion, in normal tissues.62,63
“Mechanisms whereby HERVs could influence autoimmunity include molecular mimicry, superantigen motifs that bypass the normal MHC restrictive process of T cell stimulation, aberrant expression of antigens, and the presence of neo-antigens”
BIOLOGICAL IMPORTANCE OF HERVS
Phylogenetic studies have shown that some HERVs emerged over 3 million years ago, whereas others appeared after the divergence of hominoid and ape lineages.64 Consequently, HERVs have been present in our genome for a considerable period of time and perhaps have been retained because they performed a useful biological function. Alternatively, some HERVs may have been difficult to eliminate and thus persisted during evolution. To confer a selective advantage, a premise remains that HERVs produce products and/or augment mechanisms that benefit host survival. Several possibilities are highlighted below.
Immunosuppressive peptide
The product derived from the env gene of mammalian type C retroviruses possesses motifs—for example, the fusion peptide, leucine zipper protein, and immunosuppressive peptide (ISP)—that are essential for fusion and the infection of cells. In brief, the precursor env product is cleaved into two components: a surface protein (gp70) and a transmembrane protein (p15E) that contains an immunosuppressive region.32 Of interest is an ISP, termed CKS-17, that suppresses lymphocytes and alters cytokine profiles in animal models. The ISP sequence from the murine leukaemia virus LQNRRGLDLLFLKEGGLC (single amino acid code) is reasonably well conserved in HERV-H19 (LQNRRGLDLLTAEKGGLC), HERV-R (ERV-3) (YQNRLALDYLLAQEEGVC), and HERV-E(4–1) (YQNRLALDYLLAAEGGVC), but less so in HERV-K10 (FEASKAHLNLVPGTEAIA). The presence of an ISP could be advantageous to a virally infected cell—in terms of shielding or “cloaking” itself from immunological attack—but may equally be important to a host. This is perhaps exemplified by HERV-R (ERV3), which is highly expressed in trophoblastic cells and results in high concentrations of env protein (∼ 65 kDa) in syncytiotrophoblasts.65 The immunosuppressive potential of this HERV and the fusogenic nature of placenta tissue suggests a possible involvement in normal placental function, in protecting the developing fetus from maternal immune responses.66,67 Furthermore, it is possible that HERVs may change the pattern of gene expression during embryo development by altering different rates of development of different parts of the embryo. Another endogenous retrovirus, HERV-W, has also been shown to encode a protein termed syncytin, which may have a role in placental morphogenesis.68
Antiviral resistance
Mechanisms of conferring protection against a related viral agent may include retroviral receptor blockade (by HERV products) and interference of replication by antisense mRNA. However, it is also possible that HERV peptides could prime an immune response to an undesirable agent. Intriguingly, cohorts of female sex workers in the Gambia and Kenya have been identified who remain uninfected and seronegative despite being repetitively exposed to HIV. The apparent “resistance” to HIV infection is suggested to result from MHC class I restricted cytotoxic T lymphocytes (CTLs)—HLA-B35 and HLA-A2 (A*6802)/HLA-B18—which are found in Gambians and Kenyans, respectively.69,70 The immune response to HIV infection is characterised by a vigorous HIV specific CTL response where virus-specific CTLs recognise antigen in the form of processed peptides (eight to 10 amino acids in length), which are bound in the cleft of MHC class I molecules on the surface of antigen presenting cells. In this context, recent data71 have highlighted similarities between HIV CTL peptide sequences and regions of HERV-K10. Consequently in some cases previous exposure to HERV peptides could potentially immunise certain individuals, although this argument does not explain why some females seroconvert after a reduction in sex work.72
Long terminal repeats
Retroviral genes that have been integrated into the genome are bordered by short direct repeats of host DNA and LTR sequences of about 500–600 nucleotides. These LTRs can influence neighbouring genes because they may contain transcriptional regulatory elements such as enhancers, promoters, hormone responsive elements, and polyadenylation signals. The influence on protein expression is demonstrated by proteins such as salivary amylase, ZNF80, cytochrome C1, Kruppel-like H-plk, and phospholipase A2-L, whose genes are linked to LTRs.32 Numerous solitary LTRs are also present within our genome. These sequences, which include about 10 000 copies of solitary LTRs pertaining to HERV-K, have arisen through recombination events that could have “deleted” harmful retroviral genes. It is noteworthy that LTRs are located near MHC genes and thus could modulate antigen expression, particularly in individuals who possess identical MHC genes but different LTRs.73 With reference to autoimmune diseases, the presence of an HERV-K LTR in the HLA-DQ region (DQ-LTR3) has been shown to have a strong influence on the occurrence of IDDM.74 Furthermore, the presence of DQ-LTR on HLA-DQB1*0302 and its absence on DQB1*0201 are independent risk markers for IDDM. The presence of DQ-LTR has also been shown to be a marker for increased susceptibility to RA in subjects with the HLA-DR4-DQB1*0301 and HLA-DR4-DQB1*0302 haplotypes.75 In addition, polymorphisms within LTRs could be important because changes in sequence could greatly affect promoter activity. Interestingly, three allelic forms of the LTR of HERV-R (ERV3) have been shown, although no link could be associated with disease, in this case multiple sclerosis.76 The fact that LTRs interact with chemicals (such as carbon tetrachloride), hormones, and environmental agents and function as switches for alternative splicing (for example, the short and long forms of the human leptin receptor77) highlights the importance of these elements. In addition, their location (adjacent to MHC or T cell receptor genes) and transactivation by helper viruses7 warrants further research into their potential role in autoimmune diseases and cancer.
Plasticity
HERVs constitute only a part of what are termed “transposable elements”, a generic term encompassing both DNA sequences that can be excised and reinserted at another site and retroelements. The term retroelements describes any sequence that can replicate itself by a process involving reverse transcription, and includes HERVs, retrotransposons (which mostly lack an env gene), retroposons, and retrosequences. Retroposons and retrosequences are exemplified by long stretches of related sequences (up to 6 kbp) called LINES and very short interspersed repeat elements of about 300 bp, respectively. Thus far, from being a fixed, immutable structure, the genome of a eukaryotic cell can harbour many sequences that move from one site on a chromosome to a completely different position. This phenomenon of plasticity is considered important because it permits rapid changes in our genome that could not be afforded by mutations alone. Furthermore, retroelements may carry regulatory sequences (such as enhancers and promoters) to new sites in the genome and thus alter the expression of existing adjacent genes.78 Of course, there remains a potential to disrupt genes by insertion mutation and produce defective and/or truncated products, as has been highlighted by Kazazian, who reported cases of human genetic disease caused by the random insertion of LINES.79
PREMATURE STOP CODONS
It is known that many HERVs possess premature stop codons and of course the RNA products of truncated genes may well be eliminated before protein translation.80 However, examples of premature stop codons that result in human disease by the production of a truncated protein product have been demonstrated in cases of the DNA repair disorder xeroderma pigmentosum81 and also in leukaemia.82 In addition, approximately 5% of cystic fibrosis cases are caused by premature termination codons. Interestingly, some aminoglycoside antibiotics appear to suppress stop codons in several organisms. Howard and colleagues83 demonstrated this effect in cell cultures that expressed constructs carrying two different premature stop codons within the cystic fibrosis gene (CFTR). Treatment with antibiotic produced a full length CFTR: the physiological suppression of the premature stop codon was thought to result from a mispairing of an amino-acyl tRNA that successfully bound to a stop codon. The importance of this observation is that HERVs, despite possessing premature stop codons, may have the potential to produce truncated or full length products that are fundamental to the mechanisms of molecular mimicry, aberrant expression of products, and the stimulation of T cells through superantigen motifs.84
CONCLUSIONS
This article has provided an overview of a complex topic that may have ramifications in host protection, cancer, and autoimmunity. Ultimately, are HERVs friends or foes? In conferring a biological advantage, HERVs (and solitary LTRs) may indeed be beneficial. Their role in immunological homeostasis and perhaps protection against exogenous retroviruses is intriguing. Alternatively, HERV insertion mutation, molecular mimicry, superantigen motifs, and recombination with other viruses could be responsible for the development and pathology of disease. An additional aspect is whether the presence of HERV peptides during ontogeny culminates with a hole in the immune repertoire. As a result, peptides with similarity to HIV-CTL sequences could be more dangerous to a given individual.
Take home messages.
Human endogenous retroviruses (HERVs) make up part of our genome and represent footprints of previous retroviral infection
HERVs possess a similar genomic organisation (gag–pol–env) to present day exogenous retroviruses but are not infectious
The HERV-K superfamily represents one of the most active HERVs and is capable of producing retroviral particles
HERVs may be of benefit to the host but could also be harmful, and may be involved in certain autoimmune diseases and cancers
“An additional aspect is whether the presence of HERV peptides during ontogeny culminates with a hole in the immune repertoire”
Clearly, there is a need for multicentre studies to ascertain firm associations between HERV(s) and autoimmune disease states and certain cancers. In particular “gene chip” technologies will no doubt relate HERV expression with disease and pathological progression. Transcription of individual HERVs or the coordinated expression of HERVs, although important, must be balanced against expression found in normal tissues. Consequently, studies of HERV/LTR polymorphisms, transactivation by helper viruses (or other triggers), and the role of full length or spliced transcripts may provide further knowledge of these viruses. In addition, there is a requirement for a panel of readily available antibody reagents (for example, monoclonal antibodies, recombinant phage antibodies) to determine retroviral products at the site of disease. No doubt the field of HERV research will continue to accelerate so that we can fully ascertain the consequences of renegade endogenous retroviruses and their transfer in xenotransplanation.85,86
Acknowledgments
Dr Nelson would like to thank St John’s College, Oxford for providing a visiting scholarship to initiate this article.
Abbreviations
CFTR, cystic fibrosis gene
CTL, cytotoxic T lymphocyte
EBV, Epstein-Barr virus
HERV, human endogenous retrovirus
HIV, human immunodeficiency virus
HTLV, human T cell leukaemia virus
IDDM, insulin dependent diabetes mellitus
ISP, immunosuppressive peptide
LINES, long stretches of related sequences
LTR, long terminal repeat
MHC, major histocompatibility complex
MMTV, mouse mammary tumour virus
PCR, polymerase chain reaction
PTN, pleiotrophin gene
RA, rheumatoid arthritis
SLE, systemic lupus erythematosus
SS, Sjogren’s syndrome
TGCT, testicular germ cell tumour
REFERENCES
- 1.Tristem M. Identification and characterisation of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol 2000;74:3715–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin J. Endogenous retroviruses. In: Weiss R, Teich N, Varmus H, et al, eds. Molecular biology of tumour viruses, 2nd ed. Cold Spring Harbour, USA: Cold Spring Harbour Laboratory Press, 1985:357–404.
- 3.Leib-Mosch C, Brack-Werner R, Werner T, et al. Endogenous retroviral elements in human DNA. Cancer Res 1990;50(suppl):5636–42. [PubMed] [Google Scholar]
- 4.Boller K, Konig H, Sauter M, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 1993;196:349–53. [DOI] [PubMed] [Google Scholar]
- 5.Lower R, Boller K, Hasenmaier B, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci U S A 1993;90:4480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson PN. Retroviruses in rheumatic diseases. Ann Rheum Dis 1995;55:441–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson PN, Lever AML, Smith S, et al. Molecular investigations implicate human endogenous retroviruses as mediators of anti-retroviral antibodies in autoimmune rheumatic disease. Immunol Invest 1999;28:277–89. [DOI] [PubMed] [Google Scholar]
- 8.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 2001;409:860–921. [DOI] [PubMed] [Google Scholar]
- 9.Baumforth KRN, Nelson PN, Digby JE, et al. The polymerase chain reaction demystified. Mol Pathol 1998;52:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lower R, Lower J, Kurth R. Review. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A 1996;93:5177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernhard W. Electron microscopy of tumour cells and tumour viruses. Cancer Res 1958;18:491–509. [PubMed] [Google Scholar]
- 12.Dalton AJ. Further analysis of detailed structure of types B and C particles. J Natl Cancer Inst 1972;48:1098–9. [PubMed] [Google Scholar]
- 13.Coffin J. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol 1992;176:143–64. [DOI] [PubMed] [Google Scholar]
- 14.Martin MA, Bryan T, Rasheed S, et al. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc Natl Acad Sci U S A 1981;78:4892–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr Top Microbiol Immunol 1989;148:115–32. [DOI] [PubMed] [Google Scholar]
- 16.Urnovitz HB, Murphy WH. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev 1996;9:72–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson DA, Mager DL, Leong JAC. Endogenous human retroviruses. In: Levy JA, ed. The retroviridae, Vol. 3. New York: Plenum Press, 1994:465–535.
- 18.Franklin GC, Chretien S, Hanson IM, et al. Expression of human sequences related to those of mouse mammary tumor virus. J Virol 1988;62:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson ML, Lindeskog M, Medstrand P, et al. Diversity of human endogenous retrovirus class II-like sequences. J Gen Virol 1999;80:255–60. [DOI] [PubMed] [Google Scholar]
- 20.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and B retrovirus: differential transcription in normal human tissues. J Virol 1993;67:6778–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ono M, Yasunaga T, Miyata T, et al. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol 1986;60:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dangel AW, Baker BJ, Mendoza AR, et al. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K(C4) are a molecular clock of evolution. Immunogenetics 1995;42:41–52. [DOI] [PubMed] [Google Scholar]
- 23.Tonjes RR, Lower R, Boller K, et al. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol 1996;13:S261–7. [DOI] [PubMed] [Google Scholar]
- 24.Barbulescu M, Turner G, Seaman MI, et al. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol 1999;9:861–8. [DOI] [PubMed] [Google Scholar]
- 25.Perl A, Rosenblatt JD, Chen ISY, et al. Detection and cloning of new HTLV-related endogenous sequences in man. Nucleic Acids Res 1989;17:6841–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson A, Svensson A, Rolny C, et al. Expression of human endogenous retrovirus ERV3 (HERV-R) mRNA in normal and neoplastic tissues. Int J Oncol 1998;12:309–13. [DOI] [PubMed] [Google Scholar]
- 27.Sauter M, Schommer S, Kremmer E, et al. Human endogenous retrovirus K10: expression of gag protein and detection of antibodies in patients with seminomas. J Virol 1995;69:414–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Löwer R, Löwer J, Tondera-Koch C, et al. A general method for identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology 1993;192:501–11. [DOI] [PubMed] [Google Scholar]
- 29.Schulte AM, Shoupeng L, Kurtz A, et al. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc Natl Acad Sci U S A 1996;93:14759–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bera TK, Tsukamoto T, Panda DK, et al. Defective retrovirus insertion activates c-Ha-ras proto-oncogene in an MNU-induced rat mammary carcinoma. Biochem Biophys Res Commun 1998;248:835–40. [DOI] [PubMed] [Google Scholar]
- 31.Sauter M, Roemer K, Best B, et al. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumours. Cancer Res 1996;56:4362–5. [PubMed] [Google Scholar]
- 32.Larsson E, Andersson G. Beneficial role of human endogenous retroviruses: facts and hypotheses. Scand J Immunol 1998;48:329–38. [DOI] [PubMed] [Google Scholar]
- 33.Lindeskog M, Medstrand P, Blomberg J. Sequence variation of human endogenous retrovirus ERV9-related elements in an env region corresponding to an immunosuppressive peptide: transcription in normal and neoplastic cells. J Virol 1993;67:1122–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin U, Türeci Ö, Pfreundschuh M. Serological identification of human tumor antigens. Curr Opin Immunol 1997:9:709–16. [DOI] [PubMed] [Google Scholar]
- 35.Keydar I, Ohno T, Nayak R, et al. Properties of retrovirus-like particles produced by a human breast carcinoma cell line: immunological relationship with mouse mammary tumour virus proteins. Proc Natl Acad Sci U S A 1984;81:4188–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patience C, Simpson GR, Colletta AA, et al. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol 1996;70:2654–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol 1987;61:2059–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurtz A, Schulte AM, Wellstein A. Pleiotrophin and midkine in normal development and tumour biology. Crit Rev Oncog 1995;6:151–7. [PubMed] [Google Scholar]
- 39.Rhim JS, Yang JH, Lee IH, et al. Oncogene and transgenics correlates of cancer and risk assessments. In: Zarvos C, ed. Oncogenes and human cancers. New York: Plenum Press, 1992:91–109.
- 40.Sutkowski N, Conrad B, Thorley-Lawson DA, et al. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity 2001;15:579–89. [DOI] [PubMed] [Google Scholar]
- 41.Garry RF, Fermin CD, Darenn JH, et al. Detection of a human intracisternal particle antigenically related to HIV. Science 1990;250:1127–9. [DOI] [PubMed] [Google Scholar]
- 42.Talal N, Dauphinee MJ, Dany M, et al. Detection of serum antibodies to retroviral proteins in patients with primary Sjören’s syndrome (autoimmune exocrinopathy). Arthritis Rheum 1990;33:774–81. [DOI] [PubMed] [Google Scholar]
- 43.Perl A, Colombo E, Agarwal R, et al. Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndrome: correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum 1995;38:1660–71. [DOI] [PubMed] [Google Scholar]
- 44.Ziegler B, Gay RE, Huang G, et al. Immunohistochemical localization of HTLV-I p19- and p24-related antigens in synovial joints of patients with rheumatoid arthritis. Am J Pathol 1989;135:1–5. [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson PN, Lever AML, Bruckner FE, et al. Polymerase chain reaction fails to incriminate exogenous retroviruses HTLV-I and HIV-1 in rheumatological diseases although a minority of sera cross-react with retroviral antigens. Ann Rheum Dis 1994;53:749–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Giovine F, Bailly S, Bootman J, et al. Absence of lentiviral and human T cell leukaemia viral sequences in patients with rheumatoid arthritis. Arthritis Rheum 1994;37:349–58. [DOI] [PubMed] [Google Scholar]
- 47.Herrmann M, Kalden JR. PCR and reverse dot hybridisation for the detection of endogenous retroviral transcripts. J Virol Methods 1994;46:333–48. [DOI] [PubMed] [Google Scholar]
- 48.Herrman M, Neidhart M, Gay S, et al. Retrovirus-associated rheumatic syndromes. Curr Opin Rheumatol 1998;10:347–54. [DOI] [PubMed] [Google Scholar]
- 49.Garson JA, Tuke PW, Giraud P, et al. Detection of virion-associated MSRV-RNA in serum of patients with multiple sclerosis. Lancet 1998;351:33. [DOI] [PubMed] [Google Scholar]
- 50.Christensen T, Sorensen P, Dissing P, et al. Expression of sequence variants of endogenous retrovirus RGH in particle form in multiple sclerosis. Lancet 1998;352:1033. [DOI] [PubMed] [Google Scholar]
- 51.Blond J, Beseme F, Duret L, et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol 1999;73:1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HS, Takenako O, Crow TJ. Isolation and phylogeny of endogenous retrovirus sequences belonging to the HERV-W family primates. J Gen Virol 1999;80:2613–19. [DOI] [PubMed] [Google Scholar]
- 53.Conrad B, Weissmahr RN, Boni J, et al. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell 1997;90:303–13. [DOI] [PubMed] [Google Scholar]
- 54.Katsumata K, Ikeda H, Sato M, et al. Cytokine regulation of env gene expression of human endogenous retrovirus-R in human vascular endothelial cells. Clin Immunol 1999;93:75–80. [DOI] [PubMed] [Google Scholar]
- 55.Murphy VJ, Harrison LC, Rudert WA, et al. Retroviral superantigens and type 1 diabetes mellitus. Cell 1998;95:9–11. [DOI] [PubMed] [Google Scholar]
- 56.Lower R, Tonjes RR, Boller K, et al. Development of insulin-dependent diabetes mellitus does not depend on specific expression of the human endogenous retrovirus HERV-K. Cell 1998;95:11–13. [DOI] [PubMed] [Google Scholar]
- 57.Nakagawa K, Harrison LC. The potential roles of endogenous retroviruses in autoimmunity. Immunol Rev 1996;152:194–236. [DOI] [PubMed] [Google Scholar]
- 58.Krieg AM, Gourley MF, Perl A. Endogenous retroviruses: potential etiologic agents in autoimmunity. FASEB J 1992;6:2537–44. [DOI] [PubMed] [Google Scholar]
- 59.Wallace BM, Lasker JS. UV light and HIV gene activation. Science 1992;257:1211–12. [DOI] [PubMed] [Google Scholar]
- 60.Krieg AM, Steinberg AD, Khan AS. Increased expression of novel full-length endogenous MCF-related transcript in autoimmune mouse strains. Virology 1988;162:274–6. [DOI] [PubMed] [Google Scholar]
- 61.Wu J, Zhou T, He J, Mountz JD. Autoimmune disease in mice due to integration of an endogenous retrovirus in an apoptosis gene. J Exp Med 1993;178:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Medstrand P, Lindeskog M, Blomberg J. Expression of human endogenous retroviral sequences in peripheral blood mononuclear cells of healthy individuals. J Gen Virol 1992;73:2463–6. [DOI] [PubMed] [Google Scholar]
- 63.Krieg AM, Gourley MF, Klinman DM, et al. Heterogenous expression and coordinate regulation of endogenous retroviral sequences in human peripheral blood mononuclear cells. AIDS Res Hum Retroviruses 1992;8:1991–8. [DOI] [PubMed] [Google Scholar]
- 64.Steinhuber S, Brack M, Hunsmann G, et al. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet 1995;96:188–92. [DOI] [PubMed] [Google Scholar]
- 65.Boyd MT, Bax CMR, Bax BE, et al. The human endogenous retrovirus ERV-3 is upregulated in differentiating placental trophoblast cells. Virology 1993;196:905–9. [DOI] [PubMed] [Google Scholar]
- 66.Venables PJW, Brookes SM, Griffith D, et al. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology 1995;211:589–92. [DOI] [PubMed] [Google Scholar]
- 67.Sverdlov ED. Retroviruses and primate evolution. Bioessays 2000;22:161–71. [DOI] [PubMed] [Google Scholar]
- 68.Mi S, Lee X, Li X-P, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000;403:785–9. [DOI] [PubMed] [Google Scholar]
- 69.Rowland-Jones S, Sutton J, Ariyoshi K, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med 1995;1:59–64. [DOI] [PubMed] [Google Scholar]
- 70.Rowland-Jones SL, Dong T, Fowke KR, et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Invest 1998;102:1758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roden D, Ejtehadi HD, Rowland-Jones S, et al. Use of bioinformatics to highlight antigenic regions of exogenous and endogenous retroviruses. Immunology 2001;104:S112–13. [Google Scholar]
- 72.Kaul R, Rowland-Jones SL, Kimani J, et al. Later seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J Clin Invest 2001;107:341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kambhu S, Falldorf P, Lee JS. Endogenous retroviral long terminal repeats within the HLA-DQ locus. Proc Natl Acad Sci U S A 1990;87:4927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badenhoop K, Tonjes RR, Rau H, et al. Endogenous retroviral long terminal repeats of the HLA-DQ region are associated with susceptibility to insulin-dependent diabetes mellitus. Hum Immunol 1996;50:103–10. [DOI] [PubMed] [Google Scholar]
- 75.Seidl C, Donner H, Petershofen E, et al. An endogenous retroviral long terminal repeat at the HLA-DQB1 gene locus confers susceptibility to rheumatoid arthritis. Hum Immunol 1999;60:63–8. [DOI] [PubMed] [Google Scholar]
- 76.Rasmussen HB, Geny C, Deforges L, et al. Expression of endogenous retrovirus in blood mononuclear cells in brain tissue from multiple sclerosis patients. Acta Neurol Scand 1995;169:38–44. [DOI] [PubMed] [Google Scholar]
- 77.Kapitonov VV, Jurka T. The long terminal repeat of an endogenous retrovirus induces alternative splicing and encodes an additional carboxy-terminal sequence in the human leptin receptor. J Mol Evol 1999;48:248–51. [DOI] [PubMed] [Google Scholar]
- 78.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Biol Cell 2001;21:1973–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kazazian HH. Mobile elements and disease. Curr Opin Genet Dev 1998;8;343–50. [DOI] [PubMed] [Google Scholar]
- 80.Muhlemann O, Mock-Casagrande CS, Wang J, et al. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol Cell 2001;8:33–43. [DOI] [PubMed] [Google Scholar]
- 81.Gozukara EM, Kahn SG, Metin A, et al. A stop codon in xeroderma pigmentosum group C families in Turkey and Italy: molecular genetic evidence for a common ancestor. J Invest Dermatol 2001;117:197–204. [DOI] [PubMed] [Google Scholar]
- 82.Bittorf T, Busfield SJ, Klinken SP, et al. Truncated erythropoietin receptor in a murine erythroleukemia line. Int J Biochem Cell Biol 1996:28:175–81. [DOI] [PubMed] [Google Scholar]
- 83.Howard M, Frizzell RA, Bedwell DM. Aminoglycoside antibiotics restore CFTR function by overcoming premature stop mutations. Nat Med 1996;2:467–9. [DOI] [PubMed] [Google Scholar]
- 84.Woodland DL. Immunity and retroviral superantigens in humans. Trends Immunol 2002:23:57–8. [DOI] [PubMed] [Google Scholar]
- 85.Nelson PN. Fossil renegades. New Sci 1999;161:51. [Google Scholar]
- 86.Towler EM, Gulnik SV, Bhat TN, et al. Functional characterization of the protease of human endogenous retrovirus K10: can it complement HIV-1 protease? Biochemistry 1998;37:17137–44. [DOI] [PubMed] [Google Scholar]