Abstract
Aims: Molecular genetic changes involved in tumorigenesis and malignant transformation of human tumours are novel targets of cancer diagnosis and treatment. This study aimed to analyse the expression of putative tumour suppressor genes, FHIT and WT-1, and tumour rejection genes, BAGE, GAGE-1/2, MAGE-1, MAGE-3, and HAGE (which are reported to be important in human cancers), in salivary gland neoplasms.
Methods: Gene expression was analysed by reverse transcription polymerase chain reaction (RT-PCR) in normal salivary gland tissue and 44 benign and malignant salivary gland tumours.
Results: Aberrant FHIT transcripts were found in one of 38 normal salivary glands, three of 28 adenomas, and two of 16 carcinomas. WT-1 mRNA was detectable in two adenomas and five carcinomas. Immunoblotting showed that WT-1 mRNA expression was associated with raised WT-1 protein concentrations. RT-PCR for detection of BAGE, GAGE, and MAGE gene expression was positive in two adenomas and nine carcinomas, but negative in normal salivary gland tissue. HAGE mRNA was found in two normal salivary glands, 11 benign, and eight malignant tumours.
Conclusions: FHIT mRNA splicing does not appear to be involved in the genesis of salivary gland neoplasms. The upregulation of WT-1 mRNA in tumours of epithelial/myoepithelial phenotype may imply a potential role of WT-1 in the genesis and/or cellular differentiation of these salivary gland tumours. The tumour rejection genes were more frequently, but not exclusively, expressed in malignant salivary gland tumours than in benign neoplasms, although none was suitable as a diagnostic marker of malignancy in salivary gland neoplasms.
Keywords: salivary gland neoplasms, FHIT, WT-1, BAGE, GAGE, MAGE, HAGE
There is great variation in the histopathological tumour types and malignant potential of salivary gland neoplasms. The differential diagnosis between the different tumour types is generally based on morphological criteria alone, because immunohistochemical reactions are of limited value. Molecular genetic changes in cell regulators contribute to the cellular differentiation, malignant transformation, and tumour progression of salivary gland neoplasms. Many of these changes, which may be of diagnostic, prognostic, and therapeutic importance, are still under investigation. These facts prompted us to examine the expression of the FHIT, WT-1, BAGE, GAGE-1/2, HAGE, MAGE-1, and MAGE-3 genes in different benign and malignant salivary gland tumours to determine their potential role as tumour markers in the histological and cytological diagnosis of salivary gland lesions.
“ FHIT is a candidate tumour suppressor gene, whose mRNA is alternatively or abnormally spliced in various human neoplasms”
The FHIT (fragile histidine triad) gene located on chromosome 3p14.2 is composed of 10 exons encoding a 1.1 kb mRNA that is ubiquitously expressed at relatively low levels.1 FHIT is a candidate tumour suppressor gene, whose mRNA is alternatively or abnormally spliced in various human neoplasms.2 The FHIT gene was selected for our study because of its spatial relation to chromosome locus 3p21, which is one of the most frequent chromosomal breakpoints reported in pleomorphic adenomas.3 In addition, the FHIT gene was reported to be a translocation partner in pleomorphic adenomas.4 The putative Wilms’s tumour suppressor gene 1 (WT-1) maps to chromosome 11p13. It is involved in mesenchymal to epithelial cell differentiation processes,5–8 which may be important in the histogenesis of salivary gland tumours of epithelial/myoepithelial differentiation.
The BAGE, GAGE, and MAGE genes form different gene families, all of them encoding tumour rejection antigens, which are recognised by cytotoxic T cells in the context of human leucocyte antigen molecules.9 Recently, another member of the MAGE family, termed HAGE, was identified on chromosome 6q.10 Because these genes were shown to be preferentially activated in tumour tissues of various histological types, including head and neck carcinomas, they have attracted interest as potential diagnostic markers and as targets for tumour specific gene therapy.11 However, their expression has not been investigated in salivary gland neoplasms to date.
MATERIALS AND METHODS
Sample collection
The material investigated consisted of a total of 28 benign and 16 malignant salivary gland neoplasms including two myoepitheliomas, 16 pleomorphic adenomas, 10 cystadenolymphomas (Warthin tumours), in addition to five acinic cell carcinomas, four mucoepidermoid carcinomas, three adenoid cystic carcinomas, two primary squamous cell carcinomas (SCCs), one basal cell adenocarcinoma, and one adenocarcinoma not otherwise specified. All tumours but one adenoidcystic carcinoma from the hard palate arose in the parotid gland. Tumours were classified according to the criteria of the Armed Forces Institute of Pathology classification of salivary gland tumours.12
Fresh tumour tissues from all 44 salivary gland tumours and corresponding normal salivary gland tissues from 38 cases were snap frozen separately in liquid nitrogen directly after surgical removal and stored at −80 C° until further processing.
RNA extraction and cDNA synthesis
Total RNA was isolated from the frozen tissues using the TRI ReagentTM method according to the manufacturer’s instructions (Sigma, Taufkirchen, Germany). This method is an improved version of the single step RNA isolation developed by Chomczynski and Sacchi.13 After extraction, the concentration and purity of RNA were defined photometrically and a 2 μg aliquot of total RNA was reverse transcribed using the First Strand cDNA synthesis kitTM, according to the manufacturer’s instructions (MBI Fermentas, St Leon-Rot, Germany). Random hexamers supplied by the manufacturer were used for the synthesis of cDNA.
PCR amplification
PCR was performed on a DNA thermal cycler ProgeneTM (Thermo-DUX, Wertheim, Germany) using thin walled reaction tubes. Oligonucleotide primers purified by high pressure liquid chromatography were supplied by MWG Biotech (Munich, Germany). All primers corresponded to sequences located in different exons to avoid polymerase chain reaction (PCR) amplification of genomic DNA sequences. For amplification of the β actin, FHIT, and WT-1 genes, 2 μl of cDNA was supplemented with 5 μl 10× PCR buffer, 1 μl each of 10mM dNTP, 20 pM each of primer solutions, 1 unit of thermostable Taq polymerase (Pharmacia, Freiburg, Germany), and water to a final volume of 50 μl.
All probes were prescreened using β actin primers 5′-CTACAATGAGCTGCGTGTGGC-3′ (sense strand) and 5′-CAGGTCCAGACGCAGGATGGC-3′ (antisense strand) to test the integrity of the total RNA extracted. These primers yielded a 240 bp product. Cycling conditions have been described previously.14 A seminested PCR strategy was applied to amplify FHIT cDNA using primer sequences 5′-TCCGTAGTGCTATCTACATCC-3′ (exon 3, sense primer), 5′-TCCTCTGATCTCCAAGAGGC-3′ (exon 9, antisense primer 1), and 5′-CCTCCTTGTCATGTTTCTGG-3′ (exon 9, antisense primer 2). The primers were selected from a published sequence1 and yielded a 570 bp fragment (sense primer–antisense primer 1) and a 533 bp fragment (sense primer–antisense primer 2), respectively. After an initial denaturing step at 95°C for five minutes, cDNA was amplified by 40 cycles comprising three temperature steps. In each cycle, a 30 second melting step at 95°C was followed by an annealing step for one minute at 60°C, and an extension step for two minutes at 72°C. The final PCR cycle was completed with a primer extension step for five minutes at 72°C. The cycling conditions of the second round PCR differed from the first in the annealing temperature (62°C) and the number of cycles (30 cycles).
WT-1 PCR was performed using primers 5′-GAGAGCGATAACCACACAAC-3′ (exon 6, sense primer) and 5′-GATGACCAAACTCCAGCTGG-3′ (exon 10, antisense primer). This primer pair yielded a 529 bp fragment.5 The underlying temperature cycling schedule was initiated by a melting step lasting five minutes at 95°C and terminated by a final extension step at 72°C for seven minutes. Forty PCR cycles were performed. Each run consisted of a melting step (95°C at 45 seconds), a primer annealing step (58°C at 45 seconds), and an extension step (72°C at 30 seconds).
BAGE, GAGE-1/2, HAGE, MAGE-1, and MAGE-3 gene expression was analysed using a One Step RT-PCR kitTM (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. A 100 ng aliquot of total RNA was used for reverse transcription (RT). Primer sequences for BAGE PCR were 5′-TGGCTCGTCTCACTCTGG-3′ (sense primer) and 5′-CCTCCTATTGCTCCTGTTG-3′ (antisense primer), resulting in a 162 bp product. Primer 5′-GACCAAGACGCTACGTAG-3′ (sense primer) and primer 5′-CCATCAGGACCATCTTCA-3′ (antisense primer) were used for amplification of a nucleotide sequence present in GAGE-1 and GAGE-2 RNA. These primers gave a 244 bp product. The GAGE-1 and GAGE-2 PCR procedure was performed as described by Van den Eynde et al.9
The expression of the HAGE gene was analysed using oligonucleotides 5′-CCTTTCAATGTTATCCTGAG-3′ (sense primer) and 5′-CTTCGTCAATCTGAAGAATA-3′ (antisense primer), which gave a 431 bp PCR product. HAGE cDNA was amplified with 35 cycles at 95°C for 45 seconds, 50°C for one minute, and 72°C for one minute, followed by a final extension step at 72°C for seven minutes.
MAGE-1 and MAGE-3 specific sequences were amplified using oligonucleotide pairs 5′-CGGCCGAAGGAACCTGA CCCAG-3′ (exon2, sense primer)/5′-GCTGGAACCCTCACTG GGTTGCC-3′ (exon 3, antisense), which yielded a 421 bp product and 5′-TGGAGGACCAGAGGCCCCC-3′ (exon 2, sense primer)/5′-GGACGATTATCAGGAGGCCTGC-3′ (exon 3, antisense primer), which gave a 715 bp product, respectively. Amplification of MAGE-1 and MAGE-3 cDNA was performed for 35 cycles at 95°C for 45 seconds, 64°C for one minute, and 72°C for 30 seconds, followed by a final extension step at 72°C for seven minutes.
Gel electrophoresis and cDNA sequencing
For visualisation of PCR fragments, a 10 μl aliquot from each PCR assay was separated on a 1.5% (wt/vol) agarose gel containing 0.5 μg ethidium bromide/ml. The gel was photographed using a CCD camera (Biometra, Goettingen, Germany).
For sequencing, the different FHIT PCR products were separated on 1.5% low melting agarose (Biozym, Hameln, Germany) and cut from the gel. The QIAEX II gel extraction kitTM (Qiagen) was used to purify the FHIT cDNA. The BAGE, GAGE-1/2, HAGE, MAGE-1, MAGE-2, and WT-1 amplification products were purified with the QIAquick PCR purification kitTM (Qiagen).
Isolated PCR fragments (200 ng aliquots) were labelled with the PRISM Ready Dye Deoxy terminator cycle sequencing kitTM (Applied Biosystems, Weiterstadt, Germany), according to the manufacturer’s instructions, and analysed in an Applied Biosystems DNA sequencer (ABI310). Oligonucleotides previously used for amplification of fragments served as sequencing primers.
WT-1 immunoblot
Pieces of tissue were isolated from adenoid cystic carcinoma and corresponding normal parotid gland tissue. Total protein was extracted by boiling the tissue in cracking buffer (0.125 M Tris/HCl, pH 6.8, 6% sodium dodecyl sulfate (SDS), 20% glycerin, 10% dithiothreitol, and 0.03% bromophenol blue) for five minutes at 95°C. The cell lysate was separated by standard SDS-polyacrylamide gel electrophoresis.15 The gels were electroblotted using a polyvinylidenedifluoride membrane (Millipore, Bedford, USA), as described by Gültekin et al.16 Afterwards, membranes were either stained with Coomassie blue or immunoblotted. For the detection of WT-1 protein, the blotted cell lysate was incubated with bovine serum albumin in Tris buffered saline (TBS) buffer (5% (wt/vol) for one hour, then polyclonal rabbit anti-WT-1 antibodies (Santa Cruz Biotechnology, Santa Cruz, California, USA), diluted in TBS (1/250), were added. After overnight incubation at 4°C, the membrane was washed with TBS buffer for 10 minutes. Peroxidase labelled antirabbit antibody (Dako, Hamburg, Germany) was diluted in TBS (1/250) and added to the membrane. After an incubation step for one hour at room temperature, the membrane was washed with TBS for 10 minutes and bound antibodies were detected with diaminobenzidine (DAB) liquid substrate system kit, according to the manufacturer’s protocol (Sigma, Munich, Germany).
RESULTS
Table 1 ▶ summarises the PCR results obtained for 16 malignant and 28 benign salivary gland tumours, in addition to five samples of normal salivary gland tissue adjacent to the tumour. All specimens contained non-degraded RNA according to the results of β actin RT-PCR (fig 1 ▶). For each gene, the specificity of representative PCR fragments was confirmed by automatic sequence analysis.
Table 1.
Number of salivary gland tissue samples presenting positive polymerase chain reaction results
| FHIT | ||||||||||
| Histological type | n | β Actin | Normal transcripts | Aberrant splicing | WT-1 | BAGE | GAGE-1/2 | HAGE | MAGE 1 | MAGE 3 |
| Normal salivary gland tissue | 38/38 | 38/38 | 1/38 | 0/5 | 0/5 | 0/5 | 2/5 | 0/5 | 0/5 | |
| Myoepithelioma | 2 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Pleomorphic adenoma | 16 | 16 | 16 | 3 | 1 | 0 | 2 | 5 | 0 | 0 |
| Cystadenolymphoma | 10 | 10 | 10 | 0 | 0 | 0 | 0 | 6 | 0 | 0 |
| Total of benign neoplasms | 28 | 28 | 28 | 3 | 2 | 0 | 2 | 11 | 0 | 0 |
| Acinic cell carcinoma | 5 | 5 | 5 | 1 | 2 | 0 | 0 | 3 | 1 | 0 |
| Mucoepidermoid carcinoma | 4 | 4 | 4 | 0 | 0 | 0 | 1 | 2 | 0 | 0 |
| Adenoid cystic carcinoma | 3 | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Basal cell adenocarcinoma | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Adenocarcinoma NOS | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 |
| Squamous cell carcinoma | 2 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | 1 | 1 |
| Total no. of malignant neoplasms | 16 | 16 | 16 | 2 | 5 | 2 | 4 | 8 | 2 | 1 |
NOS, not otherwise specified.
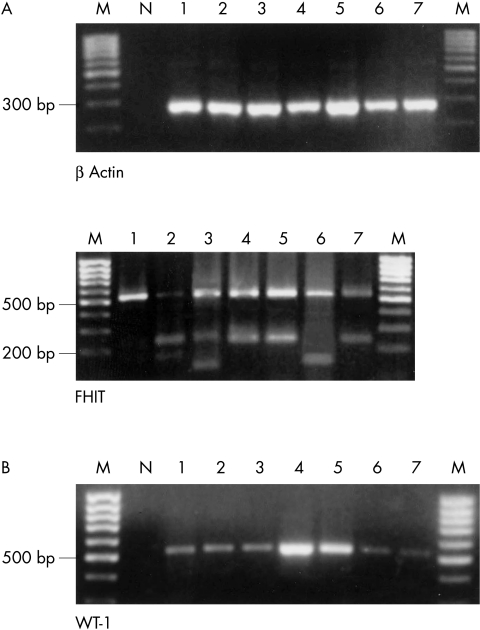
Figure 1.
(A) Agarose gel electrophoresis showing representative polymerase chain reaction (PCR) products of β actin reverse transcription PCR to demonstrate the integrity of the RNA (top panel). The analysis of FHIT (bottom panel) demonstrated in each case the full length product of 533 bp (lane 1) and in six cases various additional aberrant FHIT transcripts (lanes 2–7). The number of the lane corresponds to the histological diagnosis shown in table 2 ▶. (B) WT-1 mRNA was detectable in two benign and five malignant tumours (lanes 1–7). M, molecular weight marker; N, negative control.
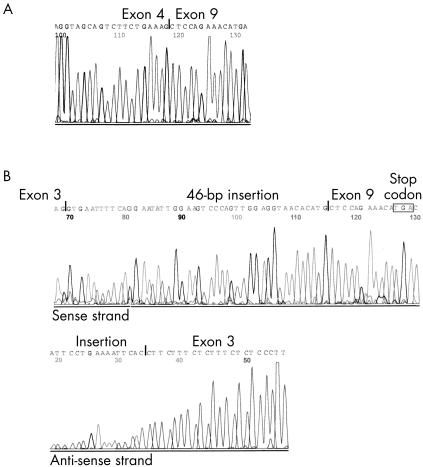
Normal FHIT transcripts were found in all benign and malignant neoplasms. Aberrantly spliced variants of the FHIT mRNA were detected in three pleomorphic adenomas and in two of the malignant tumours—one acinic cell carcinoma and one SCC (fig 1 ▶). Because of the high sensitivity of the seminested PCR applied, transcripts were detected that were present in low amounts in the specimens. The nucleotide sequences of full length FHIT cDNA (533 bp PCR product) and the various FHIT transcript variants were determined by automatic DNA sequencing (table 2 ▶). None of the tumour samples contained mutant FHIT mRNA. Sequence analysis of the splice variants revealed deletions of exons 4–7 in acinic cell carcinoma (142 bp PCR product), and exons 5–7 in SCC and three pleomorphic adenomas (235 bp PCR product). In addition, one pleomorphic adenoma contained a splice variant combining deletion of FHIT exons 4–8 with an insertion of 46 bp, which was identified as a non-coding sequence from intron 5 (119 bp PCR product). This insertion induced a premature stop codon in exon 9 at position 361 (fig 2 ▶). In one case of normal salivary gland tissue, FHIT mRNA transcripts were found that showed deletion of exons 5–7 and 5–8 (235 and 166 bp PCR products respectively).
Table 2.
Sequence analysis of aberrant FHIT transcripts
| Histological diagnosis | Exons deleted |
| (2) Normal gland tissue | 5–7, 5–8 |
| (3) Pleomorphic adenoma | 5–7, 4–8 combined with insertion of 46 bp |
| (4) Pleomorphic adenoma | 5–7 |
| (5) Pleomorphic adenoma | 5–7 |
| (6) Acinic cell carcinoma | 4–7 |
| (7) Squamous cell carcinoma | 5–7 |
Figure 2.
Automatic sequencing of two aberrant FHIT transcripts. (A) This splice variant was found in normal salivary gland tissue and lacked exons 5–8. (B) This variant was detected in a pleomorphic adenoma and demonstrated replacement of exons 4–8 by a non-coding sequence of 46 bp identified in intron 5. The insertion shifted the reading frame and introduced a premature stop codon at the beginning of exon 9.
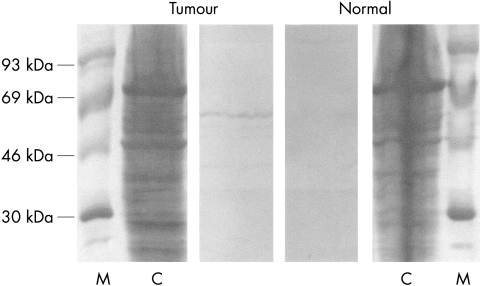
The tumour suppressor gene WT-1 was expressed in two of 28 benign tumours (one myoepithelioma and one pleomorphic adenoma), in five of 16 carcinomas (fig 1 ▶), but not in normal tissue. To test whether the expression of WT-1 mRNA correlated with the concentration of WT-1 protein, immunoblotting was performed with protein lysates from one adenoid cystic carcinoma and from the corresponding normal tissue (fig 3 ▶). The results demonstrated that WT-1 protein was detectable only in the carcinoma.
Figure 3.
Immunoblot of tissue lysate from adenoid cystic carcinoma and normal surrounding salivary gland tissue. The 52 kDa WT-1 protein is detected by a polyclonal antibody in tumour tissue lysate but not in normal salivary gland tissue lysate. C, Coomassie staining; M, molecular weight marker (Rainbow High, Amersham, Braunschweig, Germany).
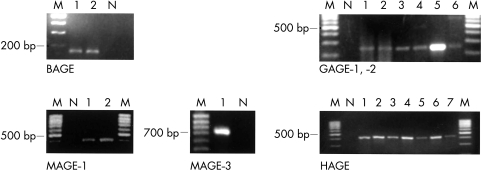
BAGE mRNA was detected in both SCCs, GAGE-1/2 mRNA was detected in two pleomorphic adenomas and four malignant neoplasms, and the MAGE-1 and MAGE-3 genes were activated in two and one malignant tumours, respectively (fig 4 ▶). In contrast, the tumour rejection genes BAGE, GAGE-1/2, MAGE-1, and MAGE-3 were not expressed in normal salivary gland tissue or in cystadenolymphomas, and none of the benign tumours contained BAGE, MAGE-1, or MAGE-3 mRNA.
Figure 4.
Reverse transcription polymerase chain reaction results of the tumour rejection genes BAGE, GAGE-1/2, MAGE 1, MAGE-3, and HAGE showing products of appropriate size for each gene. M, molecular weight marker; N, negative control.
Taken together, RT-PCR analysis of BAGE, GAGE-1/2, MAGE-1, MAGE-3, and WT-1 gene expression revealed four PCR fragments in four of 28 benign tumours and 14 PCR fragments in eight of 16 malignant tumours. When considering the malignant tumours, analysis of both SCCs alone resulted in six positive findings. In addition, MAGE-1 and MAGE-3 mRNA was expressed in the same SCC that demonstrated alternative FHIT splicing.
HAGE mRNA was detected in two of five normal salivary gland tissue samples, in 11 of 28 benign tumours, and in eight of 16 malignant tumours.
DISCUSSION
Genetic alterations of the FHIT gene, such as the absence of mRNA or, more commonly, small aberrant transcripts lacking two or more exons17 have been demonstrated in various types of cancer, benign neoplasms, and normal tissues, including endometrial carcinomas and hyperplasia,18 benign and malignant thyroid tumours,19 lung cancer,20 breast cancer,2 liver tumours,21,22 and renal neoplasms.23 However, the role of these changes in carcinogenesis (if any) and the function of FHIT as a tumour suppressor gene is a matter of controversy.24
To investigate the potential role of FHIT in the genesis and diagnosis of salivary gland neoplasms we searched for aberrant transcripts in a variety of benign and malignant tumours and found FHIT splicing in one of 38 normal salivary gland tissue probes, three of 28 benign salivary gland tumours, and two of 16 malignant salivary gland tumours. Sequence analysis revealed various losses of exons between exon 3 and exon 9, most commonly the loss of exons 5–7. The different types of splice variants found in salivary gland tissue and tumours have been described previously in tumours of other histogenesis, and were summarised as class one transcripts characterised by the deletion of exon 5.1 These aberrant FHIT mRNA transcripts are presumably non-functional because the translation start site is located in the missing exon 5.19 Notably, in FHIT mRNA obtained from a pleomorphic adenoma, exon 4–8 was replaced by a non-coding sequence from intron 5. This alteration shifts the reading frame and may lead to the synthesis of an aberrant truncated FHIT protein. Such insertions of non-coding sequences are rarely found in human carcinomas and were previously described in carcinomas from the oesophagus and the stomach,1 in Burkitt lymphoma,25 and in endometrial carcinoma.18
In conclusion, FHIT splice variants are infrequent findings in normal salivary gland tissue, benign salivary gland tumours, and malignant salivary gland tumours. Thus, these variants cannot be used as markers for the diagnosis of salivary tumours. Moreover, our results indicate that FHIT is not associated with the development or malignant transformation of salivary gland neoplasms.
The putative Wilms’s tumour suppressor gene 1 (WT-1) is physiologically expressed in the early development of the urogenital tract to activate specific genes in cellular differentiation. It is unclear whether WT-1 acts as a transcriptional activator or repressor in vivo.26 During the development of the fetal urogenital tract, WT-1 is involved in the switch from the mesenchymal cell differentiation pathway to the epithelial cell differentiation pathway.5–8 Gene expression is restricted to specific cell types during different stages of development, and WT-1 is not expressed in most normal adult epithelial tissues. WT-1 alterations, such as upregulation of the gene, alternative splicing of WT-1 mRNA and/or WT-1 gene mutations, have been found in several malignant tumours. These changes were detected in Wilms’s tumours,27 leukaemias,26 and malignant mesotheliomas.8 Recently, WT-1 expression has been described in a high proportion of epithelial tumour cell lines of different origins, including gastric, colonic, lung, ovarian, and breast carcinomas, indicating an essential role in the growth of malignant tumours. These findings suggest an oncogenic rather than a tumour suppressor gene function.28
“WT-1, which is thought to be a factor that induces the switch from mesenchymal to epithelial differentiation, may also be involved in cellular differentiation and tumorigenesis of salivary gland tumours with epithelial/myoepithelial differentiation”
In our present study, WT-1 mRNA was found in five of 16 malignant and two of 28 benign salivary gland tumours. In contrast, normal salivary gland tissue did not express the WT-1 gene. The immunoblot experiment proved that WT-1 gene expression in the tumour is associated with increased WT-1 protein concentrations in comparison with corresponding normal salivary gland tissue. Interestingly, WT-1 expression was restricted to those tumours that share a proposed histogenesis of bidirectional epithelial/myoepithelial differentiation,29 whereas both SCCs did not express WT-1. These results suggest that WT-1, which is thought to be a factor that induces the switch from mesenchymal to epithelial differentiation, may also be involved in cellular differentiation and tumorigenesis of salivary gland tumours with epithelial/myoepithelial differentiation. However, WT-1 is of limited value as a diagnostic marker of malignancy in salivary gland neoplasms.
To our knowledge, this is the first report on the expression pattern of the tumour rejection genes BAGE, GAGE, MAGE, and HAGE in salivary gland tissue and tumours of the salivary gland. These data reaffirm former studies indicating that these genes are predominantly expressed in tumours.10,30,31 Consequently, BAGE, GAGE, and MAGE mRNA was not detectable in all five samples of normal salivary gland tissue. In view of the negative RT-PCR results for BAGE, GAGE, and MAGE gene expression in all 10 cystadenolymphomas, it should be remembered that there is still ongoing discussion concerning the neoplastic or reactive nature of this lesion.32,33
Take home messages.
FHIT mRNA splicing is rare in salivary gland tumours and does not appear to be involved in the development of these neoplasms
WT-1 mRNA and protein were found in tumours of epithelial/myoepithelial phenotype so that WT-1 may have a potential role in the genesis and/or cellular differentiation of these salivary gland tumours
The tumour rejection genes MAGE, GAGE, and BAGE were more frequently, but not exclusively, expressed in malignant salivary gland tumours than in benign ones
However, these genes have no potential role as diagnostic markers of malignancy or in the immunotherapy of salivary gland carcinomas
The high incidence of HAGE gene expression in benign/malignant neoplasms, in addition to non-neoplastic salivary gland tissue, is in contrast to the data of Martelange et al,10 who found HAGE mRNA to be strictly tumour specific, with the exception of male germline cells. This contradiction may be explained by the highly sensitive RT-PCR strategy applied in our series, and indicates that HAGE gene expression is not restricted to malignant tumours, but is also present at a low rate in benign neoplasms and non-neoplastic tissue. Conventional PCR is not always suitable for detecting such differences in HAGE gene expression, which may be of tumorigenic or diagnostic importance. Therefore, in further studies the application of real time PCR will help to define those differences.
The comparison between BAGE, GAGE, and MAGE expression in pleomorphic adenomas and myoepitheliomas, on the one hand, and carcinomas of different histological types on the other revealed one or more positive PCR results in two of 18 benign and in five of 16 malignant tumours. However, this small difference in the proportion of gene expression between benign and malignant tumours is essentially based on the BAGE, GAGE, and MAGE expression detected in both SCCs. In contrast to the rare primary squamous cell carcinomas of the salivary gland, the tumour rejection genes BAGE, GAGE, and MAGE are expressed only in a minority of the most common benign and malignant salivary gland tumours and, therefore, do not appear to be suitable diagnostic markers of malignancy at this tumour site. The analysis of additional tumours will be necessary to elucidate whether the rates of BAGE, GAGE, and MAGE mRNA expression differ between primary SCCs of salivary glands and SCCs of other origin in the head and neck region.
In summary, FHIT mRNA splicing is a rare event in salivary gland neoplasms and is not restricted to malignant tumour types. Because normal salivary gland tissue sporadically reveals aberrant FHIT mRNA, splicing is probably not involved in the genesis of salivary gland neoplasms. Moreover, the detection of FHIT transcripts is not suitable for the diagnosis of malignancy in salivary gland tumours.
WT-1 mRNA and protein were demonstrated in different types of epithelial/myoepithelial salivary gland tumours, especially carcinomas. We speculate that WT-1 expression might be involved in the genesis and/or cell differentiation of salivary gland tumours.
Finally, our study showed that the tumour rejection genes of the MAGE, GAGE, and BAGE families are more frequently, but not exclusively, expressed in malignant salivary gland tumours, in particular in primary SCCs of the parotid gland. However, the overall low expression rate of these genes limits their potential role as diagnostic markers of malignancy or in the immunotherapy of salivary gland carcinomas.
Acknowledgments
The authors thank Mrs E Hottenrott for her valuable technical assistance.
Abbreviations
FHIT, fragile histidine triad
RT-PCR, reverse transcription polymerase chain reaction
SCC, squamous cell carcinomas
SDS, sodium dodecyl sulfate
TBS, Tris buffered saline
WT-1, Wilms’s tumour suppressor gene 1
REFERENCES
- 1.Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996;84:587–97. [DOI] [PubMed] [Google Scholar]
- 2.Biéche I, Latil A, Becette V, et al. Study of FHIT transcripts in normal and malignant breast tissue. Genes Chromosomes Cancer 1998;23:292–9. [PubMed] [Google Scholar]
- 3.Bullerdiek J, Takla G, Barnitzke S, et al. Relationship of cytogenetic subtypes of salivary gland pleomorphic adenomas with patient age and histologic type. Cancer 1989;64:876–80. [DOI] [PubMed] [Google Scholar]
- 4.Geurts JMW, Schoenmakers EFPM, Röijer E, et al. Expression of reciprocal hybrid transcripts of HMGIC and FHIT in a pleomorphic adenoma of the parotid gland. Cancer Res 1997;57:13–17. [PubMed] [Google Scholar]
- 5.Haber DA, Sohn RL, Buckler AJ, et al. Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci U S A 1991;88:9618–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundlos S, Pelletier J, Darveau A, et al. Nuclear localization of the protein encoded by the Wilms’ tumor gene WT1 in embryonic and adult tissues. Development 1993;119:1329–41. [DOI] [PubMed] [Google Scholar]
- 7.Kreidberg JA, Sariola H, Loring JM, et al. WT-1 is required for early kidney development. Cell 1993;74:679–91. [DOI] [PubMed] [Google Scholar]
- 8.Langerak AW, Williamson KA, Miyagawa K, et al. Expression of the Wilms’ tumor gene WT1 in human malignant mesothelioma cell lines and relationship to platelet-derived growth factor A and insulin-like growth factor 2 expression. Genes Chromosomes Cancer 1995;12:87–96. [DOI] [PubMed] [Google Scholar]
- 9.Van den Eynde B, Peeters O, De Backer O, et al. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med 1995;182:689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martelange V, De Smet C, De Plaen E, et al. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res 2000;60:3848–55. [PubMed] [Google Scholar]
- 11.Dalerba P, Frascella E, Macino B, et al. MAGE, BAGE and GAGE gene expression in human rhabdomyosarcomas. Int J Cancer 2001;93:85–90. [DOI] [PubMed] [Google Scholar]
- 12.Ellis GL, Auclair PL. Tumors of the salivary glands. In: Rosai J, Sobin LH, eds. Atlas of tumor pathology. Washington, DC: Armed Forces Institute of Pathology, 1995.
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 1987;162:156–9. [DOI] [PubMed] [Google Scholar]
- 14.Schlott T, Reimer S, Jahns A, et al. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol 1997;182:54–61. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- 16.Gültekin H, Heermann KH. The use of polyvinylidenedifluoride membranes as a general blotting matrix. Anal Biochem 1988;172:320–9. [DOI] [PubMed] [Google Scholar]
- 17.Carapeti M, Aguiar RCT, Sill H, et al. Aberrant transcripts of the FHIT gene are expressed in normal and leukaemic haemopoietic cells. Br J Cancer 1998;78:601–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozaki K, Enomoto T, Yoshino K, et al. FHIT alterations in endometrial carcinoma and hyperplasia. Int J Cancer 2000;85:306–12. [DOI] [PubMed] [Google Scholar]
- 19.Zou M, Shi Y, Farid NR, et al. FHIT gene abnormalities in both benign and malignant thyroid tumours. Eur J Cancer 1999;35:467–72. [DOI] [PubMed] [Google Scholar]
- 20.Tokuchi Y, Kobayashi Y, Hayashi S, et al. Abnormal FHIT transcripts found in both lung cancer and normal lung tissue. Genes Chromosomes Cancer 1999;24:105–11. [PubMed] [Google Scholar]
- 21.Chen Y-J, Chen P-H, Chang J-G. Aberrant FHIT transcripts in hepatocellular carcinomas. Br J Cancer 1998;77:417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlott T, Ahrens K, Ruschenburg I, et al. Different gene expression of MDM2, Gage-1,-2 and FHIT in hepatocellular carcinoma and focal nodular hyperplasia. Br J Cancer 1999;80:73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eyzaguirre EJ, Miettinen M, Norris BA, et al. Different immunohistochemical patterns of Fhit protein expression in renal neoplasms. Mod Pathol 1999;12:979–83. [PubMed] [Google Scholar]
- 24.Le Beau MM, Drabkin H, Glover TW, et al. An FHIT tumor suppressor gene? Genes Chromosomes Cancer 1998;21:281–9. [DOI] [PubMed] [Google Scholar]
- 25.Ferrer M, Lopez-Borges S, Lazo PA. Expression of aberrant functional and nonfunctional transcripts of the FHIT gene in Burkitt’s lymphomas. Mol Carcinog 1999;25:55–63. [DOI] [PubMed] [Google Scholar]
- 26.King-Underwood L, Renshaw J, Pritchard-Jones K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood 1996;87:2171–9. [PubMed] [Google Scholar]
- 27.Little MH, Prosser J, Condie A, et al. Zinc finger point mutations within the WT1 gene in Wilms tumor patients. Proc Natl Acad Sci U S A 1992;89:4791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oji Y, Ogawa H, Tamaki H, et al. Expression of the Wilms’ tumor gene WT1 in solid tumors and its involvement in tumor cell growth. Jpn J Cancer Res 1999;90:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dardick I. Histogenesis and morphogenesis of salivary gland neoplasms. In: Ellis GL, Auclair PL, Gnepp DR, eds. Surgical pathology of the salivary glands. Philadelphia: WB Saunders, 1991:108–128.
- 30.Zambon A, Mandruzzato S, Parenti A, et al. MAGE, BAGE, and GAGE gene expression in patients with esophageal squamous cell carcinoma and adenocarcinoma of the gastric cardia. Cancer 2001;91:1882–8. [PubMed] [Google Scholar]
- 31.Gillespie AM, Rodgers S, Wilson AP, et al. MAGE, BAGE and GAGE: tumour antigen expression in benign and malignant ovarian tissue. Br J Cancer 1998;78:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allegra SR. Warthin’s tumor: a hypersensitivity disease? Hum Pathol 1971;2:403–20. [DOI] [PubMed] [Google Scholar]
- 33.Aguirre JM, Echebarria MA, Martinez-Conde R, et al. Warthin tumor. A new hypothesis concerning its development. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1998;85:60–3. [DOI] [PubMed] [Google Scholar]