Abstract
Aims: To elucidate genes that participate in the process of oncogenesis, primers based on the E6 genes of genital human papillomaviruses (HPVs) were used to amplify potential expressed sequence tags (ESTs) from the MOLT-4 T lymphoblastic leukaemia cell line.
Methods: Using the polymerase chain reaction (PCR) with human papillomavirus E6 gene primers, an EST from the MOLT-4 T lymphoblastic leukaemia cell line was amplified. Via rapid amplification of cDNA ends (RACE) and cycle sequencing from MOLT-4 and fetal lung cDNA libraries, overlapping cDNAs of 2786 bp and 2054 bp of the corresponding novel human intronless gene designated MOST-1 (for MOLT-4 sequence tag-1) were characterised and assigned the symbol C8orf17 by the HUGO Nomenclature Committee.
Results: Both cDNAs contained a potential open reading frame (ORF) of 297 bp incorporating a methionine codon with an ideal Kozak consensus sequence for translation initiation, and encoding a putative hydrophilic polypeptide of 99 amino acids. Although reverse transcription PCR (RT-PCR) demonstrated MOST-1 expression in all 19 cancer and two normal cell lines tested, differential expression was seen in only nine of 16 normal tissues tested (heart, kidney, liver, pancreas, small intestine, ovary, testis, prostate, and thymus). A 388 bp fragment was amplified from the NS-1 mouse myeloma cell line, the sequence of which was identical to that within the MOST-1 ORF. The MOST-1 gene was mapped by fluorescent in situ hybridisation to chromosome 8q24.2, a region amplified in many breast cancers and prostate cancers, which is also the candidate site of potential oncogene(s) other than c-myc located at 8q24.1. Analysis of paired biopsies of invasive ductal breast cancer and adjacent normal tissue by semiquantitative and real time RT-PCR revealed average tumour to normal ratios of MOST-1 expression that were two times greater in grade 3 cancers than in grade 1 and 2 cancers. Quantitative real time PCR of archival prostatic biopsies displayed MOST-1 DNA values that were 9.9, 7.5, 4.2, and 1.4 times higher in high grade carcinomas, intermediate grade carcinomas, low grade carcinomas, and benign hyperplasias, respectively, than in normal samples.
Conclusions: These data suggest a role for MOST-1 in cellular differentiation, proliferation, and carcinogenesis.
Keywords: amplification, breast cancer, C8orf17, MOST-1, overexpression, prostate cancer
Following the publication of a working draft of the human genome sequence, the Human Genome Project faces new challenges. Major tasks ahead include the identification of the estimated 35 000 genes residing within three billion base pairs (bp) of DNA, and the characterisation of their regulatory elements and of an even greater number of transcriptional units and translated products.1 To isolate the numerous expressed genes among large tracts of non-coding genomic DNA, expressed sequence tags (ESTs) represent an efficient and economical “short cut” route for gene identification. The idea of exploiting ESTs has been established as a practical approach for the discovery of novel human genes. ESTs are instrumental in the evaluation of human gene diversity and expression patterns, in addition to the delineation of exons and alternative splicing events. These cDNA fragments are also useful for the annotation of physical maps, and function as handles for the classification of multigene families.2,3
“To isolate the numerous expressed genes among large tracts of non-coding genomic DNA, expressed sequence tags represent an efficient and economical “short cut” route for gene identification”
The search for ESTs and their corresponding genes implicated in the causation of human cancers is intensifying in the quest for better diagnostic markers and therapeutic agents.4,5 Certain viruses contribute greatly to the development of specific cancers; for example, the association between human papillomaviruses (HPV) and genital carcinomas. The E6 and E7 early genes and oncoproteins of high risk genital HPV types possess transforming abilities and are crucial in genital carcinogenesis.6–9 In an attempt to elucidate genes that participate in the process of oncogenesis, primers based on the E6 genes of genital HPVs were used to amplify potential ESTs from the MOLT-4 T lymphoblastic leukaemia cell line. Reverse transcription polymerase chain reaction (RT-PCR) of MOLT-4 RNA using primers targeting the E6 genes of HPV types 11 and 18 generated an EST, the sequence of which revealed no significant homology to a known gene in the GenBank database. Arising from this novel EST, we report here the characterisation of a novel human gene designated MOST-1 isolated via rapid amplification of cDNA ends (RACE) and cycle sequencing, its localisation to chromosome 8q24.2, its overexpression in breast cancers, and its amplification in prostatic neoplasia relative to their respective normal tissues.
METHODS
Preparation of cDNA from cell lines
Cell lines were cultured in appropriate growth media recommended by the American Type Culture Collection. Cells were harvested, washed with phosphate buffered saline (PBS), pelleted, and resuspended in 10mM Tris/HCl (pH 7.5), 0.15M NaCl, 1.5mM MgCl2, and 0.65% Nonidet-P40. After removal of nuclei by centrifugation, the cytoplasmic lysate (supernatant) was transferred to an equal volume of buffer containing 7M urea, 1% sodium dodecyl sulfate (SDS), 0.35M NaCl, 10mM Tris/HCl (pH 7.5). The mixture was extracted with phenol/chloroform/isoamylalcohol, and RNA was precipitated with cold absolute ethanol. Total cellular RNA was reverse transcribed in a reaction mix comprising first strand buffer, dithiothreitol, RNase inhibitor, random primers, four dNTPs, and SuperScript II reverse transcriptase (Gibco BRL, Rockville, Maryland, USA), incubated at room temperature for 10 minutes, 42°C for one hour, and 99°C for five minutes.
Extraction of MOLT-4 genomic DNA
Cells were centrifuged, washed with PBS, suspended in 0.3M sodium acetate, 0.5% SDS, 5mM EDTA (pH 7.0), extracted with phenol/chloroform/isoamylalcohol, and DNA precipitated with 4M LiCl and cold absolute ethanol.
Isolation of the original MOST-1 EST
The initial EST fragment representing MOST-1 was amplified from MOLT-4 cDNA using HPV primers 11q (5′-CTTCCATGCATGTTGTCCAG-3′) and 18c (5′-GGT TTCTGGCACCGCAGGCA-3′).10 The PCR was subjected to 95°C for one minute, followed by 35 cycles each of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for one minute, and the PCR products were analysed by agarose gel electrophoresis. Three selected amplified bands were excised, the DNA eluted, and reamplified for an additional 25 cycles. PCR products were purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) or GENECLEAN II kit (Qbiogene, Carlsbad, California, USA), and cycle sequenced using the ABI PRISM BigDye terminator cycle sequencing ready reaction kit, and an ABI PRISM 377 DNA sequencer (PE Applied Biosystems, Foster City, California, USA). Database searches were performed with the BLAST algorithm at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast) to authenticate that the MOST-1 EST had not been reported before.
Identification of MOST-1 cDNA by RACE
Marathon-Ready MOLT-4 and human fetal lung cDNA libraries ligated with adaptor oligonucleotides (Clontech, Palo Alto, California, USA) were used as templates for RACE reactions. Based on the original MOST-1 EST, gene specific primers TF1 (nucleotides (nt) 1108–1127) and TR4.2 (nt 1299–1277) were designed. Before RACE, PCR was performed with these primers to confirm that the cDNA libraries contained the cDNA of interest. Primers TR4.2 and TF1 were individually paired with the adaptor primer AP1 in initial RACE experiments via long distance PCR using Advantage 2 polymerase (Clontech). Nested primers TR1 (nt 1125–1102) and TF2 (nt 1136–1158) were then paired with the adaptor primer AP2 in subsequent RACE to obtain the 5′ and 3′ RACE fragments, respectively. Thermal cycling conditions were 94°C for 30 seconds, followed by 25 cycles each of 94°C for five seconds and 68°C for four minutes. The RACE products were cycle sequenced as described above. Amino acid sequence analysis and motif searches were performed using bioinformatics software (http://www.expasy.ch).
Organisation of the MOST-1 gene
To determine the presence of exon(s) or intron(s), primer pairs flanking 200–400 bp target fragments were designed based on the full length MOST-1 cDNA sequence derived by RACE. With these MOST-1 primer pairs, PCR products amplified from MOLT-4 genomic DNA and cDNA were sequenced, compared, and subjected to Grail computational analysis (http://compbio.ornl.gov/Grail-1.3).
Chromosomal localisation of MOST-1
The chromosomal position of MOST-1 was mapped by fluorescence in situ hybridisation (FISH), using a genomic probe of ∼2.6 kb generated from MOLT-4 genomic DNA and primers TF10 (nt 34–57) and TR8 (nt 2670–2648). Probe labelling, hybridisation procedures, and visualisation methods were carried out as described previously.3,11–14 The MOST-1 sequence was also subjected to computational analysis with the UniGene program (http://www.ncbi.nlm.nih.gov/UniGene).
Screening for MOST-1 expression in normal human tissues and cancer cell lines
Multiple tissue cDNA panels generated from poly(A)+ RNA of 16 human adult tissues (0.8 ng each; Clontech), and cDNAs from 19 human cancer and two normal cell lines (200 ng each) were screened for MOST-1 expression. PCR with primers TF5 (nt 860–882) and TR4.2 flanking a 440 bp fragment was performed using a profile of 95°C for one minute, followed by 25 cycles each of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. A subsequent amplification was repeated using a 50× dilution of the first PCR product as template. To serve as controls for the human adult tissues and cell lines, housekeeping genes were also subjected to PCR using glyceraldehyde-3-phosphate dehydrogenase (G3PDH) specific primers,15 and/or HUEL gene specific primers: 5R2 (5′-AAGTATGTAATGGAAAGTCGTG-3′) and R3 (5′-AAGCATAAAGGTCTCTAGTTCTTCAGG-3′).3 All reactions were performed in duplicate.
Semiquantitative and real time RT-PCR of frozen breast tissue biopsies
Total RNAs were previously isolated from 27 pairs of snap frozen invasive ductal breast carcinoma and adjacent normal tissue samples.16–18 The breast cancers were histologically graded according to modified criteria of the Bloom and Richardson system.19,20 After reverse transcription, each cDNA sample (equivalent to about 80 ng of total RNA) was subjected to PCR amplification using primers TF5 and TR4.2 at 95°C for 30 seconds, followed by 35 cycles each of 95°C for 30 seconds, 65°C for 30 seconds, and 72°C for one minute. Semiquantitative analysis of MOST-1 mRNA was facilitated by concurrent RT-PCR of the G3PDH housekeeping gene.15 A prior PCR cycle optimisation was conducted to ensure that all reactions remained in the linear region (by removal and analysis of samples at 15, 20, 25, 30, 35, and 38 cycles). The resultant RT-PCR products were electrophoresed along with DNA markers in agarose gels stained with ethidium bromide, visualised under ultraviolet light, and analysed by densitometric scanning using a GS-700 Imaging Densitometer and Quantity One software (Bio-Rad, Hercules, California, USA). MOST-1 expression was quantified by calculating the ratio of the intensity of each RT-PCR product of MOST-1 to that of G3PDH.16–18 Relative MOST-1 expression was further presented as the differential expression between corresponding tumour (T) and normal (N) tissues as follows: overexpression in tumour tissue compared with matched normal tissue (T > N); decreased expression in tumour tissue compared with matched normal tissue (T < N); and no difference (T = N).21 All reactions were performed in triplicate or duplicate, and mean values derived. Real time PCR analysis with the LightCycler-DNA Master SYBR Green I (Roche Diagnostics, Mannheim, Germany) was carried out for 12 representative sample pairs using the iCycler iQ real time PCR detection system (Bio-Rad), as described below.
Real time PCR analysis of DNAs from archival prostatic biopsies
DNA was extracted from randomly selected, paraffin wax embedded tissue sections of normal, hyperplastic, and malignant prostatic biopsies using buffer comprising 0.5M Tris, 10mM NaCl, 20mM EDTA, 1% SDS (pH 9.0), and proteinase K. This was accompanied by phenol/chloroform/isoamylalcohol extraction and ethanol precipitation. DNA samples were reconstituted to the same concentration of 100 ng/μl, and 100 ng of each sample was used for amplification. Real time PCR was performed with primers TF1 and TR4.2 using the iCycler iQ SG-1 quantitative PCR reaction buffer kit (Bio-Rad) and LightCycler-DNA Master SYBR Green I (Roche Diagnostics), at 95°C for four minutes, followed by 40 cycles each of 95°C for 30 seconds, 65°C for 30 seconds, and 72°C for one minute. Also included was the amplification of the G3PDH housekeeping gene.15 All reactions were performed in triplicate or duplicate, and mean values derived. Real time PCR conditions were optimised by varying MgCl2 concentrations between 1.5mM and 5mM. Different starting amounts of template were also used for validation of amplification efficiency. For each sample, the threshold cycle (CT) values for both MOST-1 and G3PDH were determined, and the difference between their CT values (ΔCT) was calculated as recommended by the manufacturer of the real time PCR kit (Bio-Rad). The mean ΔCT values of each category of normal samples, hyperplastic samples, low, intermediate, and high grade carcinoma samples were calculated using SPSS for Windows (release 10.0.5). These mean ΔCT values were then subjected to analysis by multiple comparisons, and the p values between their differences were computed using the Student’s paired sample t test. Acting as the reference, the mean ΔCT of normal tissue samples was compared against the mean ΔCT values of hyperplastic samples and carcinomas of varying grades to obtain the relative DNA values presented as a numerical fold change for each category of prostatic neoplasia.21,22
RESULTS
Structural analysis of MOST-1 cDNA
Amplification of the initial MOST-1 EST of ∼ 350 bp from MOLT-4 cDNA can be explained by the homology between the 3′ ends of the HPV 18c and 11q primers with nt 1036–1044 and nt 1399–1410 of the novel cDNA, respectively (fig 1A ▶). BLAST searches of the GenBank database revealed no significant homology between this EST fragment and a known gene.
Figure 1.
(A) cDNA sequence of the human MOST-1 gene (GenBank accession number AF220264). The 2786 nucleotide (nt) sequence was derived from the MOLT-4 cDNA library, whereas an overlapping and identical 2054 nt sequence (nt 664–2705) was generated from a human fetal lung cDNA library. The italicised nucleotides in bold face correspond to the 3′ segments of primers targeting human papillomavirus E6 genes. MOST-1 cDNA contains a potential open reading frame of 297 nt (nt 1115–1411), encoding 99 amino acids, with the presumed translation start and termination codons within boxes. Three potential mRNA destabilising signals (ATTTA) are shown as underlined nucleotides. (B) Deduced amino acid sequence of a putative MOST-1 polypeptide comprising 99 residues.
Screening for this novel EST in the two cDNA libraries of the MOLT-4 T lymphoblastic leukaemia cell line and of human fetal lung authenticated its presence in both libraries. We then proceeded to isolate the full length cDNA sequence corresponding to this EST from which specific primers were designed for use in RACE experiments with these libraries. From the MOLT-4 library, products of ∼ 1.7 kb and ∼ 1.2 kb were generated via 3′ and 5′ RACE, respectively. The fetal lung library yielded bands of ∼ 1.6 kb and ∼ 0.5 kb from 3′ and 5′ RACE reactions, respectively. Cycle sequencing of the overlapping 3′ and 5′ RACE fragments revealed composite cDNA sequences of 2786 bp and 2054 bp in MOLT-4 and fetal lung, respectively (fig 1A ▶). This last cDNA sequence in fetal lung was identical to nt 664–2705 of its MOLT-4 counterpart. The resultant cDNA sequence of 2786 bp was found to contain a continuous open reading frame (ORF) of 297 bp, with an in frame nonsense codon 168 bp upstream of the putative start codon. The first methionine codon lies in an extremely favourable context for translation initiation—that is, GGCACCATGG—which conforms exactly to the ideal sequence for a eukaryotic translation start site.23 This potential ORF was predicted to encode 99 amino acids (fig 1B ▶), forming a basic, hydrophilic protein (grand average of hydrophobicity of –0.417) of 11 219 kDa. This putative protein had an isoelectric point of 8.59, and was classified as unstable in nature with an instability index computed to be 76.92. No notable amino acid sequence motifs were evident. However, three ATTTA mRNA destabilising signals24 were present in the 3′ untranslated region (UTR) of the gene. This novel gene was designated MOST-1 (for MOLT-4 sequence tag-1), assigned the symbol C8orf17 by the HUGO Nomenclature Committee, and its cDNA sequence deposited in GenBank under accession number AF220264.
To search for introns, the sequences of PCR products amplified from genomic and cDNA templates using identical primer pairs were compared but showed no differences. Subjecting the MOST-1 cDNA sequence to computational analysis by exon–intron search engines such as Grail also arrived at the same conclusion—that this gene was intronless.
The MOST-1 gene is located on chromosome 8q24.2
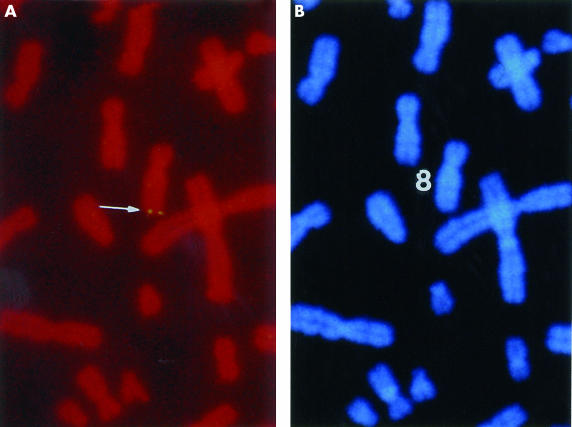
A detection efficiency of ∼ 50% was attained with the FISH probe; that is, out of 100 mitotic figures checked, 50 displayed hybridisation signals on one pair of homologous chromosomes. Using DAPI (diamidinophenylindole) banding to identify the specific chromosome, the assignment between the signal from the probe and the long arm of chromosome 8 was obtained. Based on a summary of 10 photographs, MOST-1 was localised to a more precise position at chromosome 8q24.2 (fig 2 ▶), and no other loci were detected with the probe. The MOST-1 sequence was also mapped by UniGene computational analysis to chromosome 8q24.3.
Figure 2.
Chromosomal localisation of the MOST-1 gene. (A) Double fluorescence in situ hybridisation signals detected by a MOST-1 specific probe at chromosome region 8q24.2, indicated by the arrow. (B) DAPI staining of the same mitotic figure to identify chromosome 8.
MOST-1 is ubiquitously expressed in cancer cell lines but differentially expressed in normal human tissues
RT-PCR with MOST-1 specific primers TF5 and TR4.2 yielded expected 440 bp target fragments of detectable but varying intensities from cDNAs of 19 human cancer cell lines and two normal cell lines tested (table 1 ▶). However, of the 16 normal human adult tissues tested, MOST-1 RT-PCR products were detectable in only nine tissues (heart, kidney, liver, pancreas, small intestine, ovary, testis, prostate, and thymus), but not in others (brain, lung, skeletal muscle, colon, placenta, spleen, and peripheral blood leucocytes). The G3PDH and/or HUEL housekeeping genes served as cDNA quality controls, and were successfully amplified from the tested human cell lines and adult tissues with good correlation (data not shown).
Table 1.
Comparative MOST-1 expression in human tissues, normal cell lines, and cancer cell lines
| Normal tissues | MOST-1: G3PDH ratio | Cell lines | MOST-1: G3PDH ratio |
| Pancreas | 0.61 | MRC-5 fetal lung cells | 1.35 |
| Prostate | 0.57 | Breast myoepithelial cells | 1.15 |
| Ovary | 0.55 | MDA-MB-231 breast adenocarcinoma | 2.11 |
| Testis | 0.33 | Hs578T breast ductal carcinoma | 1.14 |
| Liver | 0.33 | MCF7 breast adenocarcinoma | 1.13 |
| Heart | 0.31 | ZR-75-1 breast ductal carcinoma | 0.44 |
| Kidney | 0.31 | PC-3 prostate adenocarinoma | 1.20 |
| Thymus | 0.28 | DU145 prostate carcinoma | 1.13 |
| Small intestine | 0.26 | CaSki cervical carcinoma | 1.18 |
| Lung* | 0.17 | HeLa cervical carcinoma | 1.06 |
| Peripheral leucocytes* | 0.17 | SiHa cervical carcinoma | 0.90 |
| Skeletal muscle* | 0.14 | T24 bladder carcinoma | 0.86 |
| Placenta* | 0.08 | HepG2 hepatocellular carcinoma | 1.08 |
| Colon* | 0.06 | Hep3B hepatocellular carcinoma | 0.95 |
| Brain* | 0.02 | PLC/PRF/5 hepatocellular carcinoma | 0.69 |
| Spleen* | 0.00 | Mahlavu hepatocellular carcinoma | 0.64 |
| KATO III gastric carcinoma | 0.76 | ||
| U-937 histiocytic lymphoma | 1.59 | ||
| Raji Burkitt’s lymphoma | 0.96 | ||
| HL-60 promyelocytic leukaemia | 0.95 | ||
| MOLT-4 T-lymphoblastic leukaemia | 0.94 |
All cDNA samples were subjected to PCR with MOST-1 and G3PDH primers, and the individual ratios of the densitometric readings of their amplified products were calculated. *Samples for which no MOST-1 amplified products were visualised by agarose gel electrophoresis.
G3PDH, glyceraldehyde-3-phosphate dehydrogenase; PCR, polymerase chain reaction.
MOST-1 is overexpressed in a subset of breast cancers
MOST-1 expression in paired breast tissue biopsies was compared by first normalising densitometric readings of MOST-1 with G3PDH RT-PCR products, and then calculating the ratios between corresponding tumour and adjacent normal tissues (T : N). Of the 27 paired biopsy samples tested, relative T : N expression of MOST-1 was increased in 11 pairs, unchanged in four, and decreased in another 12. Real time RT-PCR performed on 12 randomly selected pairs of tumour and normal tissues (six grade 1 and 2 cancers and six grade 3 cancers) revealed MOST-1 expression data consistent with semiquantitative RT-PCR results (data not shown). The T : N ratio of MOST-1 expression showed a correlation with breast cancer grade; that is, mean ratios for grade 1 and 2 versus grade 3 were 0.84 versus 1.69 by semiquantitative RT-PCR, and 1.41 versus 2.92 by real time RT-PCR. However, the differences between the means for grade 3 versus grade 1 and 2 cancers were not significant (table 2 ▶).
Table 2.
Ratio of MOST-1 expression in grade 1–3 breast cancers to expression seen in the corresponding adjacent normal tissues
| Tumour:normal ratio of MOST-1 mRNA (mean (SD)) | ||
| Breast cancer grade | Semiquantitative RT-PCR | Real time RT-PCR |
| 1 and 2 | 0.8370 (0.3781) (n=14) | 1.4148 (1.2565) (n=6) |
| 3 | 1.6874 (1.5545) (n=13) | 2.9154 (5.2143) (n=6) |
The mean ratios and SD values were calculated from semiquantitative and real time reverse transcription polymerase chain rection (RT-PCR) experiments.
MOST-1 DNA copy numbers are increased in prostatic neoplasia
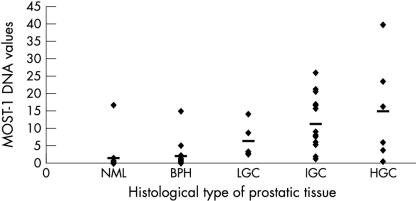
Because of the relatively low incidence of prostate cancers in Singapore, sufficient numbers of paired biopsies of prostatic cancer and adjacent normal tissues for statistical analysis were not readily available. Hence, we resorted to analysing DNA from a total of 61 histologically assessed, archival prostate biopsy sections; that is, 15 normal prostates, 21 benign prostatic hyperplasias, and 25 prostatic carcinomas. All DNA samples were subjected to real time PCR experiments to derive each individual ΔCT value—the difference between the threshold cycles for MOST-1 and G3PDH gene detection. The formula 2-ΔCT was used to measure the amount of MOST-1 DNA (fig 3 ▶). By comparing the mean ΔCT value of normal samples (as the benchmark reference) with those of prostatic neoplasia, relative MOST-1 DNA values were increased 1.4 times in hyperplasias, and 9.9, 7.5, and 4.2 times in high, intermediate, and low grade carcinomas with the corresponding Gleason scores of 8–9, 6–7, and 3–5, respectively. Multiple comparisons between the mean for normal tissues against the individual means for neoplastic tissues indicated that the differences for hyperplasias or low grade cancers were significant (p = 0.014 and p = 0.002, respectively), and for intermediate or high grade cancers were highly significant (p = 0.000 for both grades). Interestingly, only one sample each of normal and hyperplastic prostatic tissue appeared to have significant amplification of MOST-1.
Figure 3.
Correlation between histological types of prostatic tissues with MOST-1 DNA values. NML and BPH refer to normal prostate and benign prostatic hyperplasia, respectively. LGC, IGC, and HGC denote increasing grades of prostatic cancer (low, intermediate, and high), corresponding to Gleason scores of 3–5, 6–7, and 8–9, respectively. MOST-1 DNA values were calculated according to the formula 2-ΔCT, where ΔCT equals the difference between the threshold cycles for MOST-1 and G3PDH (glyceraldehyde-3-phosphate dehydrogenase) gene detection by real time polymerase chain reaction. The horizontal bars indicate mean values.
DISCUSSION
The MOST-1 gene was found to be intronless, and structural analysis of its cDNA predicted an unstable transcript, with a strong consensus sequence at the putative methionine start codon23 of a potential ORF encoding a relatively small protein. Present within the 3’ UTR of MOST-1, the AUUUA motif is a highly conserved sequence repeated three or more times in RNAs encoding many short lived cytokines and protooncogenes.24 The generally low abundance of MOST-1 mRNA in normal adult tissues was reflected by the necessity for reamplification during screening for tissue expression of MOST-1, and by the inability to detect MOST-1 transcripts by northern blot analysis (data not shown). These features are characteristic of many transiently expressed genes encoding small polypeptides involved in signalling or metabolic cascades that are being discovered at an accelerating pace. For example, human G protein coupled receptors that mediate ligand induced signalling are encoded by predominantly intronless genes.25 Interestingly, the novel PSGR prostate specific gene with homology to a G protein coupled receptor is overexpressed in prostate cancers.21
It is noteworthy that MOST-1 was expressed in all the human cancer cell lines tested, but only in specific normal human adult tissues. Another important finding was the detection of MOST-1 mRNA in human fetal lung but not in adult lung tissue, suggesting that MOST-1 expression is dependent on tissue differentiation. RT-PCR of the NS-1 mouse myeloma cell line using MOST-1 primers TF7 (nt 1000–1022) and TR4.1 (nt 1387–1368) amplified an EST, the sequence of which was completely identical to nucleotides 1000–1387 within the MOST-1 ORF (data not shown). This indicates the existence of a murine homolog of the MOST-1 gene, and implies its conservation throughout mammalian evolution.
The localisation of MOST-1 to chromosome 8q24.2 is noteworthy because this region (and its vicinity) has been shown by many comparative genomic hybridisation studies to be amplified mainly in breast and prostate cancers, and also in ovarian, testicular, renal, bladder, and colorectal tumours.26–33 A recent study using FISH in pathological organ confined prostate cancers demonstrated that an additional increase or over-representation of 8q24 together with loss of 8p22 are associated with poor prognosis.34 Although the localisation of the c-myc protooncogene at 8q24.1 has been cited to account for many poorly differentiated prostate cancers, the entire long arm of chromosome 8 is usually present at an increased copy number, suggesting that other genes may be involved.26,35 For example, a gene that is differentially expressed during prostate cancer progression has been identified on chromosome 8q11.36 Amplification of c-myc may not always correlate with 8q amplification in breast and prostate cancers.29,30 Other genes located at the 8q24.2 region are the prostate stem cell antigen (PSCA) gene, a prostate specific gene that apparently plays a role in prostate cancer progression,37 and the brain adenylyl cyclase gene HBAC1.38 This locus has also been implicated in various diseases such as partial trisomy 8q, where patients suffer from psychomotor retardation,39 and hereditary spastic paraplegia, in which the dominant HSP gene has been localised to 8q23–q24.40 Diseases associated with 8q for which the exact disease locus has yet to be discovered include autosomal recessive achromatopsia with defective photoreceptors,41 retinitis pigmentosa,42 and tibial hemimelia in Langer–Giedion syndrome.43
“It is noteworthy that MOST-1 was expressed in all the human cancer cell lines tested, but only in specific normal human adult tissues”
The amplification of the chromosomal locus coinciding with that of MOST-1 in breast and prostate malignancies prompted us to perform quantitative PCR experiments to determine MOST-1 RNA and DNA values in frozen breast cancers and archival prostate tumours compared with normal tissues. Two fifths of breast cancers exhibited MOST-1 overexpression, concurring with previous comparative genome hybridisation studies that revealed amplification of 8q24.2 in these tumours. Compared with lower grade breast cancers, those of grade 3 showed higher T : N ratios of MOST-1 transcript values of up to 13.5. Relative to normal prostate specimens, benign prostatic hyperplasia samples displayed an average 1.4-fold increase in MOST-1 DNA, whereas up to approximately a 10-fold amplification was seen in prostatic cancers, especially those of higher grades and Gleason scores.
Aberrations of MOST-1 expression and copy number appear to be associated with substantial subsets of breast and prostate carcinomas, especially those of higher grade. Taken together, these findings suggest that MOST-1 may serve as a potential marker of breast and prostate tumour progression, and also imply a possible role for MOST-1 in cellular differentiation, proliferation, and tumorigenesis.
Take home messages.
Starting with an expressed sequence tag from the MOLT-4 T lymphoblastic leukaemia cell line, we characterised a novel human, intronless gene, designated MOST-1
The MOST-1 gene was mapped by fluorescent in situ hybridisation to chromosome 8q24.2, a region amplified in many breast cancers and prostate cancers, which is also the candidate site of potential oncogene(s)
The MOST-1 gene was overexpressed in certain subsets of breast carcinoma and copy numbers were increased in prostatic neoplasia relative to the respective normal tissues; these effects appeared to correlate with grade
The MOST-1 gene identified here may play a role in cellular differentiation, proliferation, and carcinogenesis
Acknowledgments
We thank Dr BH Bay and Dr R Jin for providing the breast biopsy specimens, Dr F Dong for statistical advice, and the WHO Immunology Centre, National University of Singapore for the generous gift of some cell lines. This study was supported by a grant from the Biomedical Research Council, Singapore. JMM Tan is a recipient of a research scholarship from the National University of Singapore.
Abbreviations
CT, threshold cycle
EST, expressed sequence tag
FISH, fluorescence in situ hybridisation
G3PDH, glyceraldehyde-3-phosphate dehydrogenase
HPV, human papillomavirus
nt, nucleotides
N, normal
ORF, open reading frame
PBS, phosphate buffered saline
PCR, polymerase chain reaction
RACE, rapid amplification of cDNA ends
RT, reverse transcription
SDS, sodium dodecyl sulfate
T, tumour
UTR, untranslated region
REFERENCES
- 1.Wright FA, Lemon WJ, Zhao WD, et al. A draft annotation and overview of the human genome. Genome Biol 2001;2:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MD, Kelly JM, Gocayne JD, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 1991;252:1651–6. [DOI] [PubMed] [Google Scholar]
- 3.Sim DLC, Chow VTK. The novel human HUEL (C4orf1) gene maps to chromosome 4p12–p13 and encodes a nuclear protein containing the nuclear receptor interaction motif. Genomics 1999;59:224–33. [DOI] [PubMed] [Google Scholar]
- 4.Strausberg RL. The Cancer Genome Anatomy Project: new resources for reading the molecular signatures of cancer. J Pathol 2001;195:31–40. [DOI] [PubMed] [Google Scholar]
- 5.Onyango P. Genomics and cancer. Curr Opin Oncol 2002;14:79–85. [DOI] [PubMed] [Google Scholar]
- 6.Chow VTK, Leong PWF. Complete nucleotide sequence, genomic organization and phylogenetic analysis of a novel genital human papillomavirus type, HLT7474-S. J Gen Virol 1999;80:2923–9. [DOI] [PubMed] [Google Scholar]
- 7.Chow VTK, Loh E, Yeo WM, et al. Identification of multiple genital HPV types and sequence variants by consensus and nested type-specific PCR coupled with cycle sequencing. Pathology 2000;32:204–8. [PubMed] [Google Scholar]
- 8.Stoler MH. Human papillomaviruses and cervical neoplasia: a model for carcinogenesis. Int J Gynecol Pathol 2000;19:16–28. [DOI] [PubMed] [Google Scholar]
- 9.Mantovani F, Banks L. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 2001;20:7874–87. [DOI] [PubMed] [Google Scholar]
- 10.Tham KM, Chow VTK, Singh P, et al. Diagnostic sensitivity of polymerase chain reaction and Southern blot hybridization for the detection of human papillomavirus DNA in biopsy specimens from cervical lesions. Am J Clin Pathol 1991;95:638–46. [DOI] [PubMed] [Google Scholar]
- 11.Heng HHQ, Squire J, Tsui LC. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc Natl Acad Sci U S A 1992;89:9509–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heng HHQ, Tsui LC. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 1993;102:325–32. [DOI] [PubMed] [Google Scholar]
- 13.Quek HH, Chow VTK. Genomic organization and mapping of the human HEP-COP gene (COPA) to 1q. Cytogenet Cell Genet 1997;76:139–43. [DOI] [PubMed] [Google Scholar]
- 14.Chow VTK, Lim KM, Lim D. The human DENN gene: genomic organization, alternative splicing, and localization to chromosome 11p11.21–p11.22. Genome 1998;41:543–52. [DOI] [PubMed] [Google Scholar]
- 15.Abdel-Mageed AB, Agrawal KC. Antisense down-regulation of metallothionin induces growth arrest and apoptosis in human breast carcinoma cells. Cancer Gene Ther 1997;4:199–207. [PubMed] [Google Scholar]
- 16.Jin R, Bay BH, Chow VTK, et al. Metallothionein 1E mRNA is highly expressed in oestrogen receptor-negative human invasive ductal breast cancer. Br J Cancer 2000;83:319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin R, Bay BH, Chow VTK, et al. Metallothionein 1F mRNA expression correlates with histological grade in breast carcinoma. Breast Cancer Res Treat 2001;66:265–72. [DOI] [PubMed] [Google Scholar]
- 18.Jin R, Chow VTK, Tan PH, et al. Metallothionein 2A expression is associated with cell proliferation in breast cancer. Carcinogenesis 2002;23:81–6. [DOI] [PubMed] [Google Scholar]
- 19.Bloom HJG, Richardson WW. Histological grading and prognosis in breast cancer: a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 1957;11:359–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 1991;19:403–10. [DOI] [PubMed] [Google Scholar]
- 21.Xu LL, Stackhouse BG, Florence K, et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res 2000;60:6568–72. [PubMed] [Google Scholar]
- 22.Leong PWF, Liew K, Lim W, et al. Differential display RT-PCR analysis of enterovirus-71-infected rhabdomyosarcoma cells reveals mRNA expression responses of multiple human genes with known and novel functions. Virology 2002;295:147–59. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 1991;266:19867–70. [PubMed] [Google Scholar]
- 24.Akashi M, Shaw G, Hachiya M, et al. Number and location of AUUUA motifs: role in regulating transiently expressed RNAs. Blood 1994;83:3182–7. [PubMed] [Google Scholar]
- 25.Gentles AJ, Karlin S. Why are human G-protein-coupled receptors predominantly intronless? Trends Genet 1999;15:47–9. [DOI] [PubMed] [Google Scholar]
- 26.Visakorpi T, Kallioniemi AH, Syvanen A, et al. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995;55:342–7. [PubMed] [Google Scholar]
- 27.Forozan F, Karhu R, Kononen J, et al. Genome screening by comparative genome hybridization. Trends Genet 1997;13:405–9. [DOI] [PubMed] [Google Scholar]
- 28.Kuukasjarvi T, Tanner M, Pennanen S, et al. Genetic changes in intraductal breast cancer detected by comparative genomic hybridization. Am J Pathol 1997;150:1465–71. [PMC free article] [PubMed] [Google Scholar]
- 29.Nupponen NN, Kakkola L, Koivisto P, et al. Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998;153:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nupponen NN, Porkka K, Kakkola L, et al. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999;154:1777–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Angelis PM, Clausen OPF, Schjolberg A, et al. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer 1999;80:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knuutila S, Aalto Y, Autio K, et al. DNA copy number losses in human neoplasms. Am J Pathol 1999;155:683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loveday RL, Greenman J, Drew PJ, et al. Genetic changes associated with telomerase activity in breast cancer. Int J Cancer 1999;84:516–20. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchiya N, Slezak JM, Lieber MM, et al. Clinical significance of alterations of chromosome 8 detected by fluorescence in situ hybridization analysis in pathologic organ-confined prostate cancer. Genes Chromosomes Cancer 2002;34:363–71. [DOI] [PubMed] [Google Scholar]
- 35.Saramaki O, Willi N, Bratt O, et al. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am J Pathol 2001;159:2089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang GT, Tapsi N, Steenbeek M, et al. Identification of a gene on human chromosome 8q11 that is differentially expressed during prostate-cancer progression. Int J Cancer 1999;83:506–11. [DOI] [PubMed] [Google Scholar]
- 37.Reiter RE, Gu Z, Watabe T, et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci U S A 1998;95:1735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stengel D, Parma J, Gannage MH, et al. Different chromosomal localization of two adenylyl cyclase genes expressed in human brain. Hum Genet 1992;90:126–30. [DOI] [PubMed] [Google Scholar]
- 39.Stengel-Rutkowski S, Lohse K, Herzog C, et al. Partial trisomy 8q. Two case reports with maternal translocation and inverted insertion: phenotype analyses and reflections on the risk. Clin Genet 1992;42:178–85. [DOI] [PubMed] [Google Scholar]
- 40.Hedera P, Rainier S, Alvarado D, et al. Novel locus for autosomal dominant hereditary spastic paraplegia, on chromosome 8q. Am J Hum Genet 1999;64:563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milunsky A, Huang XL, Milunsky J, et al. A locus for autosomal recessive achromatopsia on human chromosome 8q. Clin Genet 1999;56:82–5. [DOI] [PubMed] [Google Scholar]
- 42.Inglehearn CF, McHale JC, Keen TJ, et al. A new family linked to the RP1 dominant retinitis pigmentosa locus on chromosome 8q. J Med Genet 1999;36:646–8. [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens CA, Moore CA. Tibial hemimelia in Langer–Giedion syndrome—possible gene location for tibial hemimelia at 8q. Am J Med Genet 1999;85:409–12. [DOI] [PubMed] [Google Scholar]