Abstract
Aims: To define regions of loss on the distal portion of chromosome 12q in gastric adenocarcinoma.
Methods: Microsatellite analysis on chromosome 12 was performed on 19 human gastric cancer cell lines using 77 markers, 71 of which were within or distal to 12q21; some portions of this region showed extended regions of homozygosity (ERHs) in 10 of 19 gastric cancer cell lines. In addition, microdissected tumour cells from 76 primary gastric adenocarcinomas were examined using 13 markers of interest implicated by the cell line data; 70% of these showed allelic imbalance (AI) at one or more markers in or distal to 12q21.
Results: Mapping ERHs in the cell lines and sites of AI in the tumours identified three regions that contain putative tumour suppressor genes: region A is located within 2.8 Mb between markers D12S1667 and D12S88; region B, within 1.9 Mb between markers D12S1607 and D12S78; and region C, in 0.74 Mb between markers D12S342 and D12S324. Fluorescence in situ hybridisation (FISH) analysis in two cell lines confirmed that two of the ERHs reflected deletions, not amplifications, of D12S81 in region A and D12S340 in region C. FISH analysis of marker D12S1075 within an ERH containing region B in one cell line showed neither amplification nor deletion. AI on 12q was not associated with prognosis, but was associated with ethnicity of the patient.
Conclusions: These results identify regions on chromosome 12 that appear to contain tumour suppressor genes important in the development of gastric cancer.
Keywords: chromosome 12, gastric cancer, human, fluorescence in situ hybridisation
Cancer develops as a multistep process that includes inactivation of tumour suppressor genes, each of which acts as a brake on cell proliferation, and amplification of oncogenes, which promote cell growth. Microsatellite analysis has been particularly helpful in revealing sites of tumour suppressor genes, through loss of heterozygosity (LOH) studies, using markers with high percentages of heterozygosity. When one allele of a tumour suppressor gene is mutated, the other normal allele frequently becomes lost, often accompanied by duplication of the mutant allele, in addition to surrounding markers, thus generating a homozygous state for markers in the region of the tumour suppressor gene. In the study of primary tumours, LOH is frequently incomplete because of the presence of contaminating normal cells; therefore allelic imbalance (AI) is a more accurate term. AI can also occur if one allele is highly amplified.
“Several studies of gastric adenocarcinoma reported deletions of portions of chromosome 12, but these regions were not well defined, because small numbers of markers were used”
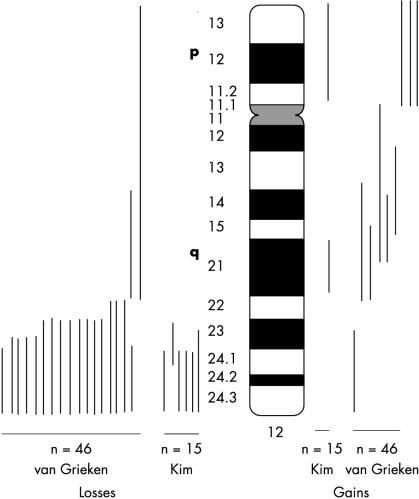
In the study of gastric cancer, the investigation of regions of AI has been a useful approach for the identification of areas of DNA containing tumour suppressor genes (reviewed in Tahara, 19981). Several studies of gastric adenocarcinoma reported deletions of portions of chromosome 12, but these regions were not well defined, because small numbers of markers were used.2–5 Additional evidence for the loss of part of chromosome 12, especially 12q, has been obtained from comparative genomic hybridisation (CGH) studies (fig 1 ▶).6–8 These studies reported losses on the distal end of chromosome 12 of 40–43% of gastric adenocarcinomas examined. We sought to prepare, using a large number of markers, a detailed map of regions of AI on the distal portion of chromosome 12q to identify regions of loss in gastric cancers.
Figure 1.
Comparative genomic hybridisation studies by van Grieken and Kim6,8 show losses in the distal portion of chromosome 12q. In the study by van Grieken et al, 17 of 46 gastric cancers showed deletions on the distal portion of 12q; in the study by Kim et al, six of 15 gastric cancers showed such deletions. Figure 1 ▶ was prepared from diagrams in published reports; however, because of the limits of resolution of CGH, the precise breakpoints on 12q cannot be determined with this technique.
In addition to the study of primary tumours, another approach for analysis is to examine gastric cancer cell lines for extended regions of homozygosity (ERHs). Because the microsatellite markers we used were highly polymorphic (with an observed average rate of heterozygosity of 66%), it is statistically unlikely that a series of adjacent markers would all be homozygous by chance. For example, in a series of five consecutive markers, each of which had a 66% probability of appearing in a heterozygous state in any individual, the probability of all five appearing in a homozygous state is (1 − 0.66)5, or 0.0045. Consequently, the appearance of such ERHs in a cell line is unlikely to be explained by chance and more likely to occur as a result of the same mechanism described for LOH: loss of a portion of DNA flanking a tumour suppressor gene. Thus, microsatellite analysis of gastric cancer cell lines can be a very useful adjunct to the study of primary tumours, for detecting regions of DNA that may contain tumour suppressor genes. We have combined this approach with the analysis of AI of primary tumours for the study of the distal end of the long arm of chromosome 12 in gastric cancer.
MATERIALS AND METHODS
Cell lines
The gastric adenocarcinoma cell lines used were: YCC-1, YCC-2, YCC-3, YCC-5, YCC-6, YCC-7, YCC-10, YCC-11,9 MKN-28, MKN-74, MKN-75,10 SNU-1, SNU-5, SNU-16, N87, KATO-III,11 AGS, RF-1, and Hs746T. The last four cell lines were obtained from the American Type Culture Collection (Rockville, Maryland, USA). Cell lines N87, SNU-1, SNU-5, and SNU-16 were provided by Dr H Oie (National Cancer Institute, Bethesda, USA), and cell lines MKN-28, MKN-74, and MKN-75 were provided by Dr R Lotan (M D Anderson Cancer Center, Houston, USA). Cell lines were maintained in Dulbecco’s minimal essential medium (DMEM), with high glucose (Hs746T), DMEM/F12 (MKN-28, MKN-74, and MKN-75), RPMI-1640 (SNU-1, SNU-5, SNU-16, and N87), Ham’s F12 (AGS), Iscove’s medium (KATO-III), Liebowitz’s L-15 medium (RF-1), or Eagle’s minimal essential medium (all YCC cell lines). All media were purchased from Life Technologies (Bethesda, Maryland, USA), and were supplemented with 10% fetal bovine serum, except for KATO-III, which was supplemented with 20% fetal bovine serum. Media were supplemented with penicillin (50 units/ml)/streptomycin (50 μg/ml) or antibiotic/antimycotic according to the manufacturer’s specifications (Life Technologies). Trypsinised cell pellets (2 × 105 cells) were digested overnight in 40 μl of a proteinase K solution (1 mg/ml, in 50mM Tris/HCl buffer, pH 8.0, with 1mM EDTA and 0.45% Tween 20) at 52°C overnight. Digested materials were heated to 95°C for 15 minutes to denature the proteinase K. This material was used as a template for the polymerase chain reaction (PCR) without additional purification.
Tumours
Formalin fixed, paraffin wax embedded gastrectomy specimens from 76 cases of gastric adenocarcinoma were obtained from hospitals in New Orleans, Louisiana, USA (n = 35), San Antonio, Texas, USA (n = 33), and Rochester, Minnesota, USA (n = 8). The ethnicity, age, and sex of the patients and anatomical sites of the tumours were obtained from medical records or tumour registries. Ethnic groups represented were non-Hispanic white (n = 38), Hispanic (mainly of Mexican descent; n = 18), and African–American (n = 19). The ethnic origin of one patient was unknown. All specimens and patient information were acquired with approval of the institutional review boards of the University of Texas Health Sciences Center at San Antonio or Louisiana State University Health Sciences Center. Survival data were available for 66 patients from Louisiana and Texas. Staging was performed using standard TNM criteria.12 Tumours were graded and classified as intestinal, diffuse, or mixed subtype by an experienced surgical pathologist (JCB) according to the criteria of Laurén.13 Tumours were included in our study only if they could be microdissected, either manually or with laser capture microdissection (LCM) with the Pixcell II or IIe microscope (Arcturus Engineering, Mountain View, California, USA) such that the preparations would include at least 70% tumour nuclei.14 The series of cases omitted diffuse tumours with a low density of tumour cells, and tumours in which profuse inflammatory cells were intimately associated with the tumour cells. According to convention,15 tumours known to display high frequency microsatellite instability, using the Bethesda panel of five markers (BAT25, BAT26, D2S123, D5S346, and D17S250), were omitted from our study, but tumours with low frequency microsatellite instability were included. The details of the analysis for microsatellite instability were reported previously.16
Of the 76 tumours, 69 were microdissected using LCM; seven tumours were manually microdissected. Sections (6 μm thick) were cut in a manner designed to avoid cross contamination from other patients’ specimens during microtomy3; slides were prepared as described previously, for either manual or LCM harvested materials.16 Tumour and benign cells were harvested by LCM using a 30 μm laser spot with 50–60 mWatt power. Cell preparations of 1000–2000 pulses were harvested and digested in 40 μl of a proteinase K solution (as described above for the cell lines) at 52°C overnight. Digested materials were heated at 95°C for 15 minutes to denature the proteinase K. This material was used as a template for PCR without additional purification.
Polymerase chain reaction
DNA templates were amplified with primers for the microsatellite markers listed in fig 2 ▶.17 Annealing temperatures were optimised for each primer pair, and details of PCR conditions are available upon request (bschne@lsuhsc.edu). DNA templates were amplified as described previously,3 except that the cycle number was increased to 40. Control reactions consisted of all components except template. PCR products were separated on 7% polyacrylamide gels with 33% formamide and 33% urea, and bands were visualised with autoradiography without intensifying screens. In all primary tumours, DNA bands amplified from tumours were compared with those amplified from benign cells (always lymphocytes, except for one case in which normal epithelium was used) from the same patient, using approximately equivalent numbers of laser pulses from tumour and normal tissue. AI was evaluated by visual inspection, using as a standard a change in band ratios of 50%, as described by MacGrogan et al.18 For the cell lines, normal tissue from the same patient from whom the cell line was derived was unavailable. Consequently, a statistical argument was used to determine whether the length of a region of homozygosity was considered significant. The product of probabilities of homozygosity for consecutive markers was calculated to determine the probability that the entire region would be homozygous by chance. In practice, the ERHs we identified, consisting of five adjacent markers, gave a probability no greater than 0.0069. Of the 77 markers examined, 71 were located in the region of 12q21 or distal to that. Marker order was determined mainly by the database at the University of California at Santa Cruz (http://genome.ucsc.edu/goldenPath). Numbers such as base position 83969109 refer to base positions on chromosome 12 in the Santa Cruz website, using the November 2002 freeze of the browser, except for marker D12S360, which was placed using the June 2002 freeze. Two markers not listed in the Santa Cruz website (D12S76 and D12S386) were positioned according to data located in the National Center for Bioinformatics website (http://www.ncbi.nlm.nih.gov/genemap98/map.cgi?CHR=12).
Figure 2.
The diagram indicates the positions of markers and extended regions of homozygosity (ERH; shown in grey) in 19 gastric cancer cell lines. 1, homozygous; 2, heterozygous. Del, marker was shown to be deleted in FISH experiments. Not del, equivalent numbers of centromeres and non-centromeric probes were seen in FISH experiments. The markers in boldface were used for the analysis of the tumours shown in fig 5 ▶.
Fluorescence in situ hybridisation
Metaphase chromosome spreads of cell lines KATO-III and MKN-28 were prepared on glass slides using a standard cytogenetic protocol.19,20 The centromere probe was chromosome enumeration probe (CEP) 12 α satellite DNA, which was labelled with Spectrum Orange (Vysis, Downers Grove, Illinois, USA). Bacterial artificial chromosome (BAC) DNA used to identify regions containing markers D12S1075 and D12S81 had addresses RP11–150F20 and RP11–389G16, respectively21; a P1 artificial chromosome (PAC) probe containing marker D12S340 had address RPCI1–157K6. The BAC and PAC DNAs were labelled by nick translation with biotin-14-dATP (Life Technologies). Chromosome spreads were denatured for two minutes in a 70% formamide/2× saline sodium citrate (SSC), pH 7.0, solution. Hybridisation solutions containing 40 ng of the labelled BAC or PAC DNA and 0.5 μl of the CEP DNA in buffer (50% formamide, 10% dextran sulfate, and 2× SSC, pH 7.0) were denatured for five minutes and hybridised overnight in a humid chamber at 37°C, washed, and incubated in avidin conjugated fluorescein isothiocyanate (FITC), at 1.5 mg/ml. Amplification was performed with biotinylated anti-avidin and avidin–FITC. Chromosomes were counterstained using DAPI II (4′,6-diamidino-2-phenylindole; Vysis) and viewed using a Zeiss Axioplan 2 fluorescent microscope equipped with FITC, DAPI, Texas Red, and triple bandpass filter sets. Images were captured using Applied Imaging Probevision software and printed using a Mitsubishi CP800DW colour printer.
Statistical analysis
The relation between AI at the three regions and categorical variables of sex or ethnicity of the patient and anatomical site, stage, TNM, and grade was established using either the χ2 test or Fisher’s exact test. For continuous variables such as age, ANOVA was used to ascertain the significance of the association between those variables and AI at the three sites. Time to event was used to calculate the relative hazard of death given AI status. Kaplan Meier curves were constructed using SPSS (SPSS, Chicago, Illinois, USA), and the Tarone-Ware test was used to test for significance. A Cox proportional hazards model was used to ascertain the significance of AI in the presence of other covariates such as age and stage.
RESULTS
ERHs were identified by the examination of 19 gastric cancer cell lines, using 77 microsatellite markers, 73 of which were on 12q, with 71 markers located at 12q21 or more distal. Beginning at 12q21, markers were located on average 0.9 Mb apart. Cell lines were scored as to whether one or two alleles were visible, and results are summarised in fig 2 ▶. The microsatellite markers used in this analysis were mostly highly polymorphic, averaging 66% observed rates of heterozygosity (or 34% homozygosity), such that five consecutive markers showing only one allele have an average probability of occurring by chance of 0.0045 or less. Therefore, regions where five or more consecutive markers showed only one allele were likely to have been deleted from the chromosome. Ten of the 19 cell lines contained ERHs on 12q; three cell lines showed ERHs over most of distal 12q. Most useful, however, were smaller ERHs, such as those found in cell lines AGS, Hs746T, and KATO-III.
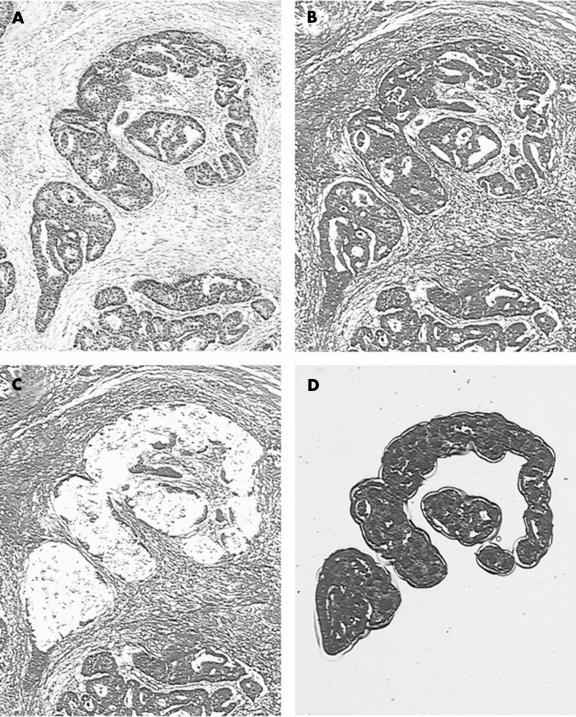
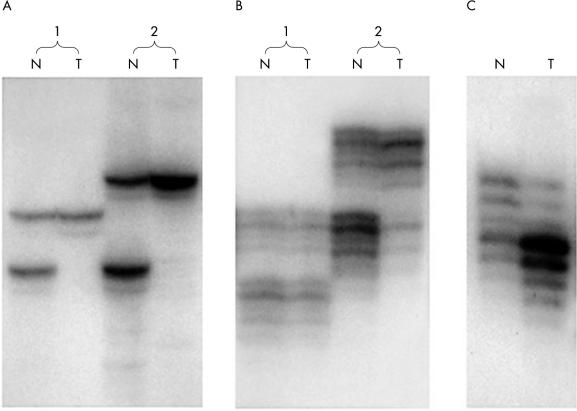
The isolation of tumour cells and lymphocytes from 69 of the 76 patients was performed with LCM (fig 3 ▶). This technique allows the isolation of very pure populations of tumour cells, so that the loss of one allele was sometimes almost complete (fig 4A ▶) Other examples of microsatellite analysis in tumour–normal pairs are shown in fig 4B,C ▶. DNA from cell lines was amplified with all 77 markers, but not all markers amplified well from templates retrieved from paraffin wax blocks. From the analyses of the ERHs in the cell lines, 13 robustly amplifying markers were selected within or near regions of interest.
Figure 3.
Laser capture microdissection images. (A) The stained and coverslipped section is used for the identification of tumour cells (haematoxylin and eosin stain). (B) The next section to that shown in (A) is dark because of the lack of a coverslip (methyl green stain). (C) Tumour cells are specifically removed. (D) The tumour cells have been captured to provide DNA as a template for the polymerase chain reaction.
Figure 4.
(A) In laser capture microdissection harvested tumours, which have very little contamination by normal DNA, near complete absence of one of the sets of bands may be seen in the tumours, as shown in the tumour lane in both cases. Marker, D12S1051. (B) Amplification of DNA from normal (N) and tumour (T) cells at marker D12S105 in two normal/tumour pairs. In case 1, tumour DNA shows no difference from normal DNA at this marker. In case 2, allelic imbalance is indicated because the lower set of bands is decreased in density. Faint reactions were not scored. For example, if light bands were visible, but neither upper nor lower bands exceeded the density seen in the lower bands of the tumour in case 2, that reaction would not be scored. (C) Two sets of bands of approximately equal density were amplified from the normal DNA, whereas we sometimes observed a decrease in the density of one band from the tumour DNA, accompanied by intensification of the density of the other band, consistent with a duplication of the portion of the chromosome containing the putative mutant gene. Marker, D12S1128.
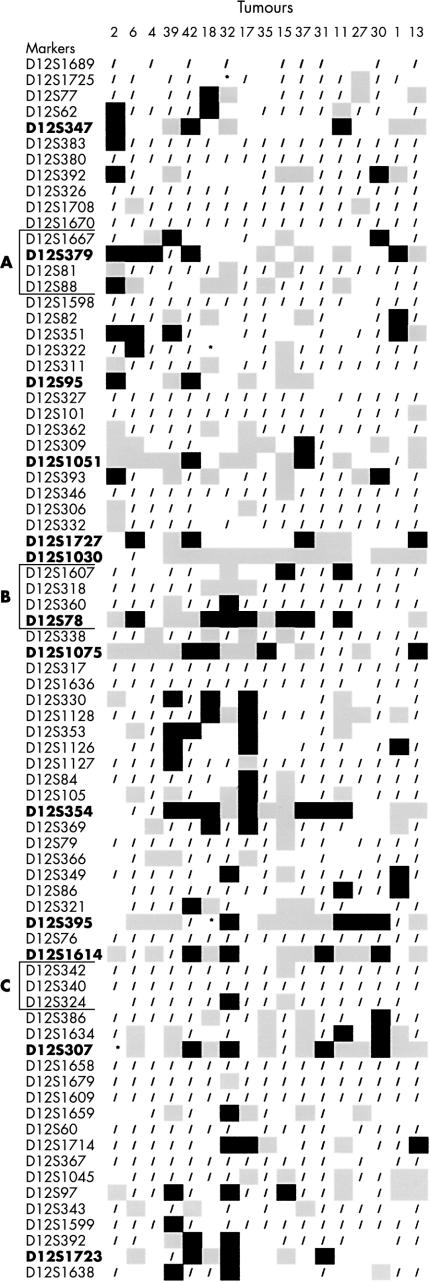
Microsatellite analysis of 76 microdissected tumours and matched lymphocyte control DNA was performed for the 13 markers, and results are summarised in fig 5 ▶. The percentage of informative cases showing AI at each marker is listed on the right side of fig 5 ▶, in addition to the numbers of tumours that are informative at that marker and percentages of observed heterozygosity. Eighteen tumours had AI at most of the markers examined, whereas 23 of the tumours had no losses at the markers examined. From the remaining tumours, in cases where abundant material was available, additional microsatellite analyses were performed (fig 6 ▶).
Figure 5.
Seventy six tumours were subjected to microsatellite analysis using 13 markers within or near regions A, B, or C. These 13 markers and their locations with respect to regions A, B, and C are identified in boldface in the chromosome diagram in fig 2 ▶. At the right side of the figure are percentages of allelic imbalance (% AI), numbers of informative cases (#inf), and observed percentages of heterozygosity (%het). Black squares, AI; open squares, heterozygous markers with no AI; grey squares, non-informative (homozygous); /, no data; *, microsatellite instability.
Figure 6.
Some of the tumours showing partial losses from data shown in fig 5 ▶ were subjected to additional analysis, where available material permitted. Black squares, AI; open squares, heterozygous markers with no AI; grey squares, non-informative (homozygous); /, no data; *, microsatellite instability.
From the analysis of tumour and cell line data, we identified three regions—called A, B, and C—which probably contain genes important in gastric cancer. Results from both tumours and cell lines were useful in narrowing down the regions. For example, in region A, data from cell lines implicates the region between markers D12S1670 and D12S1598, which is homozygous in eight of 17 cell lines, including three cell lines with small deletions of four to 11 markers each. However, tumour data suggest that the boundaries of the region of loss are markers D12S1667 (base position 83969109 on chromosome 12 from the Santa Cruz website, November 2002 freeze) and D12S88 (base position 86787581), a physical distance of 2.8 Mb. The ERH in region A in KATO-III covers only four markers, with a probability of 0.0083 of occurring by chance. However, fluorescent in situ hybridisation (FISH) analysis confirms that marker D12S81 is deleted in this cell line (fig 7A ▶). In region A, we found the greatest percentage of AI (41%) at D12S379, at base position 85247191 on 12q21.31.
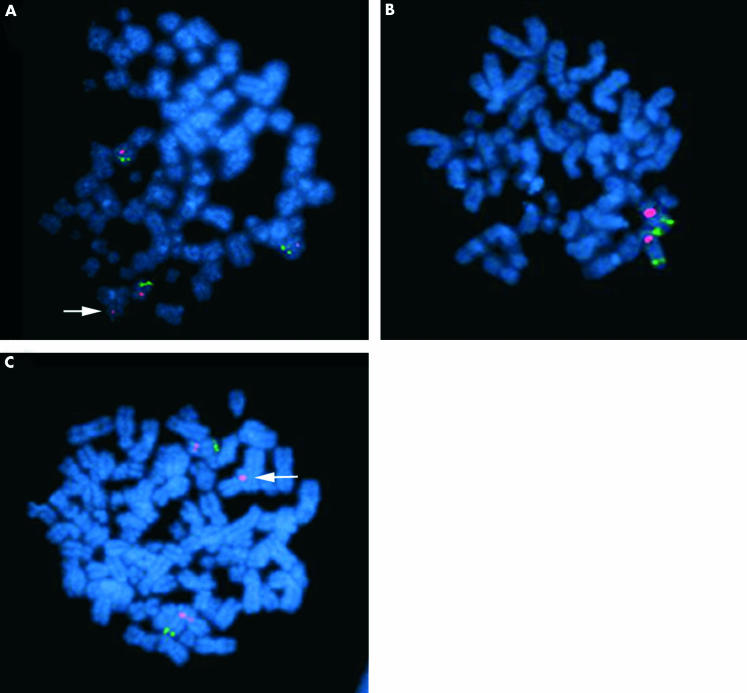
Figure 7.
(A) Fluorescence in situ hybridisation (FISH) analysis of the cell line KATO-III shows four copies of the chromosome 12 centromere (red) and three copies of sequence homologous to the bacterial artificial chromosome (BAC), which contains marker D12S81 (green), a marker contained in region A. An arrow indicates the centromere of the copy of chromosome 12 in which the marker has been deleted. (B) FISH analysis of cell line MKN-28 shows two copies of chromosome 12, indicated by the centromere probe (red). The sequence homologous to a BAC containing marker D12S1075 is present on both chromosomes 12 (green). D12S1075 is contained within an extended region of homozygosity that encompasses both regions A and B. (C) FISH analysis of cell line KATO-III indicates copies of the centromeric region of chromosome 12 stained red and copies of the DNA containing the marker D12S340, contained in region C, stained green. The arrow indicates the centromere of a copy of chromosome 12 in which the marker has been deleted.
Telomeric to region A, in 12q23, we noted three ERHs of 10 or more markers, in cell lines Hs746T, MKN-28, and SNU-16. The data from the three cell lines suggest a smallest region of overlap between D12S346 and D12S1030. However, this region is not supported by tumour data. Elimination of cell line SNU-16 from the analysis produces a pattern more consistent with that seen in the tumours (fig 6 ▶; tumours 6, 18, 32, 17, 35, 15, 37, 11, and 13). Data from these tumours and from cell lines Hs746T and MKN-28 define a region of 1.78 Mb, which we call region B, extending between D12S1607 (base position 102141863) and D12S78 (base position 104042753), a physical distance of 1.9 Mb. FISH analysis for D12S1075, which lies within the ERH containing region B in cell line MKN-28, indicated an equivalent number of centromeres and copies of the marker (fig 7B ▶). Such data are inconsistent with amplification or with deletion alone, but not with deletion followed by abnormal segregation.
In 12q24 in three cell lines (AGS, KATO-III, and YCC-5), there are ERHs that define region C, lying between D12S342 (base position 125525016) and D12S324 (base position 126266436), covering a physical distance of 0.74 Mb. In the cell line KATO-III, FISH analysis indicated that D12S340, which falls within this region (at base position 125783088), was deleted (fig 7C ▶). This interpretation was also consistent with the tumour data, although markers D12S342, D12S340, and D12S324 did not amplify well from the templates retrieved from paraffin wax. Note tumours 11, 24, and 30, which apparently have deletions spanning this region, as shown in fig 6 ▶.
There were no significant univariate associations between AI at the 12q markers examined and sex or age of the patient, stage, grade, T, N, M designation, or morphological subtype of the tumour. However, ethnicity was associated with AI on 12q (table 1 ▶). Tumours from patients from the minority ethnic groups (Hispanics and African–Americans) were more likely to show AI on 12q than were tumours from white patients (p = 0.045; table 1 ▶). This may be related to the site of the tumour because minority patients were more likely to have tumours in the antrum (p = 0.0225), and there was a trend for antral tumours to show AI at 12q (p = 0.059; table 2 ▶).
Table 1.
Allelic imbalance (AI) on chromosome 12q according to ethnicity
| AI on any portion of chromosome 12q | |||
| Negative | Positive | p Value | |
| Ethnicity (n=75) | |||
| White | 16 | 22 | |
| Hispanic | 4 | 14 | 0.045* |
| African-American | 3 | 16 | |
*Fisher’s exact test, when data are compressed into a 2×2 table of white versus minority, compared for 12q status.
Table 2.
Anatomical site of tumour compared with allelic imbalance (AI) on chromosome 12q and ethnicity
| Cardia | Body | Antrum | p Value | |
| Ethnicity (n=49) | ||||
| White | 10 | 6 | 8 | 0.0019* |
| Hispanic | 0 | 4 | 7 | |
| African–American | 1 | 3 | 10 | |
| AI on 12q (n=50) | ||||
| Absent | 4 | 6 | 4 | 0.059† |
| Present | 7 | 7 | 22 |
The anatomical site of origin of the tumour could be determined for 66% of the 76 tumours.
*Fisher’s exact test, when the table is collapsed into a 2×2 table comparing white with minority for anatomical site (cardia versus body+antrum). If the grouping is (cardia+body) versus antrum, p=0.023. †Fisher’s exact test, when table is collapsed into a 2×2 table comparing (cardia+body) versus antrum for AI on chromosome 12q.
The median survival for those patients whose tumour DNA demonstrated AI on 12q was 285 days, compared with 351 days for patients whose tumours had no AI on 12q. These numbers are not significantly different.
As expected, AI at each of the three sites on 12q was significantly associated with AI at the other two regions on chromosome 12q (p < 0.001 in each case); in addition, AI at the 12q sites was also associated with AI at any of three markers (D2S123, D5S546, or D17S250) on other chromosomes performed in a previous analysis (p = 0.036).16
DISCUSSION
The combined analysis of ERHs in cell lines and AI in tumours has been useful in refining regions that probably contain tumour suppressor genes in other tumour types, such as melanomas.22 Cell lines offer the advantage of providing virtually limitless amounts of high quality DNA, with no contamination by normal cells. Tumour analysis provides confirmation of the clinical relevance of results obtained with the artificial cell line system, and may reveal interesting associations of genetic alterations and clinical or pathological features of the tumours.
Losses of distal portions of chromosome 12q in gastric cancers have been detected in several large comparative genomic hybridisation (CGH) studies, at frequencies of 37–47%.6–8 Curiously, other CGH studies found very few or no 12q losses in gastric cancers.23–30 Whether the difference in results relate to the populations studied or to the limitations in sensitivity of CGH studies is unclear.
In studies using higher resolution techniques, such as LOH analyses, the involvement of losses on chromosome 12q in the development of some gastric cancers has been reported. Fey et al found that 55% of gastric cancers showed deletions using probes for a region of 12q24–qter.2 Sano found 31% loss at a single marker at 12q24.3–qter.4 In a different set of gastric cancers, we previously reported 38% AI at marker D12S78 (n = 48).3 Schmutte et al reported LOH of 42% at a marker internal to the thymine DNA glycosylase (TDG) gene on 12q22–q24.1.5 None of these studies used sufficient numbers of markers to define the borders of regions of LOH or AI.
Using a larger set of markers, we have identified three regions of loss on chromosome 12q in DNA from gastric cancer cell lines and gastric tumours. Some of these regions have been implicated in the development of other tumour types. Regions A and B are near regions identified by Kimura et al as being affected in pancreatic cancer.31,32 Using 40 pancreatic tumours, 19 pancreatic cancer cell lines, and 24 markers on chromosome 12q, Kimura et al reported two regions of loss: one between D12S81 and D12S1719 (67.5%) and another between D12S360 and D12S78 (60%). These overlap our regions A, at 12q21.31 and B, at 12q23.3.
A candidate gene near region B is thymine DNA glycosylase (TDG), found at base position 104240443, at 12q23.3. This gene encodes a protein that initiates T : G mismatch repair by excising the mismatched T.33 Schmutte et al found 42% LOH in gastric cancers at a marker within intron 8 of the TDG gene. No mutations were found in the TDG gene in gastric cancers.5 However, this study did not examine TDG expression or promoter methylation, and so could not exclude some other mechanism as a means of inactivation of a second allele. Yatsuoka et al reported decreases in the degree of TDG mRNA expression in pancreatic cancer cell lines.34
“A candidate gene near region B is thymine DNA glycosylase, found at base position 104240443, at 12q23.3”
Region C is located near a putative gene frequently affected in carcinoma ex pleomorphic adenomas, also called malignant mixed tumours of the salivary gland. Marker D12S395 (base position 119615138) showed LOH in 39% of these tumours.35 In a small group of paired adenomas and carcinomas, 50% of both cell types showed LOH, suggesting that loss at this site was an early event in tumour development.
When we combined Hispanic and African–American ethnic groups and compared them with non-Hispanic whites, we found an association of ethnicity with AI on chromosome 12q. We speculate that this association arises from differences in Helicobacter pylori infection patterns associated with socioeconomic status in these minority populations. In a study of Texan ethnic groups, Malaty et al found a higher incidence of H pylori infection among Hispanics and African–Americans than among whites.36 Helicobacter pylori infection is associated with damage to the lower part of the stomach.37 Association between ethnicity and tumour site has been reported in other American populations also.38 In our study, the Hispanic and black patients more frequently had tumours originating in the antrum than did white patients, and there was a strong trend for antral tumours to show AI at chromosome 12q.
Gastric cancer originating in the antrum and associated with H pylori infection appears to be a different disease to cancer of the gastric cardia; therefore, it is not surprising that these two tumour types might tend to have different genetic alterations. Other authors have previously shown differences between cardia and non-cardia gastric cancers in losses of chromosomes 4q and 14q and in gains in chromosomes 2p, 7p, 17q, and 20q.30,39 Mutations in the p53 gene occur more frequently in cardia cancers than in cancers from the lower part of the stomach.40,41
Our findings suggest that multiple tumour suppressor genes on chromosome 12 may be involved in the development of some gastric cancers. We anticipate that the identification of the specific genes involved will provide avenues for better understanding of the molecular mechanisms of gastric carcinogenesis.
Take home messages.
We identified three regions on chromosome 12 that contain putative tumour suppressor genes that are important in the development of gastric cancer
These were: region A, which is located within 2.8 Mb between markers D12S1667 and D12S88; region B, within 1.9 Mb between markers D12S1607 and D12S78; and region C, in 0.74 Mb between markers D12S342 and D12S324
The identification of the specific genes involved will hopefully improve our understanding of the molecular mechanisms of gastric carcinogenesis
Acknowledgments
We thank J Finley for skilful assistance with cell culture. We thank Drs D Bradburn, D Bostwick, W Hinchey, and E Averyt for assistance in obtaining specimens. We thank Dr R Lotan for donating cell lines MKN-28, MKN-74, and MKN-75, and Dr H Oie for providing cell lines N87, SNU-1, SNU-5, and SNU-16. We wish to acknowledge the assistance of Ms J Johnson and the staff of the tumour registries of Touro Infirmary, St Luke’s Baptist Hospital and the Audie Murphy Memorial Veterans Administration Hospital, especially M Roberts, A Burt, and B Kirk. This work was supported by NCI grant CA63308, the Health Excellence Fund of the Board of Regents of the State of Louisiana, and in part by the San Antonio Cancer Institute’s NCI Cancer Center Support Grant P30 CA54174.
Abbreviations
AI, allelic imbalance
BAC, bacterial artificial chromosome
CEP, chromosome enumeration probe
CGH, comparative genomic hybridisation
DMEM, Dulbecco’s minimal essential medium
ERH, extended region of homozygosity
FISH, fluorescence in situ hybridisation
FITC, fluorescein isothiocyanate
LCM, laser capture microdissection
LOH, loss of heterozygosity
PAC, P1 artificial chromosome
PCR, polymerase chain reaction
SSC, saline sodium citrate
REFERENCES
- 1.Tahara E. Molecular mechanism of human stomach carcinogenesis implicated in Helicobacter pylori infection. Exp Toxicol Pathol 1998;50:375–8. [DOI] [PubMed] [Google Scholar]
- 2.Fey MF, Hesketh C, Wainscoat JS, et al. Clonal allele loss in gastrointestinal cancers. Br J Cancer 1989;59:750–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider BG, Pulitzer DR, Brown RD, et al. Allelic imbalance in gastric cancer: an affected site on chromosome arm 3p. Genes Chromosomes Cancer 1995;13:263–71. [DOI] [PubMed] [Google Scholar]
- 4.Sano T, Tsujino T, Yoshida K, et al. Frequent loss of heterozygosity on chromosomes 1q, 5q, and 17p in human gastric carcinomas. Cancer Res 1991;51:2926–31. [PubMed] [Google Scholar]
- 5.Schmutte C, Baffa R, Versonese LM, et al. Human thymine-DNA glycosylase maps at chromosome 12q22–q24.1: a region of high loss of heterozygosity in gastric cancer. Cancer Res 1997;57:3010–15. [PubMed] [Google Scholar]
- 6.Van Grieken NCT, Weiss MM, Meijer GA, et al. Helicobacter pylori-related and -non-related gastric cancers do not differ with respect to chromosomal aberrations. J Pathol 2000;192:301–6. [DOI] [PubMed] [Google Scholar]
- 7.zur Hausen A, van Grieken NC, Meijer GA, et al. Distinct chromosomal aberrations in Epstein-Barr virus-carrying gastric carcinomas tested by comparative genomic hybridization. Gastroenterology 2001;121:612–18. [DOI] [PubMed] [Google Scholar]
- 8.Kim YH, Kim NG, Lim JG, et al. Chromosomal alterations in paired gastric adenomas and carcinomas. Am J Pathol 2001;158:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rha SY, Noh SH, Kwak HJ, et al. Comparison of biological phenotypes according to midkine expression in gastric cancer cells and their autocrine activities could be modulated by pentosan polysulfate. Cancer Lett 1997;118:37–46. [DOI] [PubMed] [Google Scholar]
- 10.Motoyama T, Hojo H, Watanabe H. Comparison of seven cells lines derived from human gastric carcinomas. Acta Pathol Jpn 1986;36:65–83. [DOI] [PubMed] [Google Scholar]
- 11.Sekiguchi M, Sakakibara K, Fujii G. Establishment of cultured cell lines derived from a human gastric carcinoma. Jpn J Exp Med 1978;48:61–8. [PubMed] [Google Scholar]
- 12.Fleming ID, Cooper JS, Henson DE, et al. AJCC cancer staging handbook. Philadelphia, New York: Lippincott-Ravin, 1998.
- 13.Laurén P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. Acta Pathol Microbiol Scand 1965;64:31–49. [DOI] [PubMed] [Google Scholar]
- 14.Emmert-Buck M, Bonner R, Smith PL, et al. Laser capture microdissection. Science 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 15.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–57. [PubMed] [Google Scholar]
- 16.Schneider BG, Bravo JC, Roa JC, et al. Microsatellite instability, prognosis and metastasis in gastric cancers from a low-risk population. Int J Cancer 2000;89:444–52. [DOI] [PubMed] [Google Scholar]
- 17.Dib C, Faure S, Fizames C, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 1996;380:A1–138. [DOI] [PubMed] [Google Scholar]
- 18.MacGrogan D, Levy A, Bostwick D, et al. Loss of chromosome arm 8p in prostate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer 1994;10:151–9. [DOI] [PubMed] [Google Scholar]
- 19.Moorehead PS, Nowell PC, Mellman WJ, et al. Chromosome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res 1960;20:613–16. [DOI] [PubMed] [Google Scholar]
- 20.Ikeuchi TP. Inhibiting effect of ethidium bromide on mitotic chromosome condensation and its application to high resolution chromosome banding. Cytogenet Cell Genet 1984;38:56–61. [DOI] [PubMed] [Google Scholar]
- 21.Osoegawa K, Woon PY, Zhao B, et al. An improved approach for the construction of bacterial artificial chromosome libraries. Genomics 1998;52:95–100. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg EK, Glendening JM, Karanjawala Z, et al. Localization of multiple melanoma tumor-suppressor genes on chromosome 11 by use of homozygosity mapping-of-deletions analysis. Am J Hum Genet 2000;67:417–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koizumi Y, Tanaka S, Mou R, et al. Changes in DNA copy number in primary gastric carcinomas by comparative genomic hybridization. Clin Cancer Res 1997;3:1067–76. [PubMed] [Google Scholar]
- 24.Larramendy ML, El-Rifai WE, Kokkola A, et al. Comparative genomic hybridization reveals differences in DNA copy number changes between sporadic gastric carcinomas and gastric carcinomas from patients with hereditary nonpolyposis colorectal cancer. Cancer Genet Cytogenet 1998;106:62–5. [DOI] [PubMed] [Google Scholar]
- 25.Kokkola A, Monni O, Puolakkainen P, et al. Presence of high-level DNA copy number gains in gastric carcinoma and severely dysplastic adenomas but not in moderately dysplastic adenomas. Cancer Genet Cytogenet 1998;107:32–6. [DOI] [PubMed] [Google Scholar]
- 26.Nessling M, Solinas-Toldo S, Wilgenbus KK, et al. Mapping of chromosomal imbalances in gastric adenocarcinoma revealed amplified protooncogenes MYCN, MET, WNT2, and ERBB2. Genes Chromosomes Cancer 1998;23:307–16. [DOI] [PubMed] [Google Scholar]
- 27.Sakakura C, Mori T, Sakabe T, et al. Gains, losses, and amplifications of genomic materials in primary gastric cancers analyzed by comparative genomic hybridization. Genes Chromosomes Cancer 1999;24:299–305. [DOI] [PubMed] [Google Scholar]
- 28.Guan, XY, Fu, SB, Xia, JC, et al. Recurrent chromosome changes in 62 primary gastric carcinomas detected by comparative genomic hybridization. Cancer Genet Cytogen 2000;123:27–34. [DOI] [PubMed] [Google Scholar]
- 29.Koo, SH, Kwon, KC, Shin, SY, et al. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence in situ hybridization studies. Cancer Genet Cytogenet 2000;117:97–103. [DOI] [PubMed] [Google Scholar]
- 30.Wu, M-S, Chang, M-C, Huang, S-P, et al. Correlation of histologic subtypes and replication error phenotype with comparative genomic hybridization in gastric cancer. Genes Chromosomes Cancer 2001;30:80–6. [DOI] [PubMed] [Google Scholar]
- 31.Kimura M, Abe T, Sunamura M, et al. Detailed deletion mapping on chromosome arm 12q in human pancreatic adenocarcinoma: identification of a 1-cM region of common allelic loss. Genes Chromosomes Cancer 1996;17:88–93. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M, Furukawa T, Abe T, et al. Identification of two common regions of allelic loss in chromosome arm 12q in human pancreatic cancer. Cancer Res 1998;58:2456–60. [PubMed] [Google Scholar]
- 33.Hardland U, Bentele M, Lettieri T, et al. Thymine DNA glycosylase. Prog Nucleic Acid Res Mol Biol 2001;68:235–53. [DOI] [PubMed] [Google Scholar]
- 34.Yatsuoka T, Furukawa T, Abe T, et al. Genomic analysis of the thymine-DNA glycosylase (TDG) gene on 12q22–q24.1 in human pancreatic ductal adenocarcinoma. Int J Pancreatol 1999;25:97–102. [DOI] [PubMed] [Google Scholar]
- 35.El-Naggar AK, Callender D, Coombes MM, et al. Molecular genetic alterations in carcinoma ex-pleomorphic adenoma: a putative progression model? Genes Chromosomes Cancer 2000;27:162–8. [PubMed] [Google Scholar]
- 36.Malaty HM, Evans DG, Evans DJ, et al. Helicobacter pylori in Hispanics: comparison with blacks and whites of similar age and socioeconomic class. Gastroenterology 1992;103:813–16. [DOI] [PubMed] [Google Scholar]
- 37.Forman D. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case–control studies nested within prospective cohorts. Gut 2001;49:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stemmermann GN, Nomura AMY, Kolonel LN, et al. Gastric carcinoma. Pathology findings in a multiethnic population. Cancer 2002;95:744–50. [DOI] [PubMed] [Google Scholar]
- 39.El-Rifai W, Frierson HFJ, Moskaluk CA, et al. Genetic differences between adenocarcinomas arising in Barrett’s esophagus and gastric mucosa. Gastroenterology 2001;121:592–8. [DOI] [PubMed] [Google Scholar]
- 40.Tolbert D, Fenoglio-Preiser C, Noffsinger A, et al. The relation of p53 gene mutations to gastric cancer subsite and phenotype. Cancer Causes Control 1999;10:227–31. [DOI] [PubMed] [Google Scholar]
- 41.Rugge M, Shiao YH, Busatto G, et al. The p53 gene in patients under the age of 40 with gastric cancer: mutation rates are low but are associated with a cardiac location. Mol Pathol 2000;53:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]