Abstract
Background: A novel single nucleotide polymorphism (SNP), G(−248)A, in the 5′ untranslated region of the BAX promoter and its association with reduced protein expression, progression beyond Rai stage 0, and treatment resistance in chronic lymphocytic leukaemia (CLL) has been reported previously.
Aim: To develop a restriction enzyme analysis (REA) based method for routine detection of BAX promoter SNP in a clinical laboratory.
Methods: The BAX promoter was analysed in duplicate by REA and sequencing in 90 samples (from 45 patients with CLL, 43 controls, and two cell lines). The promoter region was amplified, digested with restriction endonucleases (Aci I and Tau I), and separated by gel electrophoresis.
Results: After digestion, the normal GG genotype samples produced three distinct bands. The homozygous AA replacement abolished the cleavage site, resulting in a single band. Although the heterozygous samples produced three bands, the two smaller visible bands were reduced in intensity (> 50%). The test characteristics of Aci I REA were better than those of Tau I REA, in terms of sensitivity (100% v 77.8%), specificity (98.6% v 92.3%), positive predictive value (95.03% v 87.4%), and negative predictive value (100% v 85.83%).
Conclusions: REA using Aci I is a highly sensitive and specific method for detecting the BAX G(-248)A SNP in CLL.
Keywords: chronic lymphocytic leukaemia, Bax, single nucleotide polymorphism, restriction enzyme analysis, molecular diagnosis
Chronic lymphocytic leukaemia (CLL) is characterised by prolonged survival of CD5+ tumour B cells.1 In general, one group of patients has a relatively stable clinical course, whereas another exhibits faster disease progression.2 Approximately one third of patients with CLL exhibit treatment resistance3 during the course of their disease, although eventually all patients with this incurable disease will become unresponsive to treatment.2–4 Many attempts have been made to predict/prognosticate the disease course. This involves the assessment of traditional risk factors3 and the incorporation of more recent knowledge, such as the VH gene mutation status,5–8 CD38 expression,7–10 and chromosomal aberrations.6 Because prolonged cell survival is the key pathogenic process in CLL, it is not surprising that a relatively apoptosis resistant phenotype is associated with disease progression and inadequate response to treatment.3,11
“Mutations in the promoter and coding regions of the BAX gene have been shown to affect protein expression and function in many cancers”
The Bcl-2 family of proteins plays a central role in the pathogenesis of tumour cell longevity in CLL.11 The Bcl-2 to Bax ratio12 and Mcl-1 expression13 affect the clinical course and the response to treatment. In vitro drug induced apoptosis studies further support the role of the Bcl-2 to Bax ratio14 and Mcl-1.15 Antisense downregulation of Bcl-216 and Bax17 provides additional evidence for their role in the regulation of apoptosis in CLL.
Bax can heterodimerise with antiapoptotic Bcl-218 and Mcl-1,19 and overexpression of these last two proteins may compromise the proapoptotic ability of Bax.18,19 Mutations in the promoter and coding regions of the BAX gene have been shown to affect protein expression and function in many cancers.20,21 Approximately 20% of haemopoietic cell lines (but none from CLL) and 10% of lymphomas harbour mutations in the BAX gene.22 We were the first to identify a novel single nucleotide polymorphism (SNP), G(−248)A, in the BAX promoter—248 bp upstream of the translation site—and demonstrate its association with reduced protein expression, clinical progression beyond Rai stage 0, and treatment resistance in CLL.23 This SNP was not in the p53 binding region. No mutations were detected in the open reading frame, p53 binding sites, or the G(8) tract of exon 3.
Here, we propose a restriction enzyme analysis (REA) based approach for the detection of this BAX promoter SNP.
MATERIALS AND METHODS
Patients and controls
Approval for this research proposal was obtained from the University of Saskatchewan advisory committee on ethics in human experimentation. DNA extracted from 90 consecutive patients, who presented between 1999 and 2000, forms the basis of our study. These samples included peripheral blood samples from 45 consecutive patients with CLL from Saskatoon Health Region hospitals and registered with Saskatoon Cancer Centre, and 43 controls (healthy volunteers or patients with no previous history of cancer). The remaining two samples were two lymphoma cell lines, namely: RL, with the BAX SNP (CRL-2261; American Type Culture Collection (ATCC), Manassas, Virginia, USA), and BC-3, without the SNP (CRL-2277; ATCC). Among the patients with CLL there were 30 men and 15 women (age: range, 29 to 91 years; median, 66). CLL was diagnosed according to revised National Cancer Institute and revised European American classification of lymphoma criteria24,25; all the cases were of B cell CLL immunophenotype. At diagnosis, 25 patients were in Rai stage 0 and 20 in stages I–IV (five in stage I, nine in stage II, two in stage III, and four in stage IV).26 The control population comprised 19 men and 24 women (age: range, 42 to 86 years; median, 71).
Amplification of the BAX promoter region
The lymphocytes were separated from the peripheral blood by Ficoll-Hypaque density gradient separation using Histopaque-1077 (Sigma Diagnostics, St Louis, Missouri, USA), according to the manufacturer’s instructions. DNA was extracted from the separated lymphocytes using Qiagen kit (Qiagen Inc, Mississauga, Ontario, Canada). DNA was measured by determining the optical density (OD) at 260 nm. All DNA was of good quality (range, 1.7–1.95) as determined by the 260/280 OD ratio. The promoter region was amplified by the polymerase chain reaction (PCR) using the following set of primers22: 5′-CGGGGTTATCTCTTGGGC-3′ and 5′-GTGAGAGCC CCGCTGAAC-3′. The PCR mix contained 25 μl of Ready Mix RedTaq PCR reaction mix (20mM Tris/HCl, 100mM KCl, 3mM MgCl2, 0.4mM dNTP), with 1.5 units of Taq DNA polymerase (Sigma), 2 μl of each primer working solution (working concentration of primers: 50 pM/μl), 250 ng of extracted DNA, and PCR grade water to a final volume of 50 μl. Thirty eight cycles of amplification were performed in a Peltier thermal cycler (PTC-200; MS Research, Waltham, Massachusetts, USA). After initial denaturation at 95°C for five minutes, each cycle consisted of denaturation at 94°C for one minute, annealing at 55°C for two minutes, and extension at 72°C for three minutes. A final extension step was performed at 72°C for five minutes.
Sequencing and sequence analysis
PCR products were electrophoresed on 1.6% agarose gel, cut, and purified using the Qiaquick gel extraction kit (Qiagen, Valencia, California, USA). Sequencing was performed in duplicate from purified PCR products obtained in two different PCR reactions on the 377 DNA sequencer ABI Prism (Perkin Elmer Inc, Foster City, California, USA), according to the manufacturer’s instructions. Sequences were compared with those available at public databases; GenBank accession number U17193.
Restriction enzyme analysis
Wisconsin GCG software was used for mapping the BAX promoter. The aim was to identify possible restriction enzymes that would cleave a normal DNA allele and leave a mutant one undigested. Two restriction enzymes, Aci I (New England BioLabs Inc, Mississauga, Ontario, Canada) and Tau I (MBI Fermentas, Burlington, Ontario, Canada) were used to screen the samples for the SNP detection (fig 1 ▶). Aliquots of 10 and 20 μl of PCR products in two different tubes were digested for three hours and overnight at 37°C (for Aci I) or at 55°C (for Tau I) in 50 μl of mix consisting of 5 U of enzyme and 5 μl of buffer 3 for AciI or buffer Y+/Tango for Tau I. Aliquots of 10 and 20 μl of the PCR products were then electrophoresed on a 10% polyacrylamide gel at 165 V for 80 minutes in electrophoresis apparatus Mini-Protean II (Bio-Rad, Mississauga, Ontario, Canada). PCR products were visualised after staining with ethidium bromide using the Gel-Doc 2000 (Bio-Rad); the exact size of the band was determined using the instrument’s software program “Quantity One” (Bio-Rad). As a test for contamination, a control sample consisting of all the reagents except for the DNA was run in each experiment. In addition, for each sample, 10 μl of undigested PCR products were electrophoresed to assess the completeness of digestion. All experiments were done in duplicate. The obtained bands were visualised using Quantity One software and the Gel Doc 2000 system (BioRad).
Figure 1.
(A) Aci I and (B) Tau I restriction enzyme maps for the BAX promoter region.
Statistical analysis
The sensitivity, specificity, predictive values of the test, and the posterior probability were calculated using GraphPad software and p values of 0.05 or less were considered to be significant.
RESULTS
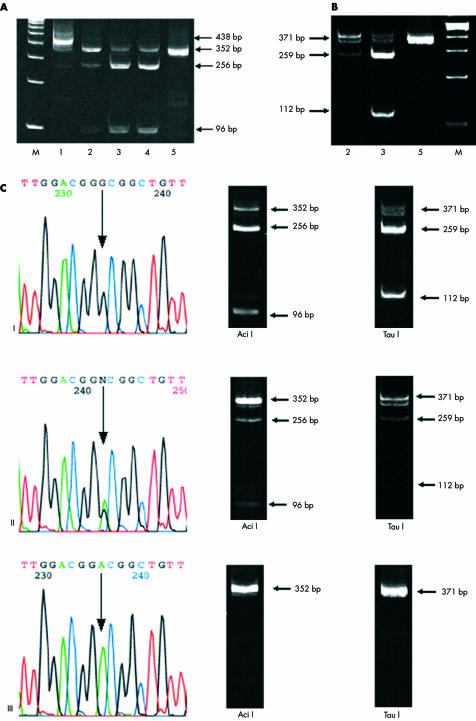
The direct sequence analysis demonstrated that 19 of the 90 samples harboured the BAX promoter SNP. In two of these 19 samples, both alleles were replaced (homozygous) and in the remaining 17 only one allele (heterozygous) was affected. The mapping analysis revealed two potential restriction enzyme sites (for Aci I, which recognises CCGC, and Tau I, which recognises GCC(G)GC) in the region within this SNP, such that the normal allele would be cut whereas the affected allele would not be digested (fig 1 ▶). Figures 2 and 3 ▶ ▶ show the results of REA analysis data using two different restriction enzymes, Aci I and Tau I, and also the direct sequence of the affected segment.
Figure 2.
Restriction enzyme analysis and parallel direct sequencing of polymerase chain reaction (PCR) products. (A) Aci I and (B) Tau I endonuclease cleavage followed by 10% polyacrylamide gel analysis. M, molecular weight marker, 100 bp DNA ladder (Gibco BRL, Burlington, Ontario, Canada). (A) Lane 1, chronic lymphocytic leukaemia (CLL) sample 1, amplified BAX promoter segment before digestion; lanes 2–5, PCR products digested with Aci I; lane 2, CLL sample 2 with a heterozygous single nucleotide polymorphism (SNP); lanes 3 and 4, CLL samples 3 and 4 with no SNP; lane 5, RL cell line with homozygous SNP. (B) PCR products digested with Tau I; lane 2, CLL sample 2 with a heterozygous SNP; lane 3, CLL sample 3 with no SNP; lane 5, RL cell line with homozygous SNP. (C) Left hand column: direct sequence of the BAX promoter region. The control sample (I) shows no alteration, whereas a CLL sample (II) shows a heterozygous SNP, and the RL cell line (III) has a homozygous SNP. Middle and right hand columns: corresponding samples digested with the Aci I and Tau I enzymes, respectively
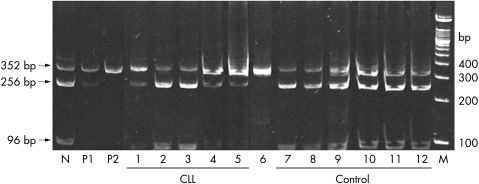
Figure 3.
Restriction enzyme analysis (with Aci I ) for the G(−248)A single nucleotide polymorphism (SNP) in the BAX gene. M, molecular weight marker (100 bp DNA ladder; Gibco BRL); N, control sample showing three distinct bands, 352 256 bp, and 96 bp; P1, positive control with heterozygous SNP showing the 352 bp (major) and 256 bp (minor) bands and an almost invisible 96 bp band; P2, positive control with a homozygous SNP, which abolishes a restriction enzyme site, resulting in a single 352 bp band; lanes 1–5, chronic lymphocytic leukaemia cases; lanes 1, 4, and 5, heterozygous SNP; lanes 2 and 3, no SNP; lane 6, RL showing the homozygous SNP; lanes 7–12, controls without the G(−248)A SNP.
Aci I REA analysis
The amplified segment was 438 bp long. When the PCR product with two normal alleles was digested with the Aci I enzyme it produced nine smaller fragments (fig 1 ▶). On 10% polyacrylamide gel analysis it was visualised as three major bands, 352 bp, 256 bp, and 96 bp in size; the other 5–26 bp segments were too small to be detected on the gel (fig 2A,C ▶). It should be noted that the 256 bp band had the highest intensity.
The promoter G(−248)A SNP resulted in the loss of a restriction site for the Aci I enzyme, and the PCR product from the heterozygous samples showed three bands, with the 352 bp band being the most intense, whereas the 96 bp band became almost invisible (fig 2A,C ▶). Homozygous samples (with both alleles replaced) showed only one 352 bp size band (fig 2A,C ▶).
By means of Aci I REA analysis, the presence of the G(−248)A SNP was detected in 18 of 18 positive cases, and its absence was determined in 68 of 69 negative cases. The results of three of 90 cases were not readable using this analysis. The sensitivity was 100% and the specificity was 98.6%, positive predictive value was 95.03%, negative predictive value was 100%, and the likelihood ratio was 71.429. Figure 3 ▶ shows an example of Aci I REA analysis routinely carried out in our laboratory.
Tau I REA analysis
The Tau I restriction enzyme cut the BAX promoter with two normal alleles into five fragments (fig 1 ▶). These could be visualised on a gel as three major bands (371, 259, and 112 bp), whereas the remaining 16–35 bp bands were too small to be detected; the 259 bp band was most intense (fig 2B,C ▶). Although the heterozygous samples, where only one allele is affected, produced three bands, similar to the normal samples, the 112 band was only just visible (fig 2B ▶). In samples homozygous for the The BAX SNP, the Tau I restriction enzyme site was abolished in both alleles, and only one band, of 371 bp, was seen on the gel (fig 2B,C ▶).
The presence of the G(−248)A SNP was detected in seven of nine positive cases by means of Tau I REA analysis, and its absence was confirmed in 12 of 13 negative cases. The results of five of 27 cases were not readable using this analysis. The sensitivity was 77.8%, specificity was 92.3%, positive predictive value was 87.04%, negative predictive value was 85.83%, and the likelihood ratio was 10.104.
DISCUSSION
CLL is an incurable disease and the treatment of patients with CLL is generally based on specific clinical indications.3 The characterisation of parameters linked to prognosis and clinical outcomes is currently an active research area.4–10,23,27 The identification of patients who will probably experience disease progression and treatment resistance is a useful clinical assessment, because these patients could be targeted for more appropriate treatment strategies. We have demonstrated an association between a novel G(−248)A polymorphism in the BAX promoter and reduced protein expression and certain clinical characteristics; notably, progression beyond Rai stage 0 and resistance to conventional prednisone/chlorambucil based treatment.23 The BAX gene in the patient samples was analysed by sequencing.
The gold standard method for point mutation/polymorphism detection is direct sequencing. Although sequencing is an accurate and a highly reproducible technique, it is labour intensive, expensive, and time consuming. In addition, at present, it is not available in all molecular diagnostics laboratories. Restriction enzyme analysis is a cheaper alternative and relies on the creation/abolition of restriction enzyme sites subsequent to nucleotide changes in the DNA sequence.28 A restriction endonuclease that would cut a normal allele and leave the affected one uncut is preferable to one that would cut the mutant allele, because the use of such an enzyme might give false results owing to incomplete digestion.28
“Although sequencing is an accurate and a highly reproducible technique, it is labour intensive, expensive, and time consuming”
The proposed test includes PCR amplification followed by incubation with an enzyme and then electrophoresis to separate the products. The time required for incubation is three hours. Purification of PCR products is not required because PCR with the above mentioned primers produces only one band, 438 bp long. After digestion with restriction endonucleases, there was no band in the 438 bp size position, indicating that digestion was complete (fig 2A ▶, samples 2 to 5; fig 3 ▶). In addition, the band in the 352 bp location in normal samples after digestion did not represent incomplete digestion because it was seen when either an excess of endonuclease (10 U) was used or digestion was prolonged (48 hours) (data not shown). As with any other laboratory test, positive and negative controls with known sequences should be included when using this molecular technique (fig 3 ▶).
Aci I performed better in the REA than did Tau I, in view of its better sensitivity (100% v 77.8%), specificity (98.6% v 92.3%), positive predictive value (95.03% v 87.4%), and negative predictive value (100% v 85.83%). The proposed REA test with the Aci I enzyme to detect the BAX promoter G(−248)A polymorphism is a highly sensitive, specific, and reproducible test; it is inexpensive, simple, and can be done in any diagnostic molecular laboratory.
Take home messages.
After enzymatic cleavage, the normal GG genotype of the BAX gene produced three distinct bands, the homozygous AA mutation produced a single band, and the heterozygous samples could also be distinguished as a result of band differences
The Aci I enzyme performed better than did Tau I, in terms of sensitivity (100% v 77.8%), specificity (98.6% v 92.3%), positive predictive value (95.03% v 87.4%), and negative predictive value (100% v 85.83%)
Thus, restriction enzyme analysis using Aci I is a highly sensitive, specific, simple, and inexpensive method for detecting the BAX G(−248)A single nucleotide polymorphism in chronic lymphocytic leukaemia, and might be useful for identifying those patients at high risk of clinical progression and treatment resistence
Acknowledgments
This work was financially supported by a grant from the Health Services Utilization and Research Commission, Saskatchewan, Canada.
Abbreviations
ATCC, American Type Culture Collection
CLL, chronic lymphocytic leukaemia
OD, optical density
PCR, polymerase chain reaction
REA, restriction enzyme analysis
SNP, single nucleotide polymorphism
REFERENCES
- 1.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: a bird of a different feather. J Clin Oncol 1999;17:399–408. [DOI] [PubMed] [Google Scholar]
- 2.Molica S. Progression and survival studies in early chronic lymphocytic leukemia. Blood 1991;78:895–9. [PubMed] [Google Scholar]
- 3.Kipps TJ. Chronic lymphocytic leukemia. Curr Opin Hematol 1997;4:268–76. [DOI] [PubMed] [Google Scholar]
- 4.Kay NE, Hamblin TJ, Jelinek DF, et al. Chronic lymphocytic leukemia: hematology 2002. American Society of Hematology Education Program Book. Washington, DC: American Society of Hematology, 2002:193–213. [DOI] [PubMed]
- 5.Hamblin TJ, Davis Z, Gardiner A, et al. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999;94:1848–54. [PubMed] [Google Scholar]
- 6.Krober A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood 2002;100:1410–16. [PubMed] [Google Scholar]
- 7.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999;94:1840–7. [PubMed] [Google Scholar]
- 8.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood 2002;100:1177–84. [PubMed] [Google Scholar]
- 9.Domingo-Domenech E, Doming-Claros A, Gonzalez-Barca E, et al. CD38 expression in B-chronic lymphocytic leukemia: association with clinical presentation and outcome in 155 patients. Haematologica 2002;87:1021–7. [PubMed] [Google Scholar]
- 10.Zupo S, Isnardi L, Megna M, et al. CD38 expression distinguishes two groups of B-cell chronic lymphocytic leukemia with different responses to anti-IgM antibodies and propensity to apoptosis. Blood 1996;88:1365–74. [PubMed] [Google Scholar]
- 11.Bannerji R, Byrd JC. Update on the biology of chronic lymphocytic leukemia. Curr Opin Oncol 2000;12:22–9. [DOI] [PubMed] [Google Scholar]
- 12.Pepper C, Hoy T, Bentley P. Elevated Bcl-2/Bax are a consistent feature of apoptosis resistance in B-cell chronic lymphocytic leukaemia and are correlated with in vivo chemoresistance. Leuk Lymphoma 1998;28:355–61. [DOI] [PubMed] [Google Scholar]
- 13.Kitada S, Anderson J, Akar S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: correlations with in vitro and in vivo chemoresponses. Blood 1998;91:3379–89. [PubMed] [Google Scholar]
- 14.Pepper C, Hoy T, Bentley DP. Bcl-2/Bax ratios in chronic lymphocytic leukaemia and their correlation with in vitro apoptosis and clinical resistance. Br J Cancer 1997;76:935–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osorio LM, De Santiago A, Aguilar-Santelises M, et al. CD6 ligation modulates the Bcl-2/Bax ratio and protects chronic lymphocytic leukemia B cells from apoptosis induced by anti-IgM. Blood 1997;89:2833–41. [PubMed] [Google Scholar]
- 16.Pepper C, Thomas A, Hoy T, et al. Antisense-mediated suppression of Bcl-2 highlights its pivotal role in failed apoptosis in B-cell chronic lymphocytic leukaemia. Br J Haematol 1999;107:611–15. [DOI] [PubMed] [Google Scholar]
- 17.Pepper C, Thomas A, Hoy T, et al. Antisense oligonucleotides complementary to Bax transcripts reduce the susceptibility of B-cell chronic lymphocytic leukaemia cells to apoptosis in a bcl-2 independent manner. Leuk Lymphoma 2002;43:2003–9. [DOI] [PubMed] [Google Scholar]
- 18.Zha H, Aime Sempe C, Sato T, et al. Proapoptotic protein Bax heterodimerizes with Bcl-2 and homodimerizes with Bax via a novel domain (BH3) distinct from BH1 and BH2. J Biol Chem 1996;271:7440–4. [DOI] [PubMed] [Google Scholar]
- 19.Bodrug SE, Aime-Sempe C, Sato T, et al. Biochemical and functional comparisons of Mcl-1 and Bcl-2 proteins: evidence for a novel mechanism of regulating Bcl-2 family protein function. Cell Death Diff 1995;2:173–82. [PubMed] [Google Scholar]
- 20.Meijerink JP, Mensink EJBM, Wang K. Hematopoietic malignancies demonstrate loss-of-function mutations of bax. Blood 1998;91:2991–7. [PubMed] [Google Scholar]
- 21.Rampino N, Yamamoto H, Ionov Y, et al. Somatic frameshift mutations in the Bax gene in colon cancers of the microsatellite mutator phenotype. Science 1997;275:967–9. [DOI] [PubMed] [Google Scholar]
- 22.Peng H, Aiello A, Packham G, et al. Infrequent bax gene mutations in B-cell lymphomas. J Pathol 1998;186:378–82. [DOI] [PubMed] [Google Scholar]
- 23.Saxena A, Moshynska O, Sankaran K, et al. Association of a novel single nucleotide polymorphism, G(−248)A, in the 5′-UTR of BAX gene in chronic lymphocytic leukemia with disease progression and treatment resistance. Cancer Lett 2002;187:199–205. [DOI] [PubMed] [Google Scholar]
- 24.Cheson BD, Bennett JM, Grever, et al. National Cancer Institute-sponsored working group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996;87:4990–7. [PubMed] [Google Scholar]
- 25.Harris NL, Jaffe ES, Stein H, et al. A revised European American classification of lymphoid neoplasms: a proposal from the international lymphoma study group. Blood 1994;84:1361–92. [PubMed] [Google Scholar]
- 26.Rai KP, Patel DV. Chronic lymphocytic leukemia. In: Hoffman R, Benz EJ, Jr, Shattil SJ, et al. Hematology basic principles and practice, 3rd ed. Philadelphia: Churchill Livingstone, 2000:1350–62.
- 27.Ghia P, Guida G, Stella S, et al. The pattern of CD38 expression defines a distinct subset of chronic lymphocytic leukemia (CLL) patients at risk of disease progression. Blood (online) 2002, Oct 24. [DOI] [PubMed]
- 28.Thornhill AR, Snow K. Molecular diagnostics in preimplantation genetic diagnosis. J Mol Diagn 2002;4:11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]