Abstract
Aims: To compare the molecular genetic changes in the Ki-ras and adenomatous polyposis coli (APC) genes between colorectal carcinomas and synchronous metastases, and then to compare and contrast those changes with previously reported changes in the two genes between these carcinomas and accompanying adenomas. This expanded comparison would provide greater understanding of the progression of molecular changes in neoplastic tissue during the development of malignancy from a benign adenoma to carcinoma and then to metastatic spread of the malignancy.
Methods: DNA was extracted from paraffin wax embedded tissue. This was followed by polymerase chain reaction and gel electrophoresis for mutations in the Ki-ras gene using single stranded conformational polymorphism analysis. Amplification of a CA repeat marker was used to assess loss of heterozygosity (LOH) at the APC gene.
Results: The findings for the Ki-ras gene in 42 paired carcinomas and synchronous metastases were identical, regardless of whether or not the carcinoma and its companion adenoma had identical Ki-ras findings. The results of APC LOH for 39 paired carcinomas and synchronous metastases were also identical, whether or not the carcinoma and its companion adenoma had identical APC LOH findings. Results were uninformative for three pairs.
Conclusions: With respect to these two genes, a carcinoma may be discordant from its companion adenoma, but the metastasis remains consistent with the colonic carcinoma.
Keywords: Ki-ras, adenomatous polyposis coli, metastasis, colonic adenoma, colon carcinoma
Molecular genetic changes have been identified in several tumour suppressor genes and oncogenes in colorectal neoplasms. These mutations are postulated to accumulate over time within a particular neoplasm as normal mucosa develops into an adenoma, which may then develop into a carcinoma. This process is referred to as the adenoma–carcinoma sequence.1 Genetic changes found in neoplasms may be either of direct relevance to the growth of the tumour or they may be secondary changes resulting from more crucial growth promoting alterations. The circumstantial evidence for the genetic changes throughout the adenoma–carcinoma sequence are based on sampling groups of lesions with similar histology representing the various stages of progression. Individual lesions have not been observed and sampled over time, because neoplasms are always removed when detected. In general, greater numbers of genetic changes are found as the pathology of a lesion advances.
The observations indicate that: (1) there is not a strict linearity of accumulated molecular changes in this progression for a particular neoplasm; and (2) not all adenomas and carcinomas removed from the same person have identical genetic changes at the same locus. Regarding the first point, we have shown that the incidence of Ki-ras mutations in in situ carcinomas is higher in the adenomatous portion than in the carcinomatous portion.2 In addition, 20% of in situ carcinomas contain a Ki-ras mutation different from the Ki-ras mutation found in the adenomatous portion of the same neoplasm.3 With regard to the second point, we have examined paired adenomas and carcinomas from the same bowel but not adjacent to each other.4 We found no association between the presence of a Ki-ras mutation in the adenoma and a mutation in the carcinoma. Furthermore, when both lesions contained a Ki-ras mutation, the mutation was different in 56% of paired lesions. Moreover, when both paired lesions demonstrate loss of heterozygosity (LOH) of the adenomatous polyposis coli (APC) gene, this will involve LOH of different alleles in 50% of the paired lesions, consistent with random loss of each copy of the APC gene rather than preferential loss of a particular chromosome.
“In general, greater numbers of genetic changes are found as the pathology of a lesion advances”
These inconsistencies in paired lesions could reflect variable responses of different groups of cells within the colon to a particular environment. However, it is possible that certain genetic changes within the cells of a particular lesion are necessary for some progression, but may not necessarily then be carried beyond that point. Therefore, we have analysed a cohort of paired carcinomas and metastases for mutations in the Ki-ras gene and for LOH of the APC gene to assess whether the variability noted in lesions earlier in the adenoma–carcinoma sequence was also present at this later stage. Our present study is an extension of an earlier study of paired adenomas and carcinomas, with the inclusion of an adenoma, a carcinoma, and a metastasis from each of the studied individuals.3,4
MATERIALS AND METHODS
Patients with a carcinoma and concurrent metastasis were identified through screening interviews with patients attending the endoscopy suite of a large community hospital between 1994 and 2000. Our initial focus was on patients with adenomas and these patients all had at least one adenoma. Patients with a family history suggestive of familial adenomatous polyposis or hereditary nonpolyposis colorectal carcinoma were excluded. All patients had undergone a primary resection. Tissue samples were processed by coded number. Pathology review of all samples was performed by one of two clinical pathologists before the molecular studies. The study design was reviewed and approved by the hospital institutional review board.
All specimens were formalin fixed and paraffin wax embedded and were processed for DNA extraction and purification as reported previously.4 A haematoxylin and eosin stained slide of each specimen was inspected microscopically to ascertain the most representative region, and then the corresponding area was manually scraped for DNA extraction. This method ensured that at least 75% of the scraped area contained neoplastic cells. LOH of the APC gene was determined through the polymerase chain reaction (PCR) amplification of a CA repeat marker within the D5S346 locus of the DP1 gene,5 using the primer set: 5′-ACTCACTCT AGTGATAAATCGGG-3′ (sense) and 5′-AGCAGATAAGACAA GTATTACTAGTT-3′ (antisense).
Those samples homozygous for this primer set were analysed with a second primer set: 5′-AGCCTGTGTGCAACAG AAGTGA-3′ (sense) and 5′-GAAGGTGAACAAAGGACAT GAAC-3′ (antisense), which amplifies an AAAT repeat within the D5S1385 locus. All samples were amplified and analysed as reported previously.4
Single stranded conformational polymorphism (SSCP) analysis was used to screen for mutations within the Ki-ras oncogene. The codon 12/13 region of exon 1 in the Ki-ras oncogene was amplified using the primer set 5′-CCTGCTGAAAATGACTGAAT-3′ (sense) and 5′-TGTTGGA TCATATTCGTCCA-3′ (antisense). Samples were amplified and analysed as reported previously.4 Samples showing Ki-ras mutation bands by SSCP were sequenced to verify the point mutation. PCR based DyeDeoxy Terminator sequencing was performed using an ABI Prism 377 DNA sequencing system (Perkin Elmer, Foster City, California, USA). PCR products were generated and subsequently sequenced using the same primers as above.
RESULTS
The paired adenomas and carcinomas with associated metastases were from 24 men and 18 women. They ranged in age from 41 to 95 years, with an average of 69.7 years. Molecular results on 38 of the 42 adenoma and carcinoma pairs were reported previously as part of a larger cohort.4 Of these 42 cases, the adenoma was removed simultaneously with the carcinoma from 38 patients. The time intervals between removal for the other four patients were: for three patients, the adenoma was removed after the carcinoma at two, five, and five years; and for one pair removal of the adenoma preceded the carcinoma by 12 years. Twenty three adenomas were tubular, 13 were tubulovillous, and six were villous in histological appearance. All metastases were removed simultaneously with the carcinomas and were located in regional lymph nodes for 39 patients, the liver for two patients, and a mesenteric implant in one patient.
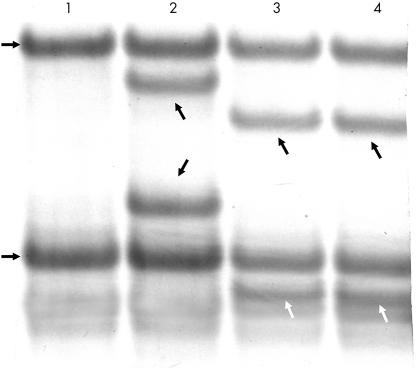
Ki-ras analysis was available for adenomas, carcinomas, and metastases in all 42 cases. The Ki-ras results were identical for 17 pairs of adenomas and carcinomas (table 1 ▶). For each associated metastasis, the Ki-ras mutation profile was also identical. However, the Ki-ras finding differed between the adenoma and carcinoma for the remaining 25 pairs. For all 25 pairs, the Ki-ras finding in the metastasis was identical to that in the carcinoma. Of note, this was also the finding for five pairs in which the adenoma and carcinoma had different Ki-ras mutations; the metastases consistently showed the specific Ki-ras mutation of the carcinoma (table 1 ▶; fig 1 ▶). Table 2 ▶ shows the frequencies and types of Ki-ras mutations.
Table 1.
Ki-ras results in primary colorectal carcinomas and synchronous metastases compared with an associated adenoma
| Ki-ras results | |||
| N | Adenoma | Carcinoma | Metastasis |
| Adenoma and carcinoma similar (17) | |||
| 12 | None | None | None |
| 5 | Mutation | Mutation | Mutation |
| Adenoma and carcinoma different (25) | |||
| 12 | None | Mutation | Mutation |
| 8 | Mutation | None | None |
| 5 | Mutation* | Mutation* | Mutation* |
None, no mutation detected; mutation, mutation detected.
*The allele showing LOH differed between the adenoma and the other two lesions.
Figure 1.
Autoradiograph showing Ki-ras mutations of codon 12 detected by single stranded conformational polymorphism. Lane 1, normal mucosa. The two horizontal arrows at the left of lane 1 indicate the two normal bands. Lane 2, caecal tubulovillous adenoma. In addition to the two normal bands, the oblique arrows indicate the bands corresponding to a GGT to GAT mutation. Lane 3, ascending carcinoma. In addition to the normal bands, the oblique arrows (black at top, white below) indicate the bands corresponding to a GGT to TGT mutation. Lane 4, lymph node metastasis with the same GGT to TGT mutation as seen in lane 3.
Table 2.
Specific types of K-ras mutations identified and their frequencies
| Mutation type (all codon 12) | Adenomas (n=18)* | Carcinomas (n=22) |
| GGT→GAT | 9 | 9 |
| GGT→GTT | 9 | 5 |
| GGT→TGT | 1 | 3 |
| GGT→AGT | 1 | 0 |
| GGT→CGT | 0 | 2 |
| Other | 0 | 3 |
The three “other” mutations in the carcinomas were: codon 12, GGT→GCT; codon 13, GGC→GAC; codon 9, a 6 bp insert.
*Two adenomas each had two different Ki-ras mutations.
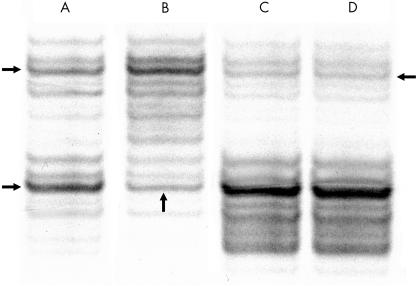
Microsatellite analysis of the tumours permits evaluation of the allele or haplotype deleted in the lesion. APC LOH analysis was available for 39 cases because two cases were uninformative with both the APC microsatellite markers used and one lymph node contained too few metastatic cells for sufficient APC LOH determination. The APC LOH results were identical for 24 pairs of adenomas and carcinomas, and each associated metastasis also gave the same result. There was no LOH for 22 of these 24 pairs; and LOH of the same allele for the adenoma, carcinoma, and metastasis in two cases. The APC LOH findings differed for 15 pairs of adenomas and carcinomas. In all 15 of these cases, the APC LOH finding in the associated metastasis was identical to that of the carcinoma. Of note, four pairs of adenomas and carcinomas had LOH of different alleles, yet in each case the associated metastasis showed LOH of the same allele as the carcinoma (table 3 ▶; fig 2 ▶).
Table 3.
APC loss of heterozygosity (LOH) results in primary colorectal carcinoma and synchronous metastases compared with an associated adenoma
| APC LOH results | |||
| N | Adenoma | Carcinoma | Metastasis |
| Adenoma and carcinoma with identical LOH patterns (24 patients) | |||
| 22 | None | None | None |
| 2 | LOH | LOH | LOH |
| Adenoma and carcinoma with different LOH patterns (15 patients) | |||
| 7 | None | LOH | LOH |
| 4 | LOH | None | None |
| 4 | LOH* | LOH* | LOH* |
None, no LOH detected; LOH, LOH detected; *the allele showing LOH differed between the adenoma and the other two lesions.
Figure 2.
Autoradiograph showing DP1 microsatellite analysis used to determine loss of heterozygosity (LOH). Lane A, normal mucosa. The two horizontal arrows to the left of lane A indicate the two normal bands reflecting the two alleles. Lane B, descending tubular adenoma. The vertical arrow points directly to the lower allele with reduced intensity compared with the normal allele, reflecting LOH of this allele. Lane C, sigmoid carcinoma; lane D, liver metastasis. The horizontal arrow to the right of lane D points to the upper allele with greatly reduced intensity compared with the normal, reflecting LOH of this allele for both the carcinoma and the metastasis.
DISCUSSION
All 42 of the metastases we report showed the same molecular profile for Ki-ras as the primary carcinoma, and none acquired additional Ki-ras mutations. Thirty nine of the 42 showed the same APC pattern in both of these tissues, and none developed LOH of the second allele in the metastases. APC analysis was uninformative in three cases.
Take home messages.
The mutations at the Ki-ras gene were identical in the 42 paired carcinomas and synchronous metastases, regardless of whether or not the carcinoma and its companion adenoma had identical Ki-ras findings
Similarly loss of heterozygosity at the adenomatous polyposis coli (APC) locus was identical for 39 paired carcinomas and synchronous metastases
Thus, for the K-ras and APC genes, a carcinoma may be discordant from its companion adenoma, but the metastasis remain consistent with the colonic carcinoma
A Medline medical literature search revealed no previous publication specifically describing APC changes in pairs of primary colorectal carcinomas with synchronous metastases in regional lymph nodes. One publication compared LOH for 5q in subsequent liver metastases from eight cases.6 Six of the eight had LOH in both paired samples. One had LOH in the primary but not in the liver metastasis, and one had LOH in the metastasis but not in the primary. There are two reports of Ki-ras mutations in primary carcinomas and metastases to pericolonic lymph nodes, or subsequently to distant organs. The authors report the presence of the same Ki-ras mutation in 35 and 21 paired samples, respectively.7,8 Others have shown identical Ki-ras mutations in 25 cases of primary colorectal carcinoma and subsequent metastases.9
However, it is probable that metastatic colorectal carcinoma cells do show some molecular genetic changes not present in the primary tumour. For instance, the development of LOH of chromosome arm 3p in colorectal metastases has been reported.6 Others have shown additional genetic gains and losses in lymph nodes and liver metastases beyond those seen in the primary carcinomas.10,11
“Thirty nine of the 42 metastases we report showed the same molecular profile for Ki-ras and APC loss of heterozygosity as the primary carcinoma”
It is clear from our data regarding in situ carcinoma plus adenoma and carcinoma pairs that different groups of neoplastic cells, even when present simultaneously within a patient’s colon, may have different molecular profiles with respect to Ki-ras mutations and APC LOH. This may reflect an equivalence of different genetic changes for the growth advantage of a developing neoplasm, or the importance of the specific changes seen in the carcinoma (recognising that we cannot infer which adenomas or in situ cancers would have developed into carcinomas). Therefore, for most neoplasms, molecular changes in these two genes are not necessarily a consistent pancolonic or lesional feature. However, with respect to these two genes, metastases do reflect the colonic neoplasm thought to be the origin of the metastasis. The lack of further genetic changes at these two loci indicates that these changes were either sufficient for metastasis to occur, or, at least, they were not greatly disadvantageous to the occurrence of metastasis. Of course, it is not possible to tell at what stage the differing LOH patterns occurred because the carcinomas were only excised at a time when metastases were present. Thus, the metastasising property of these cells may be related to these precise genetic changes or to other unmeasured genetic or biochemical changes.12
Acknowledgments
The authors would like to thank Dr E Berman for review of the histological slides. The authors acknowledge the financial support of the H Nussbaum Foundation of the Saint Barnabas Medical Center and Cancer Research, UK.
Abbreviations
APC, adenomatous polyposis coli
LOH, loss of heterozygosity
PCR, polymerase chain reaction
SSCP, single stranded conformational polymorphism
REFERENCES
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 2.Zauber NP, Sabbath-Solitare M, Bishop DT. Ki-ras mutations in adenomas from cancer-bearing and cancer-free bowel [letter]. J Pathol 1998;184:340. [DOI] [PubMed] [Google Scholar]
- 3.Zauber NP, Sabbath-Solitare M, Marotta SP, et al. Ki-ras mutation and loss of heterozygosity of the adenomatous polyposis coli gene in patients with colorectal adenomas with in situ carcinoma. Cancer 1999;86:31–6. [DOI] [PubMed] [Google Scholar]
- 4.Zauber NP, Sabbath-Solitare M, Marotta SP, et al. Molecular changes in the Ki-ras and APC genes in colorectal adenomas and carcinomas arising in the same patient. J Pathol 2001;193:303–9. [DOI] [PubMed] [Google Scholar]
- 5.Spirio L, Nelson L, Joslyn G, et al. A CA repeat 30–70 kb downstream from the adenomatous polyposis coli (APC) gene. Nucleic Acids Res 1991;19:6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaker H, Graf M, Rieker RJ, et al. Comparison of losses of heterozygosity and replication errors in primary colorectal carcinomas and corresponding liver metastases. J Pathol 1999;188:258–62. [DOI] [PubMed] [Google Scholar]
- 7.Finkelstein SD, Sayegh R, Christensen S, et al. Genotypic classification of colorectal adenocarcinoma. Cancer 1993;71:3827–38. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mulla F, Going JJ, Sowden ETHH, et al. Heterogeneity of mutant versus wild-type Ki-ras in primary and metastatic colorectal carcinomas, and association of codon-12 valine with early mortality. J Pathol 1998;185:130–8. [DOI] [PubMed] [Google Scholar]
- 9.Losi L, Benhattar J, Costa J. Stability of K-ras mutations throughout the natural history of human colorectal cancer. Eur J Cancer 1992;28A:1115–20. [DOI] [PubMed] [Google Scholar]
- 10.Zaglul AP, Kang JJ, Essig YP, et al. Analysis of colorectal cancer by comparative genomic hybridization: evidence for induction of the metastatic phenotype by loss of tumor suppressor genes. Clin Cancer Res 1998;4:879–86. [PubMed] [Google Scholar]
- 11.Al-Mulla F, Keith WN, Pickford IR, et al. Comparative genomic hybridization analysis of primary colorectal carcinomas and their synchronous metastases. Genes Chromosomes Cancer 1999;24:306–14. [DOI] [PubMed] [Google Scholar]
- 12.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001;410:50–6. [DOI] [PubMed] [Google Scholar]