Abstract
Aims: To describe mutations in the PAX6 gene in five patients with aniridia from three unrelated families.
Methods: The PAX6 gene was analysed using single stranded conformational polymorphism analysis and direct sequencing.
Results: In one family, three individuals from two generations had aniridia, whereas in each of the other families only one member was affected. The first patient had the heterozygous Q221X (1023C → T) nonsense mutation in exon 8. The same mutation was found in his mother and sister. Another patient had a heterozygous Q297X (1252C → T) mutation in exon 10. The third patient carried a heterozygous IVS5+2T → C mutation leading to aberrant splicing of mRNA.
Conclusions: These findings provide further examples of haploinsufficiency of PAX6 in aniridia.
Keywords: aniridia, PAX6 mutations, eye malformations
A niridia is a panocular disorder denominated after noticeable iris hypoplasia. Complications include cataracts, corneal vascularisation, and glaucoma, and affected patients often have severe visual impairment. The features may range from almost complete absence of the iris, through enlargement and irregularity of the pupil mimicking a coloboma, to small defects in the anterior layer noticeable only on transillumination. Aniridia may occur as an isolated feature or in combination with various syndromes. The disease can be inherited as an autosomal dominant trait and has an incidence of about 1/80 000.1 About one third of all cases of aniridia are sporadic.
Mutations of the PAX6 gene, located on chromosome 11p13, are responsible for about 80% of cases of aniridia in humans.2 In addition, PAX6 mutations are also found in patients with a variety of clinically distinct autosomal dominantly inherited congenital eye malformations,3 such as heterogeneous anterior segment malformations related to Peter’s anomaly and Rieger’s anomaly,4 keratitis,5 and foveal hypoplasia.6 Loss of function of the mutant allele is found in almost all mutations and more than 80% of exonic substitutions result in nonsense codons. About 30% of identified PAX6 mutations are deletion or insertion events, 28% are C → T changes at CpG dinucleotides, and 20% are splicing errors.3 The PAX6 mutation database (http://pax6.hgu.mrc.ac.uk) contains more than 200 different mutations.
“Mutations are found throughout the gene, so that extensive investigation is required in each case”
The PAX6 gene encodes a transcriptional regulator containing two DNA binding domains, a paired domain and a paired-type domain, a link domain, and a proline/serine/threonine rich domain.7 The human PAX6 gene consists of 14 exons, the initiation codon is in exon 4 and the termination codon in exon 13. The protein is 422 amino acids in length and a second PAX6 isoform, derived from alternative splicing, has a short insertion in the paired domain.8 Mutations are found throughout the gene, so that extensive investigation is required in each case.3
MATERIAL AND METHODS
DNA samples
We analysed PAX6 mutations in five patients from three families with aniridia after informed consent. These were the only patients with this clinical condition and therefore no affected patient was excluded from investigation. Genomic DNA was prepared by a standard procedure from isolated leucocytes.
PCR-SSCP assay and sequencing
Mutations for PAX6 were detected by single stranded conformational polymorphism (SSCP) analysis and direct sequencing. Polymerase chain reaction (PCR) primers used for the amplification of the 14 exons of PAX6 were synthesised according to the sequence reported by Glaser and colleagues7 and Love et al.9 The PCR conditions and SSCP analysis were as described by Tadokoro et al.10
RESULTS
Table 1 ▶ summarises the mutations detected in the patients with aniridia from three unrelated families. All patients had complete aniridia. All mutations are new and probably lead to the formation of a truncated PAX6 protein.
Table 1.
Summary of six PAX6 mutations in patients with aniridia
| Patient | Sex | Status | Exon | DNA outcome | Protein outcome | Domain |
| DU | Male | Familial | 8 | 1023C→T | Q221X | HD |
| NU | Female | Familial | 8 | 1023C→T | Q221X | HD |
| IU | Female | Familial | 8 | 1023C→T | Q221X | HD |
| LG | Male | Sporadic | 10 | 1252C→T | Q297X | PST |
| DL | Male | Sporadic | Intron 5 | IVS5+2T→C | Unknown | PD |
HD, homeodomain; PD, paired domain; PST, proline/serine/threonine rich domain.
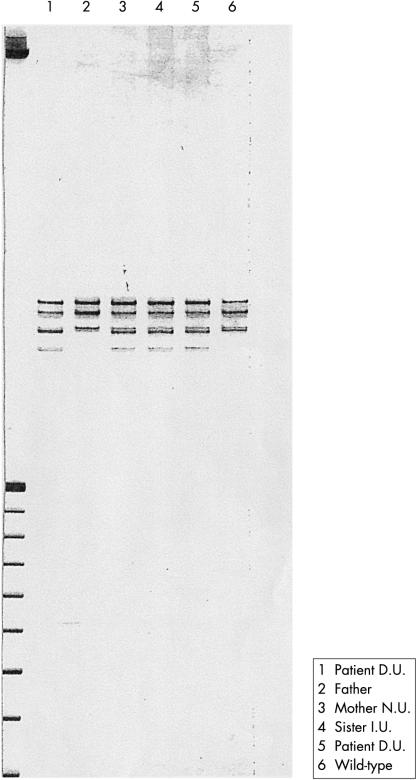
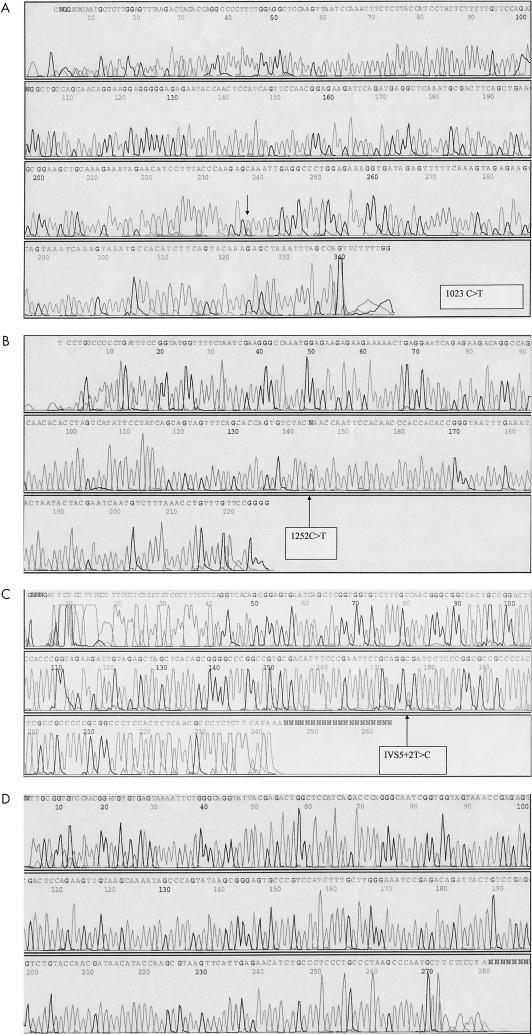
Patient DU carries a heterozygous C → T transition (1023C → T) that changes codon 221 (CAA for glutamine) into a stop codon (TAA) (Q221X), predicting a functional “null allele”. The same mutation was also detected in the patient’s mother and sister, but not in his father (figs 1, 2A ▶ ▶).
Figure 1.
Single stranded conformational polymorphism analysis of exon 8 of the PAX6 gene from patient DU (lanes 1 and 5, mutation 1023C → T), father (lane 2, normal), mother NU (lane 3, mutation 1023C → T), sister IU (lane 4, mutation 1023C → T), and wild type (lane 6).
Figure 2.
(A) Detection of nonsense mutation 1023C → T (Q221X) in patient DU by single stranded conformational polymorphism (SSCP) analysis and direct sequencing. (B) Detection of nonsense mutation 1252C → T (Q297X) in patient LG by SSCP analysis and direct sequencing. (C) Detection of intron mutation IVS5+2T → C in patient DL by SSCP analysis and direct sequencing. (D) SSCP analysis and direct sequencing of a normal control sample.
Patient LG had a heterozygous C → T transition (1252C → T) in exon 10 that alters codon 297 (CAA for glutamine) into a stop codon (TAA) (Q297X) (fig 2B ▶). The mutation was undetectable in the parents, brother, and sister, suggesting that it represents a de novo mutation.
In patient DL, the mutation IVS5+2T → C was detected (fig 2B ▶). This mutation introduces a novel restriction site for the restriction enzyme Cac8 I. Genotyping of DNA samples from the patient’s parents and sister showed no mutation. Thus, the patient probably carries a de novo mutation.
DISCUSSION
Aniridia is a human eye malformation caused by heterozygous null mutations of PAX6, a paired box transcription factor, or microdeletions of chromosome 11p13 that encompass PAX6 and are associated with WAGR syndrome (Wilms’s tumour, aniridia, genitourinary abnormalities, and mental retardation). The inheritance is autosomal dominant with high penetrance but variable expressivity.3 PAX6 mutations have been found in about 80% of patients with aniridia (both sporadic and familial).11 Normal eye development is highly susceptible to the degree of PAX6 expression; haploinsufficiency causes aniridia, and overexpression also leads to developmental defects (microphthalmia) of the eye.12, 13
The PAX6 gene exhibits a very high sequence conservation throughout evolution and as yet undiscovered missense mutations might be associated with unidentified phenotypes.3 PAX6 is involved in the development of the Rathke pouch and early anterior pituitary gland, and its expression controls the established boundaries of somatotrope, lactotrope, and thyrotrope cell types.14 The PAX6 gene is also expressed during the early stages of pancreatic development in the mouse.15 In addition, PAX6 was found to be a key regulator of pancreatic islet hormone gene transcription and pivotal for normal islet development.16 Moreover, it transactivates the glucagon and insulin promoters.
The human PAX6 gene is involved in ocular morphogenesis, and PAX6 mutations have been detected in various types of ocular anomalies, such as aniridia, corneal dystrophy, congenital cataract, and foveal hypoplasia. The gene encodes a transcriptional regulator and produces two alternative splice isoforms that have distinct DNA binding specificities.17, 18 These splice variants are found only in vertebrates.8, 19
Take home messages.
Three patients from one family had the same heterozygous Q221X (1023C → T) nonsense mutation in exon 8
Another patient had a heterozygous Q297X (1252C → T) mutation in exon 10, which leads to a prematurely truncated protein, and the third patient carried a heterozygous IVS5+2T → C mutation, which leads to aberrant mRNA splicing
These findings provide further examples of haploinsufficiency of PAX6 in aniridia
The sole detection of PAX6 mutations in these patients excludes a high risk for the development of Wilms’s tumours/WAGR syndrome
“The detection of PAX6 mutations facilitates comprehensive genetic counselling”
PAX6 mutations are found in a high proportion of patients with aniridia and new technology such as chip sequencing will facilitate high throughput and efficient mutation detection. The Wilms’s tumour susceptibility gene (WT1), which is adjacent to the PAX6 gene, can also be deleted, resulting in a predisposition to Wilms’s tumour as part of the WAGR syndrome. The sole detection of PAX6 mutations in our patients thus excludes a high risk for the development of Wilms’s tumours/WAGR syndrome. Last but not least, the detection of PAX6 mutations facilitates comprehensive genetic counselling.
Acknowledgments
We are grateful to the patients and their families for taking part in this investigation.
Abbreviations
PCR, polymerase chain reaction
SSCP, single stranded conformational polymorphism
WAGR, Wilms’s tumour, aniridia, genitourinary abnormalities, and mental retardation
REFERENCES
- 1.Nelson LB, Spaeth GL, Nowinski TS, et al. Aniridia: a review. Surv Ophthalmol 1984;28:621–42. [DOI] [PubMed] [Google Scholar]
- 2.Ton CCT, Hirvonen H, Miwa H, et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell 1991;67:1059–74. [DOI] [PubMed] [Google Scholar]
- 3.Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mutat 1998;11:93–108. [DOI] [PubMed] [Google Scholar]
- 4.Hanson IM, Fletcher JM, Jordan T, et al. Mutations at the Pax6 locus are found in heterogeneous anterior segment malformations including Peters’ anomaly. Nat Genet 1994;6:168–73. [DOI] [PubMed] [Google Scholar]
- 5.Mirzayans F, Pearce WG, MacDonald IM, et al. Mutation of the PAX6 gene in patients with autosomal dominant keratitis. Am J Hum Genet 1995;57:539–48. [PMC free article] [PubMed] [Google Scholar]
- 6.Azuma N, Nishina S, Yanagisawa H, et al. Pax6 missense mutation in isolated foveal hypoplasia. Nat Genet 1996;13:141–2. [DOI] [PubMed] [Google Scholar]
- 7.Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutation in the human PAX6 gene. Nat Genet 1992;2:232–9. [DOI] [PubMed] [Google Scholar]
- 8.Epstein JA, Glaser T, Cai J, et al. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev 1994;8:2022–34. [DOI] [PubMed] [Google Scholar]
- 9.Love J, Axton R, Churchill A, et al. A new set of primers for mutation analysis of the human PAX6 gene. Hum Mutat 1998;12:128–34. [DOI] [PubMed] [Google Scholar]
- 10.Tadokoro K, Oki N, Sakai A, et al. PCR detection of 9 polymorphisms in the WT1 gene. Hum Mol Genet 1993;2:2205–6. [DOI] [PubMed] [Google Scholar]
- 11.Axton RA, Hanson IM, Love J, et al. Combined SSCP/heteroduplex analysis in the screening for PAX6 mutations. Mol Cell Probes 1997;11:287–92. [DOI] [PubMed] [Google Scholar]
- 12.Fisher E, Scambler P. Human haploinsufficiency—one for sorrow, two for joy. Nat Genet 1994;7:5–7. [DOI] [PubMed] [Google Scholar]
- 13.Schedl A, Ross A, Lee M, et al. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell 1996;86:71–82. [DOI] [PubMed] [Google Scholar]
- 14.Kioussi C, O’Connell S, St-Onge L, et al. Pax6 is essential for establishing ventral–dorsal cell boundaries in pituitary gland development. Proc Natl Acad Sci U S A 1999;96:14378–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St-Onge L, Sosa-Pineda B, Chowdhury K, et al. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 1997;387:406–9. [DOI] [PubMed] [Google Scholar]
- 16.Sander M, Neubuser A, Kalamaras J, et al. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev 1997;11:1662–73. [DOI] [PubMed] [Google Scholar]
- 17.Glaser T, Ton CCT, Mueller R, et al. Absence of PAX6 gene mutations in Gillespie syndrome (partial aniridia, cerebellar ataxia, and mental retardation). Genomics 1994;19:145–8. [DOI] [PubMed] [Google Scholar]
- 18.Glardon S, Callaerts P, Halder G, et al. Conservation of Pax-6 in a lower chordate, the ascidian Phallusia mammillata. Development 1997;124:817–25. [DOI] [PubMed] [Google Scholar]
- 19.Walther C, Gruss P. Pax6, a murine paired box gene, is expressed in the developing CNS. Development 1991;113:1435–49. [DOI] [PubMed] [Google Scholar]