Abstract
Background: The application of lymphoscintigraphy followed by sentinel lymph node (SN) biopsy to patients with primary melanoma has revolutionised the ability to identify accurately, yet conservatively, those patients who harbour occult nodal metastases. The molecular detection of SN micrometastases facilitates the cost effective analysis of the entire SN using multiple markers. Currently, a lack of marker specificity is the main barrier preventing the molecular evaluation of SN tissue from becoming clinically applicable.
Aims: To develop a reproducible multimarker reverse transcription-polymerase chain reaction (RT-PCR) assay, with the emphasis on achieving high specificity for the accurate detection of melanoma metastases in nodal tissue.
Methods: Three pigment cell specific (PCS) markers—tyrosinase, Pmel-17, and MART-1—and one cancer testis antigen (CTA)—MAGE-3—were selected for use in a multimarker RT-PCR assay. The conditions for this assay were optimised.
Results: High specificity was achievable for each marker by optimising the PCR cycle number such that unwanted transcripts (that is, illegitimate transcripts and/or specific transcripts from other low abundance nodal cell types) remained undetectable in appropriate controls (normal visceral nodes). Tyrosinase was 100% specific at 40 PCR cycles, MAGE-3 and MART-1 at 35 PCR cycles, and Pmel-17 at 30 PCR cycles. Tyrosinase proved to be the most sensitive marker, detecting 10 melanoma cells in 0.1 g of nodal tissue.
Conclusions: Excellent reproducibility of the entire nodal processing and RT-PCR protocol for the detection of very low numbers of melanoma cells in nodal tissue was shown, although there is a risk of false positives using the PCS markers alone, because of an approximate 4–8.5% incidence rate of nodal nevi in melanoma draining SNs (these nevi being absent in all other normal nodes). MAGE-3 was shown to be the only marker that is not expressed by melanocytes. However, because not all melanomas express MAGE-3, it is recommended that more emphasis should be placed on the development of a panel of CTA markers to ensure a zero false positive rate and to provide optimum detection.
Keywords: melanoma, prognosis, reverse transcription-polymerase chain reaction, sentinel lymph node, specificity, metastasis
Despite the rapidly increasing worldwide incidence of cutaneous melanoma,1 the likelihood of death from this malignancy has been declining over the past few years. This dramatic improvement in the survival rate of patients with melanoma has mainly been the result of early diagnosis and surgical intervention in primary disease.2 Nevertheless, a subgroup of patients who undergo so called curative primary tumour surgery will still develop metastases, implying that these patients had already developed subclinical/occult/micrometastases at the time of primary tumour excision. In most of these patients, the first evidence of recurrence will be in the regional nodal basin; that is, the first lymph node basin draining the primary tumour.3 Once nodal involvement has become clinically evident, the five year survival rate decreases from 75–98% (for stage I and II patients) to 20–40%.4, 5 Of these patients, 70–100% will harbour distant and widespread micrometastases,6 and there is little hope for curative surgery. This significant decrease in the survival rate associated with metastases to the regional nodes emphasises the need to identify those patients who at primary tumour presentation already have occult nodal metastases. A more accurate subclinical nodal staging would allow earlier and potentially more effective patient management for the following reasons: (1) patients with confirmed micrometastases may represent the most appropriate group to study potentially effective immunotherapies because large tumour burdens have been shown to inhibit antitumour immunity7; (2) those patients who do not have micrometastases would be spared the morbidity and costs involved in surgical removal of the regional lymph nodes and the toxicity and costs of radiation, chemotherapy, and immunotherapy. Therefore, it is crucial that we can identify those patients with cancer who have progressive disease and to confirm metastasis of the primary tumour—this strategy being the foundation of sound cancer management principles. Thus, the challenge is to develop prognostic markers and techniques that will allow the identification of those patients who, at the time of primary tumour diagnosis, already have micrometastases.
“The significant decrease in the survival rate associated with metastases to the regional nodes emphasises the need to identify those patients who at primary tumour presentation already have occult nodal metastases”
Until recently, the detection of progressive melanoma disease has involved regular clinical and radiological follow up examinations, even though these are of limited sensitivity. Several serological markers (such as lactate dehydrogenase, melanoma inhibiting activity (MIA), and S100) have been evaluated for there ability to predict melanoma progression,8 but none has been shown to have value in predicting metastasis of the primary tumour. Recently, we have shown that a reduction in the active fraction of plasma plasminogen activator inhibitor type 1, the main regulator of fibrinolytic activity in blood, could possibly indicate increased risk for metastatic melanoma.9 Initially, the detection of circulating melanoma cells (CMCs) by sensitive single or multimarker reverse transcription-polymerase chain reaction (RT-PCR) seemed promising, but more recent studies pointed to its limited use.10 It is probable that the detection of CMCs fails as a prognostic technique mainly because of sampling error.11
It is now evident that the analysis of anatomical compartments other than blood, such as bone marrow12 and lymph nodes,5, 12–14 might be better tissue targets to confirm metastasis. The sentinel lymph node (SN), the first node of the lymphatic basin that drains a primary tumour, can be identified with 98% accuracy using radioguided surgery.15 The histopathological status of the SN accurately reflects that of the remaining nodes in the lymphatic bed.16 Considering this, and the fact that the regional lymph node status (defined as the number and extent of nodal involvement) is currently the most powerful prognostic factor for predicting survival,4 the SN has become an attractive target to confirm lymphatic metastasis of the primary tumour at an early stage. It has now clearly been shown that the status of the SN impacts greatly on prognosis: patients who are SN positive by histopathological evaluation have a significantly poorer prognosis than those found to be metastasis free (histologically), even though the former undergo a potentially curative selective completion lymph node dissection.13, 17–20 An average five year survival rate of 82% for SN negative patients versus 50% for SN positive patients has been reported.21 Thus, with the advantages of SN histopathological evaluation now being so clear, the World Health Organisation has recently issued a consensus statement recommending that sentinel lymphadenectomy should become the new standard of care for patients with primary melanoma.22 Both the American Joint Committee on Cancer (AJCC) and the International Union Against Cancer have endorsed this decision.21 The revised melanoma staging system, which now incorporates the SN status, will become official with publication of the AJCC Cancer Staging Manual of 2002.
However, histopathological evaluation of the SN has been shown to underestimate the presence of clinically relevant metastases,23 emphasising the need to develop techniques that are more sensitive and have a higher detection rate. The molecular detection of micrometastases in SNs is considerably more sensitive,5 less labour intensive, and more cost effective24 than current standard pathological evaluation. However, a lack of specificity25, 26 and variable tumour detection rates27, 28 are valid criticisms preventing the molecular evaluation of SN tissue from becoming clinically applicable, although similar limitations apply to the pathological evaluation of SN tissue.
The primary aim of our study was to develop a reproducible RT-PCR assay, with the emphasis on achieving high specificity for the accurate detection of melanoma metastases in nodal tissue. To compensate for the heterogeneity of melanoma cells,27, 29–31 a multimarker approach was followed, with the further aim of improving the detection rate of the assay. Three pigment cell specific (PCS) markers were selected: tyrosinase (a key enzyme in the melanin biosynthetic pathway), Pmel-17 (which encodes the melanosomal matrix glycoprotein, gp100—the antigenic target for the HMB45 monoclonal antibody in standard melanoma pathology), and MART-1 (which encodes the melanosomal differentiation antigen, Melan-A). One cancer testis antigen (CTA) was included, namely MAGE-3, because it is expressed in neoplastic tissues and testes only.30 Particular attention has been placed on the improvement of specificity by excluding the detection of “unwanted” transcripts (illegitimate transcripts and/or specific transcripts from other low abundance nodal cell types), in addition to differentiating between melanocytic nevi and malignant melanoma transcripts in lymph nodes. The sensitivity of the optimised multimarker assay for the presence of melanoma cells in nodal tissue was assessed. We also describe a rapid, cost effective, and clinically applicable homogenisation method for RNA extraction from nodal tissues.
MATERIALS AND METHODS
Tissue culture
The melanoma cell lines UCT-Mel-1 to UCT-Mel-8 were established in the laboratory of clinical science and immunology (University of Cape Town) from patient tumours, as described previously.32 The cells were maintained in RPMI 1640 medium supplemented with 5% fetal calf serum, 50 IU/ml penicillin and 20 μg/ml streptomycin at 37°C under 5% CO2/95% air and 90% humidity. UCT-Mel-1 and UCT-Mel-2 were classified as pigmented and UCT-Mel-3 to UCT-Mel-8 as non-pigmented adherent cell lines, according to the visual presence of pigment. The UCT-Mel-1 cell line was used to optimise the RNA extraction and RT-PCR assay, and to assess the sensitivity of each marker. All eight melanoma cell lines were used to assess the in vitro detection rate of each marker.
Lymphatic tissues
Our study was approved by the ethics and research committee of the University of Cape Town. Informed and written consent was obtained from all patients for tumour involved nodes, and from the organ donor’s next of kin for normal lymph nodes. Four melanoma involved lymph nodes (each weighing more than 1.5 g) were obtained from four patients with stage III melanoma. These nodes were confirmed to contain malignant melanoma cells by standard haematoxylin and eosin (H&E) staining. Twenty normal lymph nodes (each weighing less than 0.2 g) were obtained from the visceral region from two organ donors. One skin biopsy was obtained from the forearm of a normal control subject.
Handling, homogenisation, and RNA extraction of nodal tissue
After surgery, lymph nodes were transported on ice to the laboratory. Extranodal fat was removed and the tissue cut into 0.1–0.15 g pieces. Each piece was placed in a sterile 2 ml polypropylene cryovial and snap frozen in liquid nitrogen. The maximum time to elapse between surgical removal of the node and the freezing of the tissue was two hours. On resuming the nodal processing, a sterile 8 mm stainless steel ball was added to the frozen tissue, followed by 1.5 ml TriPure RNA extraction reagent (Roche Diagnostics, Mannheim, Germany) for every 0.15 g of tissue. After securing the cryovial in an Express Homogeniser (international patent pending; http://www.tekniva.com), high speed oscillation was carried out for a total of 30 seconds. This consisted of six cycles of five seconds “on” followed by 10 seconds “off”. The remaining procedure for total RNA extraction was essentially as described by the manufacturer (a standard guanidinium thiocyanate/phenol/chloroform method). The purity and yield of the total RNA was measured spectrophotometrically at 260 nm and 280 nm. The RNA yield was relatively consistent, being approximately 200 μg total RNA for every 0.15 g of nodal tissue. The A260/A280 ratio was typically 1.7 for RNA purity.
RT-PCR
Total RNA (1 μg) was reverse transcribed into cDNA in a 20 μl reaction containing 1× PCR buffer (10mM Tris, pH 8.3 and 50mM KCl), 5mM MgCl2, 1 mM of each dNTP, 50 pmoles of Oligo(dT)20 primer, 20 IU of MuLV reverse transcriptase, and 10 IU of RNAse inhibitor. RNA samples were thawed once only, preheated for five minutes at 90°C, and snap cooled on ice before being added to the RT mix, which was then overlaid with mineral oil. Complimentary DNA was synthesised for 30 minutes at 42°C, followed by five minutes at 95°C to stop the reaction. The cDNA was stored at −20°C or directly used for PCR. Aliquots of cDNA (5 μl) were transferred to 20 μl PCR reactions containing 1× PCR buffer, 1.5mM MgCl2, 25 pmoles of each forward and reverse primer, and 2 IU AmpliTaq GoldTM DNA polymerase. All reagents were from Perkin Elmer Biosystems (Foster City, California, USA), except when otherwise indicated. Additional dNTPs were not added to the PCR mix because the required amount was transferred from the reverse transcription reaction, providing a final dNTP concentration of 200 μM. Each 25 μl PCR sample was overlaid with mineral oil and then amplified on a Robocycler (Stratagene, La Jolla, California, USA) as follows: the first PCR cycle consisted of denaturation at 96°C for 10 minutes to activate the DNA polymerase, annealing at 65°C for 2.5 minutes, and extension at 72°C for two minutes. This was followed directly by 29–49 PCR cycles (depending on the molecular marker and the experimental purpose) at 96°C for 30 seconds, 65°C for 2.5 minutes, and 72°C for two minutes. PCR products were separated electrophoretically on a 2% agarose minigel, containing 0.5 μg/ml ethidium bromide, and photographed under ultraviolet light. The node preparation, RNA extraction, and RT-PCR reactions were all carried out in a separate room from that where gel electrophoresis was performed. Opening of tubes containing PCR products and gel electrophoresis were always performed under negative atmospheric pressure to prevent subsequent amplicon contamination of nodal/RNA samples or RT-PCR reagents.
Oligonucleotide primers
The primer sequences for tyrosinase, MART-1, porphobilinogen deaminase (PBGD),33 Pmel-17,29 and MAGE-334 were synthesised and purified at the department of cellular and molecular biology, University of Cape Town, South Africa. Table 1 ▶ lists the primer sequences and the product size generated for each primer pair. The primer pairs for each of the markers are positioned in different exons and are designed, where possible, to lie on either side of a fairly large intron (> 1 kb) to exclude the amplification of genomic DNA that may be present in the RNA sample. cDNA specificity controls were performed for each primer set to exclude the possibility of genomic DNA amplification, which might cause false positives or non-specific bands. This was done by omitting the reverse transcriptase from the reverse transcription step, followed by 40 PCR cycles for each marker.
Table 1.
Primers used in the reverse transcription-polymerase chain reaction protocol designed to detect melanoma metastases and their product size
| cDNA target | Primer | Sequence from 5′ to 3′ end | Nucleotide position (exon number) | Product size (bp) | Ref |
| Tyrosinase | >Htyr3 | GTC TTT ATG CAA TGG AAC GC | 864–883 (2) | 207 | 33 |
| <Htyr2 | AGG CAT TGT GCA TGC TGC TT | 1083–1102 (3) | |||
| MART-1 | >MART-1 | CAC TCT TAC ACC ACG GCT GA | 215–234 (2) | 299 | 33 |
| <MART-1 | AGG TGA ATA AGG TGG TGG TGA | 494–514 (5) | |||
| Pmel-17 | >Pmel-17* | TGC TGG AGA GGT GGT CAA GTG | 189–209 (2/3) | 699 | 29 |
| <Pmel-17 | CTC CAG GTA AGT ATG AGT GAC | 837–857 (6) | |||
| MAGE-3 | >MAGE-3 | GAA GCC GGC CCA GGC TCG | 438–455 (1) | 413 | 34 |
| <MAGE-3 | GGA GTC CTC ATA GGA TTG GCT CC | 2703–2725 (3) | |||
| PBGD | >PBGD | CTG GTA ACG GCA ATG CGG CT | 32–51 (1) | 339 | 33 |
| <PBGD | GCA GAT GGC TCC GAT GGT GA | 350–369 (5) |
Nucleotide positions are based on the sequences with the following GenBank accession numbers: Y00819 for tyrosinase, U06654 for MART-1, M77348 for Pmel-17, U03735 for MAGE-3, and NM_000190 for PBGD. The exon information for tyrosinase is based on GenBank accession numbers AF237808 and AF237809; for MART-1 on U06654; for Pmel-17 on U31797, U31798, U31799, U31807, and U31808; for MAGE-3 on U03735; and for PBGD on M18799, M18800, D12722, X68018, and X04217.
*Unpublished forward primer for Pmel-17, designed by Hanekom and co-workers using DNAMAN 5.1.0.0 (Lynnon Biosoft).
PBGD, porphobilinogen deaminase.
Negative and positive PCR controls
A negative control (water instead of RNA template) for each marker was included with each batch of samples to exclude the possibility of false positive results that might result from the contamination of RT-PCR reagents with DNA amplicons, these being generated from previous PCR reactions. A positive control (RNA from a normal lymph node containing 100 UCT-Mel-1 melanoma cells) for each marker was included with each batch of samples to verify the efficiency of the RT-PCR assay. The integrity of each RNA sample, in addition to the RT efficiency, was verified by RT-PCR for the housekeeping gene PBGD. PBGD has been reported to be expressed in lower amounts for each cell than the commonly used housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).33
Marker specificity
Each marker was initially tested on 10 normal lymph nodes using 50, 40, and 30 PCR cycles. The PCR cycle number at which no false positives were generated was chosen for each marker. If false positives were detected at 40 PCR cycles, the nodes were re-evaluated at 35 cycles (as opposed to 30 cycles), so that the sensitivity was not compromised. A further 10 normal lymph nodes were evaluated to confirm the specificity of each marker at the chosen PCR cycle number.
Marker sensitivity
The sensitivity assessment was based on the degree of expression of the four markers in the UCT-Mel-1 cell line. Serial dilutions of UCT-Mel-1 cells were prepared in a microtitre plate to provide 104, 103, 102, and 101 cells in each well. The cells were allowed to settle for 30 minutes at 37°C. The lowest dilution (10 cells/well) was plated in quadruplet to allow the accurate selection of a well containing 10 viable cells, as verified by phase contrast microscopy. Concurrently, normal lymph node tissue (the equivalent of 0.4 g in total) was homogenised according to the previously described protocol. The homogenates were pooled and four 1 ml aliquots were prepared. The growth medium of each microtitre well (containing the four serial dilutions of the UCT-Mel-1 cells) was carefully aspirated. The cells were then lysed with 200 μl of the respective nodal homogenate and pooled with the remaining 800 μl of homogenate. Each well was rinsed with 200 μl of fresh TriPure reagent and transferred back to the respective microcentrifuge tube. The RNA extractions were completed as described previously. This resulted in RNA from a known number of melanoma cells being present in background RNA corresponding to 0.1 g of normal lymph node tissue. Each sample was analysed by RT-PCR as described above by using the optimised cycle number for each marker (as for every experiment thereafter).
Marker detection rate
The in vitro and in vivo detection rates for melanoma cells were assessed for each marker by evaluating eight melanoma cell lines and four melanoma involved lymph nodes, respectively. This was done using multiplex PCR for MART-1 and MAGE-3 at 35 PCR cycles, multiplex PCR for Pmel-17 and PBGD at 30 cycles, and single marker PCR for tyrosinase at 40 cycles.
Expression profile of melanoma cells versus melanocytic nevi
To determine whether any of the markers would differentiate between melanocytic nevi (reported to be present in some skin draining lymph nodes) and melanoma cells, the expression of the markers in a melanoma involved node and skin melanocytes was compared. Skin melanocytes were used because skin draining lymph nodes were not available.
RESULTS
Marker specificity
To ensure 100% specificity of the markers for the presence of melanoma cells in nodal tissue, it was necessary to optimise the assay such that unwanted transcripts (that is, illegitimate transcripts and/or specific transcripts from other low abundance nodal cell types) remain undetectable. Very low amounts of gene expression, termed illegitimate transcription, have been described for many genes in most tissues and are detected usually after more than 40 PCR cycles.35 In addition, low numbers of Schwann cells (known to express tyrosinase) are present in all normal lymph nodes13 and could also account for false positives over and above illegitimate transcripts. Therefore, if high assay sensitivity is required, as was the case in our study, it is vital to determine (and then not to exceed) the “cut off” PCR cycle number for each marker, above which unwanted transcripts are detectable in the control tissues. By performing decreasing PCR cycles on the cDNA derived from 10 normal lymph nodes as described earlier, the optimal cycle number (to the closest denominator of five or 10) was determined for each marker. The specificity was then confirmed by evaluating another 10 normal nodes.
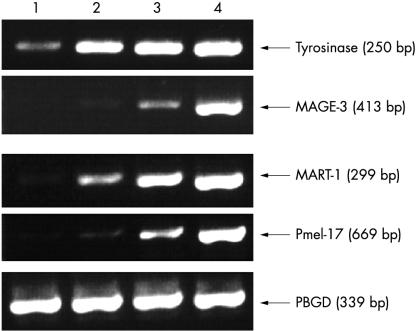
Tyrosinase was found to be 100% specific at 40 PCR cycles, MAGE-3 and MART-1 at 35 PCR cycles, and Pmel-17 at 30 PCR cycles (fig 1 ▶). Of note is that the rate of detection of unwanted transcripts in the nodal tissues did not increase at the same rate with increasing cycle numbers for each marker. In particular, although Pmel-17 was highly specific for melanoma cells at 30 PCR cycles, unwanted transcripts were detectable in all 10 normal nodes at 35 cycles. In contrast, tyrosinase was highly specific for melanoma cells at 40 PCR cycles and unwanted transcripts were detectable in only one of 10 normal nodes at 50 cycles. This last point highlights the need to evaluate sufficient numbers of control nodes to determine the cut off PCR cycle number for each marker accurately. The PBGD RT-PCR product was consistently detectable in the 20 normal nodes at 30 PCR cycles, but significantly weaker compared with 35 cycles (fig 1 ▶). Therefore, 30 PCR cycles was chosen as the optimal cycle number for this internal control because this would provide a much more sensitive assessment of RNA integrity than higher cycle numbers.
Figure 1.
Optimisation of the PCR cycle number for each marker. Lanes 1–10, 10 normal (donor) lymph nodes. Lanes +C, positive controls (UCT-Mel-1) for each marker. The optimal PCR cycle number for tyrosinase, MAGE-3, MART-1, Pmel-17, and porphobilinogen deaminase (PBGD) was 40, 35, 35, 30, and 30, respectively. Using higher PCR cycle numbers than these for each marker resulted in the detection of unwanted transcripts. The PCR product sizes for tyrosinase, MAGE-3, MART-1, Pmel-17, and PBGD are 250, 413, 299, 669, and 339 bp, respectively. The results are representative of two independent experiments.
Marker sensitivity
To assess the sensitivity of each marker for the detection of melanoma cells in lymph node tissues, decreasing numbers of UCT-Mel-1 cells were spiked into 0.1 g of normal lymph node tissue as described earlier. Tyrosinase proved the most sensitive by allowing the detection of as few as 10 melanoma cells in 0.1 g of normal nodal tissue (fig 2 ▶). MART-1 was almost as sensitive (100 melanoma cells in 0.1 g of normal nodal tissue), with MAGE-3 and Pmel-17 confidently detecting 1000 melanoma cells in 0.1 g of normal nodal tissue (although the 100 melanoma cell dilution was also weakly detectable for both markers). Overall, excellent sensitivities were achieved with all the markers, even with the reduced PCR cycle numbers needed to achieve 100% specificity. However, less sensitive markers, such as Pmel-17, are more likely to produce false negative results when evaluating SNs for micrometastases.
Figure 2.
Assessing the sensitivity of each marker by using normal nodal tissue spiked with decreasing numbers of UCT-Mel-1 cells. Lanes 1 to 4: 101, 102, 103, and 104 melanoma cells spiked into 0.1 g of normal nodal tissue, respectively. The results are representative of two independent experiments. PBGD, porphobilinogen deaminase.
To minimise the cost of the assay, we decided to investigate whether markers with the same optimal PCR cycle number (MART-1 and MAGE-3 at 35 cycles, Pmel-17 and PBGD at 30 cycles) could be combined in the same reaction (that is, multiplexed). This was facilitated by the fact that the relevant primer combinations had the same optimal annealing temperatures (65°C) and their product sizes were different enough to allow easy distinction by agarose gel electrophoresis. However, it was first necessary to assess whether multiplexing of primer sets would affect PCR sensitivity and specificity. No significant reduction in the sensitivities of each marker was observed with multiplexing and no non-specific bands were observed (results not shown).
Reproducibility
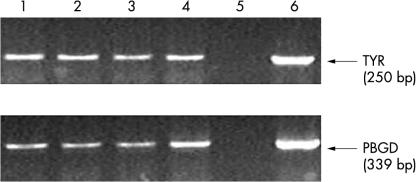
To obtain an accurate assessment of the reproducibility of the nodal processing protocol and the RT-PCR assay, quadruplet normal nodal tissue samples (each 0.1 g) were spiked with 10 UCT-Mel-1 cells. This was done essentially as described earlier, except that the melanoma cell lysate was also subjected to the nodal homogenisation process. Excellent reproducibility of the entire protocol for the detection of very low numbers of melanoma cells in nodal tissue was shown (fig 3 ▶).
Figure 3.
Reproducibility of the entire protocol used in our study. Lanes 1–4, tyrosinase and porphobilinogen deaminase (PBGD) yields from four independent nodal tissue samples each spiked with 10 UCT-Mel-1 cells and subjected to the entire protocol (nodal processing and reverse transcription-polymerase chain reaction (RT-PCR)). Lane 5, negative controls (H2O) for tyrosinase and PBGD. Lane 6, positive controls for tyrosinase and PBGD (UCT-Mel-1 RNA as RT template). The results are representative of two independent RT-PCR assays for each spiked node.
Marker detection rate
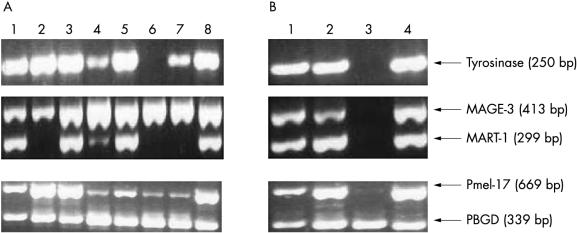
Heterogeneity of marker expression occurs among tumour cells, particularly at increasing stages of tumour progression.34, 36, 37 Therefore, another objective of our study was to determine whether a multimarker assay as opposed to a single marker assay would improve the detection rate. Therefore, the in vitro and in vivo expression profile of each marker (at the optimum PCR cycle number) was determined by evaluating eight melanoma cell lines and four melanoma involved lymph nodes (all enlarged), respectively (fig 4A, B ▶). It has been shown that melanoma cell lines, like melanocytes, tend to lack marker heterogeneity.37 In accordance with this, MAGE-3 and Pmel-17 both showed 100% detection rates for the melanoma cell lines. Only MART-1, and to a lesser degree tyrosinase, showed lower detection rates among the eight cell lines. Of note is that there was no correlation between the expression of the PCS markers (tyrosinase, MART-1, and Pmel-17) and the presence or absence of visible pigment in the cultured cell. Thus, a single marker assay with either MAGE-3 or Pmel-17 would have provided as high an in vitro detection rate as the multimarker assay. Although the number of melanoma involved nodes (all confirmed positive by H&E examination) available for our study was limited, it was surprising that the lymph nodes also showed lack of heterogeneity in marker expression, with nodes 1, 2, and 4 expressing all four markers (fig 4B ▶). However, node number 3 was not detectable with the markers (except for a very faint Pmel-17 band) and was one of two lymph nodes that were involved with unpigmented melanoma cells. Nodes number 1 and 2 contained metastatic deposits with at least focal areas of pigmentation/hyperchromasia. Thus, in our study the use of multiple markers did not result in a better in vivo detection rate than a single marker assay.
Figure 4.
Assessing the in vitro and in vivo detection rates for each marker. (A) Lanes 1–8, UCT-Mel-1 to UCT-Mel-8 cell lines, respectively. (B) Lanes 1–4, melanoma involved lymph nodes numbers 1 to 4, respectively. MART-1 and MAGE-3, in addition to Pmel-17 and porphobilinogen deaminase (PBGD) were multiplexed. The results are representative of two independent experiments.
Expression profile of melanoma cells versus melanocytic nevi
Because nodal nevi are probably derived from dermal/epidermal melanocytes, we used pigmented skin to compare the expression of the four molecular markers in melanoma involved lymph nodes with those in skin melanocytes (with skin melanocytes being representative of nodal nevi). As expected, MAGE-3 was found to be the only marker that is not expressed by melanocytes (fig 5 ▶, lane B). Thus, the three coexpressed PCS markers might contribute to false positives when analysing melanoma draining lymph nodes (for example, the SN).
Figure 5.
Differentiating between melanoma cells and melanocytic nevi. Lanes A and B, melanoma involved node and pigmented skin, respectively. The results are representative of two independent experiments. PBGD, porphobilinogen deaminase.
DISCUSSION
The early detection of occult metastases remains an immense challenge to oncologists. The improved detection of micrometastases holds the key to many basic questions on the initiation of metastasis and, importantly, would open the doorway for the testing, and ultimately, the application of tumour immunotherapy. The molecular detection of SN micrometastases offers a more sensitive and cost effective approach to the analysis of the entire SN compared with the standard histopathological examination of the SN. However, the lack of marker specificity currently represents a major stumbling block to progress in this field. Our study focuses on the improvement of specificity by excluding the detection of unwanted transcripts and differentiating between melanocytic nevi and malignant melanoma transcripts in lymph nodes.
“The presence of unwanted transcripts in the tissue being analysed poses the most serious technical barrier to the elimination of false positives”
We have considered several ways in which false positive results can arise during the analysis of SNs. Given that a small amount of genomic DNA will undoubtedly be present in every RNA extract, primer pairs for each marker should be designed such that they are specific for the cDNA template. Thereafter, a cDNA negative control should be performed at least once (at the outset of assay optimisation) for each marker. The cDNA specificity of each primer set was confirmed by the absence of non-specific bands on agarose gels (results not shown). Importantly, this cDNA negative control will also exclude the presence of pseudogenes. If present, they would be coamplified by the primers that were originally designed to amplify cDNA only, producing a PCR product indistinguishable from that derived from the cDNA template.38 To complicate the matter further, because pseudogenes for GAPDH and β actin exist, a false impression of good RNA quality could be obtained if these are used as internal controls.38 Because we have found no evidence for a PBGD pseudogene, and given that it is expressed at lower amounts in each cell than the other commonly used housekeeping genes, we strongly advise its use as an internal control.
The presence of unwanted transcripts in the tissue being analysed poses the most serious technical barrier to the elimination of false positives. Optimising the PCR cycle number, by making use of appropriate control tissues, is the key to overcoming this barrier. In our study, visceral nodes were used to achieve marker specificity. These nodes are known to be free of nodal nevi (histologically), and therefore it is reasonable to assume that the false positives detectable at high PCR cycle numbers for each of the PCS markers (and probably also for MAGE-3) are caused by illegitimate and/or Schwann cell transcripts, and not by the presence of nodal nevi. Because non-melanoma tumour involved nodes are easily available, they are often used as controls for assessing marker specificity. A few studies have shown that non-melanoma tumour involved nodes, in addition to non-neoplastic lymph nodes (for example, nodes with chronic unspecified lymphadenitis) do not have detectable amounts of PCS markers, even though most of these investigators used the more sensitive nested PCR approach.13, 27, 37, 39, 40 In contrast, others found detectable amounts of PCS markers in non-melanoma tumour involved lymph nodes using nested PCR.25, 41 To confirm the specificity of the PCS markers, 13 enlarged lymph nodes from patients with breast cancer were evaluated for PCS marker expression at the optimised PCR cycle numbers. Five of these 13 nodes were tyrosinase positive, four were MART-1 positive, and three were Pmel-17 positive (results not shown). Therefore, it can be concluded that the detection of PCS markers in the breast cancer nodes was the result of aberrant PCS marker expression by breast cancer cells. Consequently, if breast cancer involved nodes had been used in our study to assess PCS marker specificity, lower PCR cycle numbers would probably have been necessary to achieve specificity, and some of these markers would probably have been too non-specific and therefore inadequate. Thus, it is vital to use appropriate control tissues (that is, cancer free nodes) for the necessary optimisation of PCR cycle number. In addition, our study conclusively shows that nested PCR is superfluous, because unwanted marker transcripts become detectable in nodal tissues even with limited, single stage PCR cycle numbers.
Because of the presence of unwanted marker transcripts in normal nodes, the sensitivity of a marker should merely be a default of its specificity. Therefore, to achieve maximum specificity for the molecular analysis of nodal tissue (as for other target tissues), a certain degree of sensitivity will have to be forfeited. H&E staining and immunohistochemical analyses are typically capable of detecting one cancerous cell in a background of 104 and 105 normal cells, respectively.5 With a sensitivity of 10 melanoma cells for each 0.1 g of normal nodal tissue, as determined here for tyrosinase, it can be calculated that molecular analysis is 200 times more sensitive than H&E staining and 20 times more sensitive than standard immunohistochemistry. This estimation is based on the fact that there are about 2 × 108 human diploid fibroblasts (16–18 μm in diameter) in each gram of wet weight.42 This compares favourably with reported RT-PCR sensitivities of one cancerous cell in a background of 106–107 normal cells.5 It should be noted that the sensitivity assessment is based on the expression of these markers in the UCT-Mel-1 cell line. Furthermore, the expression of markers is often upregulated or downregulated in cultured cells,37 so that this in vitro sensitivity assessment is not necessarily representative of the in vivo situation. Therefore, it would be more appropriate to spike melanoma cells derived from several different melanoma involved lymph nodes into normal nodal tissue to gain a more accurate assessment of marker sensitivity. However, such an experiment would be technically challenging. Nevertheless, our approach is more representative than the sensitivity assessments of other investigators, who merely spiked decreasing numbers of melanoma cells into a fixed number of lymphocytes.25, 26, 43
The risk of producing a false negative RT-PCR result is greatly increased when there are low numbers of specific mRNA template molecules (for example, as for melanoma nodal micrometastases). Even minor variations in RNA extraction and RT-PCR efficiency, and/or minor degrees of RNA degradation may contribute to false negatives, because these variations usually go undetected by the conventional controls (for example, housekeeping gene). An additional problem is that the specific RNA aliquot evaluated may not always contain sufficient mRNA copies to produce a positive result (sampling error).33 Therefore, the accurate detection of micrometastases necessitates high assay reproducibility and appropriate controls. We have shown excellent reproducibility of the entire protocol for the detection of very low numbers of melanoma cells in nodal tissue. Furthermore, our novel homogenisation method (international patent pending) has been developed with the aim of improving reproducibility in RNA extraction. The technique breaks down tissue by providing high speed (50 Hz), single plain harmonic motion to the metal ball located in the tube. The diameter of the ball was carefully selected to give maximum disruption of tissue between the ball and sidewalls of the vial. By making use of disposables only, the risk of intersample contamination and RNA degradation is reduced. Importantly, the protocol is rapid and cost effective, which further facilitates implementation of this protocol for routine clinical purposes.
Because tumour marker heterogeneity could result in false negatives, it is important to develop a multimarker approach to improve the detection rate. The lack of marker expression in many amelanotic nodal metastases (and as seen in node 3, fig 4B ▶) suggests the need for a marker(s) that is/are capable of more accurately detecting amelanotic melanoma metastases.37 The CTA markers are good candidates because their expression is unrelated to the pigmentation pathway. Unfortunately, MAGE-3 was not detected in node 3, but there are other members of the MAGE-A family (such as MAGE-6 and MAGE-12), which have recently been shown to have better detection rates than MAGE-3.31 However, contradictory results regarding these detection rates have been published.30
Unfortunately, the in vivo expression profile of the markers, as assessed on macrometastases, does not permit a final conclusion because larger numbers of these nodes still need to be analysed. However, the use of such nodes can lead to a false impression of a high detection rate in nodes containing micrometastases. For example, markers with low sensitivity, such as Pmel-17, might appear to provide a high detection rate when analysing nodes containing macrometastases, but would actually have a low detection rate if nodes containing micrometastases were analysed. Its lower sensitivity and its high false positive rate at 35 PCR cycles, makes it unsuitable as a marker for the detection of SN micrometastases. In addition, it has been shown that the marker expression profile of nodal micrometastases differs from that of macrometastases, presumably because the micrometastases have been shown to be in a dormant state.44–47 Therefore, ideally multiple SNs containing micrometastatic deposits (confirmed by histopathology) should be used for a clinically accurate assessment of the in vivo detection rate.
Neval cells and Schwann cells, both present in lymph nodes, have been shown to have similar immunohistochemistry expression profiles to skin melanocytes.41, 48, 49 This is problematical for the specific molecular detection of melanoma cells in nodal tissue. Although the numbers of these cells within a lymph node are minimal, the extreme sensitivity of RT-PCR means that these “non-melanoma” transcripts could become detectable with PCR. Because the cellular origin of the specific transcripts cannot be confirmed as they can with H&E, false positives using RT-PCR might arise. Fortunately, by optimising the PCR cycle number on normal control lymph nodes, false positives caused by the presence of Schwann cells (present in all normal lymph nodes) can be eliminated.
Benign nodal neval clusters are reported to be present in 4–8.5% of melanoma draining SNs, whereas 1% of non-SNs within a melanoma draining regional lymph node basin contain nevi.49, 50 Only approximately 0.1% of nodes draining non-melanoma tumours (as evaluated mainly on breast cancer draining nodes) have been found to contain nevi. No nevi have been detected in non-skin draining nodes.51 The 8.5% incidence rate of nodal nevi in melanoma draining SNs (that have been shown to be free of metastases by histopathology) is of concern, because it could result in false positives in up to 8.5% of patients with melanoma undergoing SL. Unfortunately, it is not possible to determine this false positive RT-PCR risk by using normal nodes as controls (even skin draining nodes), because nodal nevi are present almost solely in melanoma draining SNs. Although most investigators mention the existence of nodal nevi as a possible cause of false positive results,27, 39 most studies nevertheless continue to make use of PCS markers for the specific detection of melanoma micrometastases. There are probably two reasons for this: (1) there is limited marker choice for the detection of melanoma cells, and (2) it is still not clear whether the SN neval load is high enough to be detectable by optimised RT-PCR for these markers.
“It is recommended that more emphasis should be placed on the development of a panel of cancer testis antigen markers, which will be useful for melanoma and other carcinomas”
In our study, normal visceral nodes were used as controls. Unfortunately these and other negative control tissue cannot exclude the detection of nodal nevi in melanoma draining SNs. Therefore, the specificities achieved for each marker in these control nodes might be lower in melanoma draining SNs. In future, true marker specificity should be assessed by making use of melanoma draining SNs shown (by histology) to contain nevi, but to be free of metastases. If such nodes were shown to be RT-PCR negative for the PCS markers using the optimised parameters, this would prove that SN nevi would not produce false positives with these markers. Neval cell containing, melanoma draining SNs would be the most appropriate control tissue, but there is a major limitation to developing these controls: if any of these SNs were shown to be RT-PCR positive for any of the PCS markers it would be reasonable to conclude that these nevi would cause false positives. However, the coexistence of metastatic deposits that were missed by pathological examination would first have to be excluded in these control nodes (for example, by performing RT-PCR for a CTA marker), because these metastatic deposits could be producing the detectable PCS transcripts, and not the nevi. This in itself is a problem because there are as yet no CTA markers with 100% detection rates. Therefore, the development of adequate controls to ensure that PCS markers will not cause false positives is essentially not possible at present.
We have shown that a highly specific and sensitive RT-PCR assay for detecting nodal metastases is achievable by careful and systematic optimisation of essential assay parameters. However, the use of PCS markers carries a risk of false positivity because of an approximate 4–8.5% incidence rate of nodal nevi in melanoma draining SNs (these nevi being absent in normal visceral nodes).49, 51 In addition, the use of breast epithelial markers for the detection of breast metastases in SNs will carry a similar risk of false positivity, as a result of the presence of benign epithelial inclusions in breast cancer draining SNs.46 In light of this fact, it is recommended that more emphasis should be placed on the development of a panel of CTA markers, which will be useful for melanoma and other carcinomas. The MAGE gene family offers attractive candidates.52 Once such a panel has been shown to provide specific and optimum detection of melanoma SN metastases, the PCS markers can be excluded to ensure a zero false positive rate. In addition, the CTA markers are more likely to have prognostic value than the PCS markers, because CTA molecules are essentially involved in the metastatic process, whereas PCS molecules are solely involved in the pigmentation pathway.
Take home messages.
We report an optimised reverse transcription-polymerase chain reaction (RT-PCR) protocol for the detection of very low numbers of melanoma cells in nodal tissue, for which excellent reproducibility of the entire nodal processing and RT-PCR protocol was seen
However, it should be noted that there is a risk of false positives using pigment cell specific markers alone, because of an approximate 4–8.5% incidence rate of nodal nevi in melanoma draining sentinel nodes (these nevi being absent in all other normal nodes)
Tyrosinase was the most sensitive marker; it was 200 times more sensitive than haematoxylin and eosin staining and 20 times more sensitive than standard immunohistochemistry
MAGE-3 was shown to be the only marker that is not expressed by melanocytes, but because not all melanomas express MAGE-3, it is recommended that more emphasis should be placed on the development of a panel of CTA markers to ensure a zero false positive rate and to provide optimum detection
In conclusion, it is worthwhile at this stage giving thought to the issue of how worldwide standardisation could be achieved should molecular evaluation of the SN be shown to be of clinical value. Ideally, every laboratory analysing SN tissue for the detection of micrometastases should use the same RT-PCR reagents, molecular markers, PCR primers, and parameters (and even the same brand of PCR machine), in addition to quality controls, because this would facilitate the achievement of similar sensitivities and specificities. However, achieving this would be extremely difficult and even unrealistic. Instead, we suggest the use of an international set of control lymph node tissue RNA: each laboratory should ensure that their optimised RT-PCR assays do not detect unwanted transcripts in this set of control tissues (a significant number) to confirm the specificity of their multimarker assays.
Acknowledgments
The authors thank Dr Kahn, Sr Bromberg, and Sr McCurdie (Groote Schuur Hospital) for providing normal visceral lymph nodes; Dr Strover (Kingsbury Hospital), Dr Pienaar, Dr Bruce-Chwatt, and Dr Hudson (Groote Schuur Hospital) for providing melanoma involved lymph nodes; Dr Dent, Dr Panieri (Groote Schuur Hospital), and Dr Ndhluni (Kingsbury Hospital) for providing breast cancer involved lymph nodes; Mr Emile Brys and Ms Caroline Algar for their technical assistance; and Ms Paterson for proofreading the manuscript. This study was supported by a grant from the Geriatric Society of South Africa (David and Freda Becker Trust) and a postintern fellowship from the Medical Research Council of South Africa.
Abbreviations
AJCC, American Joint Committee on Cancer
CMC, circulating melanoma cells
CTA, cancer testis antigen
GAPDH, glutaraldehyde-3-phosphate dehydrogenase
H&E, haematoxylin and eosin
PBGD, porphobilinogen deaminase
PCS, pigment cell specific
RT-PCR, reverse transcription-polymerase chain reaction
SN, sentinel lymph node
REFERENCES
- 1.Reeves ME, Coit DG. Melanoma. A multidisciplinary approach for the general surgeon. Surg Clin North Am 2000;80:581–601. [DOI] [PubMed] [Google Scholar]
- 2.Halpern AC, Schuchter LM. Prognostic models in melanoma. Semin Oncol 1997;24:S2–7. [PubMed] [Google Scholar]
- 3.Balch CM, Milton GW, Cascinelli N, et al. Elective lymph node dissection: pros and cons. In: Balch CM, Houghton AN, Milton GW, et al, eds. Cutaneous melanoma. Philadelphia: Lippincott, 1992:345–66.
- 4.Reintgen D, Balch CM, Kirkwood J, et al. Recent advances in the care of the patient with malignant melanoma [see comments]. Ann Surg 1997;225:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shivers SC, Wang X, Li W, et al. Molecular staging of malignant melanoma: correlation with clinical outcome. JAMA 1998;280:1410–15. [DOI] [PubMed] [Google Scholar]
- 6.Balch CM, Soong SJ, Shaw HM, et al. An analysis of prognostic factors in 8500 patients with cutaneous melanoma. In: Balch CM, Houghton AN, Milton GW, et al, eds. Cutaneous melanoma. Philadelphia: Lippincott, 1992:165–87.
- 7.Staveley OCK, Sotomayor E, Montgomery J, et al. Induction of antigen-specific T cell anergy: an early event in the course of tumor progression. Proc Natl Acad Sci U S A 1998;95:1178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlagenhauff B, Schittek B, Ellwanger U, et al. Significance of serum protein S100 levels in screening for melanoma metastasis: does protein S100 enable early detection of melanoma recurrence? Melanoma Res 2000;10:451–9. [DOI] [PubMed] [Google Scholar]
- 9.Hanekom GS, Stubbings HM, Kidson SH. The active fraction of plasmatic plasminogen activator inhibitor type 1 (PAI1) as a possible indicator of increased risk for metastatic melanoma. Cancer Detect Prev 2002;26:50–9. [DOI] [PubMed] [Google Scholar]
- 10.Schittek B, Blaheta HJ, Ellwanger U, et al. Polymerase chain reaction in the detection of circulating tumour cells in peripheral blood of melanoma patients. Recent Results Cancer Res 2001;158:93–104. [DOI] [PubMed] [Google Scholar]
- 11.Hanekom GS, Stubbings HM, Johnson CA, et al. The detection of circulating melanoma cells correlates with tumour thickness and ulceration but is not predictive of metastasis for patients with primary melanoma. Melanoma Res 1999;9:465–73. [DOI] [PubMed] [Google Scholar]
- 12.Blaheta HJ, Paul T, Sotlar K, et al. Detection of melanoma cells in sentinel lymph nodes, bone marrow and peripheral blood by a reverse transcription-polymerase chain reaction assay in patients with primary cutaneous melanoma: association with Breslow’s tumour thickness. Br J Dermatol 2001;145:195–202. [DOI] [PubMed] [Google Scholar]
- 13.Bostick PJ, Morton DL, Turner RR, et al. Prognostic significance of occult metastases detected by sentinel lymphadenectomy and reverse transcriptase-polymerase chain reaction in early-stage melanoma patients. J Clin Oncol 1999;17:3238–44. [DOI] [PubMed] [Google Scholar]
- 14.Blaheta HJ, Ellwanger U, Schittek B, et al. Examination of regional lymph nodes by sentinel node biopsy and molecular analysis provides new staging facilities in primary cutaneous melanoma. J Invest Dermatol 2000;114:637–42. [DOI] [PubMed] [Google Scholar]
- 15.Morton DL, Thompson JF, Essner R, et al. Validation of the accuracy of intraoperative lymphatic mapping and sentinel lymphadenectomy for early-stage melanoma: a multicenter trial. Multicenter selective lymphadenectomy trial group. Ann Surg 1999;230:453–63; discussion: 463–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershenwald JE, Colome MI, Lee JE, et al. Patterns of recurrence following a negative sentinel lymph node biopsy in 243 patients with stage I or II melanoma. J Clin Oncol 1998;16:2253–60. [DOI] [PubMed] [Google Scholar]
- 17.Gershenwald JE, Thompson W, Mansfield PF, et al. Multi-institutional melanoma lymphatic mapping experience: the prognostic value of sentinel lymph node status in 612 stage I or II melanoma patients. J Clin Oncol 1999;17:976–83. [DOI] [PubMed] [Google Scholar]
- 18.Clary BM, Brady MS, Lewis JJ, et al. Sentinel lymph node biopsy in the management of patients with primary cutaneous melanoma: review of a large single-institutional experience with an emphasis on recurrence. Ann Surg 2001;233:250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Stall A, Shivers SC, et al. Clinical relevance of molecular staging for melanoma: comparison of RT-PCR and immunohistochemistry staining in sentinel lymph nodes of patients with melanoma. Ann Surg 2000;231:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadd MA, Cosimi AB, Yu J, et al. Outcome of patients with melanoma and histologically negative sentinel lymph nodes. Arch Surg 1999;134:381–7. [DOI] [PubMed] [Google Scholar]
- 21.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American joint committee on cancer staging system for cutaneous melanoma. J Clin Oncol 2001;19:3635–48. [DOI] [PubMed] [Google Scholar]
- 22.McMasters KM, Reintgen DS, Ross MI, et al. Sentinel lymph node biopsy for melanoma: controversy despite widespread agreement. J Clin Oncol 2001;19:2851–5. [DOI] [PubMed] [Google Scholar]
- 23.Clary BM, Mann B, Brady MS, et al. Early recurrence after lymphatic mapping and sentinel node biopsy in patients with primary extremity melanoma: a comparison with elective lymph node dissection. Ann Surg Oncol 2001;8:328–37. [DOI] [PubMed] [Google Scholar]
- 24.van der Velde-Zimmermann D, Schipper ME, de Weger RA, et al. Sentinel node biopsies in melanoma patients: a protocol for accurate, efficient, and cost-effective analysis by preselection for immunohistochemistry on the basis of Tyr-PCR. Ann Surg Oncol 2000;7:51–4. [DOI] [PubMed] [Google Scholar]
- 25.Bieligk SC, Ghossein R, Bhattacharya S, et al. Detection of tyrosinase mRNA by reverse transcription-polymerase chain reaction in melanoma sentinel nodes [see comments]. Ann Surg Oncol 1999;6:232–40. [DOI] [PubMed] [Google Scholar]
- 26.Calogero A, Timmer-Bosscha H, Schraffordt Koops H, et al. Limitations of the nested reverse transcriptase polymerase chain reaction on tyrosinase for the detection of malignant melanoma micrometastases in lymph nodes. Br J Cancer 2000;83:184–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatta N, Takata M, Takehara K, et al. Polymerase chain reaction and immunohistochemistry frequently detect occult melanoma cells in regional lymph nodes of melanoma patients. J Clin Pathol 1998;51:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cormier JN, Hijazi YM, Abati A, et al. Heterogeneous expression of melanoma-associated antigens and HLA-A2 in metastatic melanoma in vivo. Int J Cancer 1998;75:517–24. [DOI] [PubMed] [Google Scholar]
- 29.de Vries TJ, Fourkour A, Wobbes T, et al. Heterogeneous expression of immunotherapy candidate proteins gp100, MART-1, and tyrosinase in human melanoma cell lines and in human melanocytic lesions. Cancer Res 1997;57:3223–9. [PubMed] [Google Scholar]
- 30.Basarab T, Picard JK, Simpson E, et al. Melanoma antigen-encoding gene expression in melanocytic naevi and cutaneous malignant melanomas. Br J Dermatol 1999;140:106–8. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs P, Hutchins AM, Dorian KT, et al. MAGE-12 and MAGE-6 are frequently expressed in malignant melanoma. Melanoma Res 2000;10:259–64. [PubMed] [Google Scholar]
- 32.Hoal-Van Helden EG, Wilson EL, Dowdle EB. Characterization of seven human melanoma cell lines: melanogenesis and secretion of plasminogen activators. Br J Cancer 1986;54:287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vries TJ, Fourkour A, Punt CJ, et al. Reproducibility of detection of tyrosinase and MART-1 transcripts in the peripheral blood of melanoma patients: a quality control study using real-time quantitative RT-PCR. Br J Cancer 1999;80:883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoon DS, Wang Y, Dale PS, et al. Detection of occult melanoma cells in blood with a multiple-marker polymerase chain reaction assay. J Clin Oncol 1995;13:2109–16. [DOI] [PubMed] [Google Scholar]
- 35.Schoenfeld A, Luqmani Y, Smith D, et al. Detection of breast cancer micrometastases in axillary lymph nodes by using polymerase chain reaction. Cancer Res 1994;54:2986–90. [PubMed] [Google Scholar]
- 36.Fidler IJ, Hart IR. Biological diversity in metastatic neoplasms: origins and implications. Science 1982;217:998–1003. [DOI] [PubMed] [Google Scholar]
- 37.Sarantou T, Chi DD, Garrison DA, et al. Melanoma-associated antigens as messenger RNA detection markers for melanoma. Cancer Res 1997;57:1371–6. [PubMed] [Google Scholar]
- 38.Kreuzer KA, Lass U, Landt O, et al. Highly sensitive and specific fluorescence reverse transcription-PCR assay for the pseudogene-free detection of beta-actin transcripts as quantitative reference. Clin Chem 1999;45:297–300. [PubMed] [Google Scholar]
- 39.Lukowsky A, Bellmann B, Ringk A, et al. Detection of melanoma micrometastases in the sentinel lymph node and in nonsentinel nodes by tyrosinase polymerase chain reaction. J Invest Dermatol 1999;113:554–9. [DOI] [PubMed] [Google Scholar]
- 40.Blaheta HJ, Schittek B, Breuninger H, et al. Detection of melanoma micrometastasis in sentinel nodes by reverse transcription-polymerase chain reaction correlates with tumor thickness and is predictive of micrometastatic disease in the lymph node basin. Am J Surg Pathol 1999;23:822–8. [DOI] [PubMed] [Google Scholar]
- 41.Battayani Z, Xerri L, Hassoun J, et al. Tyrosinase gene expression in human tissues. Pigment Cell Res 1993;6:400–5. [DOI] [PubMed] [Google Scholar]
- 42.Freshney RI. Culture of animal cells, 4th ed. New York: A John Wiley & Sons, Inc, 2000.
- 43.Blaheta HJ, Schittek B, Breuninger H, et al. Lymph node micrometastases of cutaneous melanoma: increased sensitivity of molecular diagnosis in comparison to immunohistochemistry. Int J Cancer 1998;79:318–23. [DOI] [PubMed] [Google Scholar]
- 44.Holmgren L, MS OR, Folkman J. Dormancy of micrometastases: balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med 1995;1:149–53. [DOI] [PubMed] [Google Scholar]
- 45.Tsao H, Cosimi AB, Sober AJ. Ultra-late recurrence (15 years or longer) of cutaneous melanoma. Cancer 1997;79:2361–70. [PubMed] [Google Scholar]
- 46.Goeminne JC, Guillaume T, Symann M. Pitfalls in the detection of disseminated non-hematological tumor cells. Ann Oncol 2000;11:785–92. [DOI] [PubMed] [Google Scholar]
- 47.Barnhill RL. The biology of melanoma micrometastases. Recent Results Cancer Res 2001;158:3–13. [PubMed] [Google Scholar]
- 48.Carson KF, Wen DR, Li PX, et al. Nodal nevi and cutaneous melanomas. Am J Surg Pathol 1996;20:834–40. [DOI] [PubMed] [Google Scholar]
- 49.Yu LL, Flotte TJ, Tanabe KK, et al. Detection of microscopic melanoma metastases in sentinel lymph nodes. Cancer 1999;86:617–27. [PubMed] [Google Scholar]
- 50.Baisden BL, Askin FB, Lange JR, et al. HMB-45 immunohistochemical staining of sentinel lymph nodes: a specific method for enhancing detection of micrometastases in patients with melanoma. Am J Surg Pathol 2000;24:1140–6. [DOI] [PubMed] [Google Scholar]
- 51.Cochran AJ. Melanoma metastases through the lymphatic system. Surg Clin North Am 2000;80:1683–93. [DOI] [PubMed] [Google Scholar]
- 52.Chomez P, De Backer O, Bertrand M, et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res 2001;61:5544–51. [PubMed] [Google Scholar]