Abstract
Background/Aims: Rearrangement of immunoglobulin gene segments, leading to B cells with functional receptors, is thought to be largely restricted to developing immature B cells in bone marrow. However, accumulating evidence suggests that mature B cells occasionally modify their antigen specificity by VH segment replacements during the germinal centre reaction to enhance antigen affinity, or to overcome self reactive antigen receptors. Although malignant B cells maintain the features of their normal counterparts in most instances, to date, such replacements have not been described for human B cell lymphomas.
Methods: Rearranged immunoglobulin heavy chain genes from two extranodal marginal zone B cell lymphomas were amplified, cloned, and sequenced. Sequences with identical CDR3 regions were selected and aligned to each other and public databases.
Results: VH replacements were seen in two extranodal marginal zone B cell lymphomas. In line with the hypothesis that in mature B cells these replacements are associated with active somatic hypermutation, in addition to VH replacement, different mutation patterns were seen in the revised VH portions. In the remaining common 3′-VH regions, these mutations could be used to establish a phylogenetic relation between the sequences, rendering the possibility of artefactual chimaeric polymerase chain reaction products very unlikely.
Conclusions: These results support the view that VH replacements are a further mechanism for reshaping antigen affinity and specificity, and indicate that these receptor modifications are not restricted to normal and reactive germinal centre B cells, but may also occur in close association with the development of malignant B cell lymphomas.
Keywords: B lymphocytes, VH replacement, immunoglobulin gene rearrangement, antibodies
The human immune system efficiently protects us against a wide variety of potential pathogens. One means of achieving this is with the generation of antibodies. These proteins bind to non-self molecules and in this way mark them out for neutralisation. To fulfill this task, the multiplicity of foreign substances has to be matched by a diverse but also specific repertoire of antibodies. Therefore, several mechanisms have evolved to create diversity and increase affinity during the development of the antibody producing B cells. To build the variable, antigen binding region of an antibody molecule, two (V and J) or three (V, D, and J) different gene segments are joined to produce light or heavy chains, respectively. For each of these segments, multiple, but differing, copies exist in the human genome. Thus, the random selection and combination of the individual segments already leads to a large number of different antibody molecules.1 Another major contribution to antibody diversity is the insertion of random nucleotides at the segment junctions during the joining process.2,3
The production of a functional antigen receptor is verified several times during these procedures and is essential for the survival of the respective B cells. Immature B cells bearing non-productive rearrangements of either chain can be rescued by new rearrangements on the other allele or, in the case of the light chains, by switching to the other isotype.
On contact with antigen, the variable region of mature B cells is further modified by the introduction of somatic point mutations during the course of the germinal centre reaction.4,5 These modifications are intended to increase the affinity of the antigen binding domain. Because this process is to a certain degree random, most cells die from apoptosis because of their insufficient ability to bind antigen.
“The expression receptor revision is used when mature B cells alter their receptors in response to antigenic stimulation”
Recently, however, evidence has been accumulating to suggest that, despite expressing functional antibodies on their cell surface, B cells can continue to rearrange their immunoglobulin (Ig) genes, and that these rearrangements can occur both on the same allele and on the second allele. Although this phenomenon was originally described for the heavy chain locus of mouse lymphoma cell lines,6,7 most subsequent studies have focused on the light chain.8–11 Because the heavy chain (IgH) gene locus is organised in a different way to that of the light chains, ongoing rearrangements at this region were considered to be unlikely and irrelevant for human B cell development. It was argued that after the primary rearrangement, no more D segments are left to join new V and J segments, yet, through transgenic mouse models12–14 and observations of human B cells,15,16 it became evident that the heavy chain can be edited by exchanging only the VH portion of the rearranged VDJ gene with a new VH segment (VH replacement). Dependent on the developmental status of the B cell, these secondary rearrangements seem to serve different purposes. In immature B cells, they enable the editing of self reactive receptors and thus play a role in mediating tolerance.10,17–19 For this, the term “receptor editing” has been introduced. The expression “receptor revision” is used when mature B cells alter their receptors in response to antigenic stimulation.20–22 The purpose of this kind of secondary recombination is not yet clear. It might serve as an additional mechanism to create diversity within the antibody repertoire, or to eliminate unproductive or self reactive rearrangements generated through the introduction of somatic mutations.
Here, we describe VH replacement in the in situ tumour cells of two human mucosa associated lymphoid tissue (MALT) lymphomas. Based on their immunophenotypical features,23 MALT lymphomas are thought to derive from post-germinal centre memory B cells.24 However, the detection of intraclonal diversity in the clonally rearranged IgH genes of the tumour cells25,26 is suggestive of a close relation to germinal centres. The observation of Ig receptor revision in the two MALT lymphomas described here is in accordance with such a setting of mutational activity, and corresponds with the diversity driven mechanism of this phenomenon. Moreover, it demonstrates that receptor revision is not only active in normal germinal centre B cells, but also in germinal centre related B cell lymphomas.
METHODS
Patient material
Both tumours were diagnosed as low grade MALT lymphomas according to the World Health Organisation classification.23 They displayed the same immunophenotype (CD19+, CD23−, CD5−, IgD−, IgM+), except that the tumour in case 1 used a κ light chain and the case 2 tumour was Igλ+. In case 1 the tumour was of gastric origin, whereas the tumour in case 2 was localised in the thyroid.
Preparation of nucleic acids for the polymerase chain reaction
Genomic DNA and total RNA were isolated from frozen tissue sections with the QIAamp DNA mini kit and the RNeasy mini kit, respectively, according to the manufacturer‘s protocol (Qiagen, Hilden, Germany). For amplification, 1 μg of RNA was then transcribed into cDNA using random hexamers or OligodT15 primers and reverse transcriptase (GeneAmp RNA PCR core kit; Perkin Elmer, Foster City, California, USA).
Polymerase chain reaction
To identify the rearranged segments of the variable heavy chain genes, a nested polymerase chain reaction (PCR) was initially performed for each case (GeneAmp PCR System 9600; Perkin Elmer Cetus, Foster City, California, USA). In the first round of amplification, a mix of six VH family specific framework 1 (FR1) primers and a consensus JH primer (LJH) were used.27 For the reamplification, an aliquot of the first PCR from each case was amplified with a family specific VH primer set for framework 2 and a nested JH primer (VLJH). PCRs were carried out in a final volume of 100 μl with 0.2 mM of each dNTP (Amersham, Piscataway, New Jersey, USA), 1.5 mM MgCl2, AmpliTaq buffer II, and 2.5 units of AmpliTaq Gold (all from Applied Biosystems, Darmstadt, Germany). Cycling parameters for the primary amplifications consisted of an initial incubation at 95°C for 10 minutes; followed by five cycles of 95°C for 25 seconds, 63°C for 30 seconds, and 72°C for 30 seconds; an additional 35 cycles of 95°C for 25 seconds, 57°C for 30 seconds, and 72°C for 30 seconds; and a final elongation step of 72°C for seven minutes. Reamplifications were carried out, after 10 minutes at 95°C, for 25 cycles of 95°C for 25 seconds, 63°C for 30 seconds, and 72°C for 30 seconds, followed by a final elongation step of 72°C for seven minutes.
To gain more sequence information of the clonal rearrangements, additional PCRs were performed involving family specific FR1 primers in conjunction with the LJH primer. The reaction conditions were the same as for the primary amplification described above. To exclude PCR artefacts, these family specific FR1 PCRs were performed three times for each case. Each of these PCRs resulted in the detection of revised IgH sequences.
To verify the existence of the revised Ig rearrangements, clone specific PCRs were carried out for case 1, using individual forward primers for the CDR2 of each of the four revised Ig rearrangements. Because the CDR3 was shared by all revised Ig rearrangements, a common clone specific reverse primer was used.
Group 1: VH3–07CDR2, 5′-GCCAACATAAAGAAAGATGGGAGT GAG-3′.
Group 2: VH3–23CDR2, 5′-GCTATTAGTGGGAGTGGTGGGAG-3′.
Group 3.1: VH3–30.1CDR2, 5′-CTGGAGTGGGTGGCAGTTATAT CATATG-3′.
Group 3.2: VH3–30.2, 5′-GTGGGTGGCAGTTATATGGTATGA TGG-3′.
Common clone specific CDR3, 5′-GGCGCTCCCCGAAACGTC-3′.
Amplification for each primer pair was performed in a volume of 100 μl with 0.2 mM of each dNTP, 1.5mM MgCl2, AmpliTaq buffer II, 2.5 units of AmpliTaq Gold, and 20 pmol of each primer. Cycling parameters consisted of 10 minutes at 95°C, followed by 95°C for 25 seconds, 60°C for 30 seconds, and 72°C for 30 seconds for 40 cycles, with a final elongation step of 72°C for 10 minutes.
GeneScan analysis
GeneScan analysis (ABI 310; Applied Biosystems) was performed to demonstrate the presence of clonal VDJ rearrangements and to determine their size. For this purpose, reamplification was carried out with a fluorescence (FAM) labelled JH primer (LJH) and the final elongation step was extended to 45 minutes. Fluorescence labelled PCR products were separated on a sequencing gel and evaluated by the GeneScan software 672 (Applied Biosystems).
Cloning and sequence analysis of PCR products
Unlabelled PCR products were separated by polyacrylamide gel electrophoresis and bands representing clonal VDJ rearrangements were cut out, eluted in distilled water overnight, and cloned into the pCRII-TOPO vector (TOPO TA cloning kit; Invitrogen, Breda, The Netherlands). Plasmid DNA from several clones (Quantum Prep plasmid miniprep kit; BioRad, Hercules, California, USA) was sequenced in both directions (Big Dye Terminator cycle sequencing kit, Applied Biosystems), using an automated DNA sequencer (DNA Sequencing Automat 377; Applied Biosystem). The sequences obtained were compared with each other and with our own and published databases (IMGT-International ImMunoGeneTics database, Marie-Paule Lefranc, Montpellier, France; http://imgt.cines.fr:8104) to determine the corresponding germline segments and the number and location of somatic mutations.
RESULTS
Detection of clonal IgH rearrangements
Dominant amplificates of 233 bp (case 1) and 245 bp (case 2) were detectable after nested primer IgH PCR and GeneScan analysis. In addition to the prominent PCR product, a moderate polyclonal B cell background was present. The clonal IgH PCR products were cloned and subsequently sequenced.
Sequence analysis of case 1
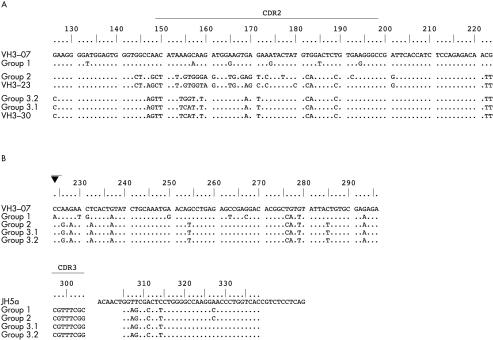
Fifty seven sequences from case 1 were used for analysis of the IgH rearrangements. These sequences were obtained from three different independent PCR experiments, each startingfrom a new aliquot of DNA. Twenty six of these rearrangements shared the same CDR3 portion and JH segment, in addition to a part of their rearranged VH segment (fig 1 ▶, positions 224 to 296, numbering according to Chothia and colleagues28). The remaining 31 IgH rearrangements displayed no similarities to each other, or to the 26 other sequences of this case. This indicated that they originated from the polyclonal B cell background present in this case.
Figure 1.
Comparison of the immunoglobulin heavy chain (IgH) rearrangements of case 1 and alignment to the corresponding germline VH segments. (A) Alignment of the revised 5′ portions of the rearranged VH segments and the corresponding VH germline segments. Differences between the revised VH rearrangements, their most homologous germline segments, and the initial VH3–07 rearrangement, respectively, are shown in capital letters. This comparison demonstrates that these 5′-VH segments derive from different VH germline segments as a consequence of the VH receptor revision in this case. (B) Alignment of the common 3′ portions of the rearranged IgH genes to the corresponding VH germline segment (VH3–07) and JH germline segment (JH5a). All subclones of this case display the highest homology to the initial VH germline segment VH3–07, although the high mutation rate might mimic another germline segment. In addition, there are common and different somatic mutations, indicative of an ongoing somatic mutation process. These results prove that all subclones of this case carry the same 3′-VH rearrangement, which was not affected by the receptor revision. The closed triangle indicates the probable breakpoint for hybrid formation, just behind the last region of homology of the 5′-VH regions.
The 26 rearrangements with identical CDR3 and JH sequences could be divided into three groups (fig 1 ▶). One group comprised 23 sequences that shared, in addition to their common CDR3/JH portion, the same VH segment (VH3–23), and varied by only a few single nucleotide substitutions (group 2). Sequences belonging to this group could be detected in each of the three independently performed PCR experiments in this case. The other two groups were represented by one and two sequences, respectively. These three sequences all derived from different PCRs. Although identical in their CDR3/JH sequences, and containing the same 3′-VH portion (positions 224–296) as the sequences of group 2, the 5′-VH parts (positions 126–223) of group 1 and 3 were clearly different to each other and to the sequences from group 2. The 5′-VH region of group 1 disclosed the highest homology to VH3–07, whereas the 5′-VH sequences of group 3 were most closely related to VH3–30.
Analysis of the mutations in these 26 sequences showed the same five somatic mutations in all 3′-VH portions when compared with the germline VH segment VH3–07 (figs 1 and 2 ▶ ▶). Another five base substitutions were identical in group 3 and in the 23 sequences of group 2. The 3′-VH3–07 portion of group 1 harboured, in addition to the five common mutations, six individual somatic mutations not present in all the other sequences of this case. This pattern of shared and differentially acquired somatic mutations (ongoing mutations) demonstrates a phylogenetic relation between the sequences. The alignment of the differing 5′-VH portions to their most closely related germline segments revealed seven further mutations in the 23 sequences of group 2, resulting in a total of 17 somatic mutations. Group 1 contained six further mutations in the 5′-VH3–07 part, in addition to the 11 mutations in its 3′-VH portion. The two sequences of group 3, which displayed 10 mutations in the 3′-VH3–07 portion, contained no (group 3.1) or two (group 3.2) further somatic mutations in the 5′-VH3–30 portion, respectively.
Figure 2.
Schematic representation of the relation between the sequence groups of case 1. Group 2: consensus of 23 sequences; groups 1, 3.1, and 3.2: one sequence each; vertical strokes represent somatic mutations; bold strokes highlight mutations that are shared between groups; black triangles indicate the probable breakpoints for the hybrid formation.
The results of the clone specific PCR, using individual primers specific for the CDR2 of the differing 5′-VH portions and a reverse primer for the common clone specific 3′ CDR3/JH portion, confirmed the presence of revised IgH rearrangements, as found with standard IgH PCR. Each primer pair gave rise to a dominant PCR product, and sequence analysis disclosed concordance with the respective rearrangement found by the standard FR1 PCR.
Sequence analysis of case 2
Sixty three sequences from case 2, obtained from three different independent PCR experiments using separate DNA aliquots, were used to compare the IgH rearrangements. Fifty three of these sequences contained the same CDR3 and JH region, in addition to an identical VH segment portion (fig 3 ▶; positions 190–298). The remaining 10 IgH sequences were not similar to one another or to the other sequences in this case. Thus, they arguably reflected the polyclonal background present in this case.
Figure 3.
Comparison of the immunoglobulin heavy chain (IgH) rearrangements of case 2 and alignment to the corresponding germline VH segments. (A) Alignment of the revised 5′ portions of the rearranged VH segments and the corresponding VH germline segments. Differences between the revised VH rearrangements, their most homologous germline segments, and the initial VH4–59 rearrangement, respectively, are shown in capital letters. These results demonstrate that these 5′-VH segments derive from different VH germline segments, as a consequence of the VH receptor revision in this case. (B) Alignment of the common 3′ portions of the rearranged IgH genes to the corresponding VH germline segment (VH4–59) and JH germline segment (JH6b). All subclones of this case display the highest homology to the initial VH germline segment, VH4–59. In addition, there are common and different somatic mutations, indicative of an ongoing somatic mutation process. Taken together, this proves that all subclones of this case carry the same 3′-VH rearrangement, which was not affected by the receptor revision. The black triangle indicates the probable breakpoint for hybrid formation.
Based on the germline VH segment used, the 53 IgH rearrangements with common CDR3 and JH segments could be divided into three groups (fig 3 ▶). The first group consisted of 49 sequences, which shared the same CDR3, JH region, and VH segment (VH4–59), and varied only at a few nucleotide positions, indicating ongoing somatic mutations. Sequences belonging to this group were present in all three PCR experiments. The second and the third groups were composed of three sequences (groups 2.1–2.3) and one sequence (group 3), respectively. Comparison of the VH segments disclosed that only the 3′ portions (positions 190–298) were identical to the VH4–59 segment of group 1. However, the 5′-VH portions (positions 73–189) of both sequence groups differed significantly both from one another and from group 1. Comparisons to known germline segments disclosed that group 3 was most similar to VH4–04, and the three sequences of group 2 had the highest homology to VH4–61, but used different alleles (group 2.1, VH4–61*07; group 2.2 and 2.3, VH4–61*02). One sequence of group 2 and the sequence of group 3 were obtained from one PCR experiment, the remaining two sequences of group 2 originate from the other two PCRs.
All sequences of case 2 shared one mutation in their common 3′-VH portion when compared with the VH4–59 germline segment (figs 3 and 4 ▶ ▶; position 255). In addition, 26 of the sequences of group 1, group 2.2, and group 3 displayed a further common base substitution (position 240), leading to the conclusion that, as in case 1, these sequences are derived from different individual rearrangements and are not generated by PCR artefacts.
Figure 4.
Schematic representation of the relation between the sequence groups of case 2. Group 1: consensus of 49 sequences; groups 2.1. 2.2, 2.3, and 3: one sequence each; vertical strokes represent somatic mutations; bold strokes highlight mutations that are shared between groups; the dashed stroke indicates a mutation not present in all sequences of this group; black triangles indicate the probable breakpoints for the hybrid formation.
Comparison of the discordant 5′-VH portions of the three sequence groups to the corresponding germline segments displayed two further somatic mutations in the VH4–59 part of group 1. The sequences of group 2 harboured six (group 2.1), eight (group 2.2), and two (group 2.3) nucleotide substitutions in their VH4–61 portion, respectively, and group 3 showed 13 additional mutations compared with VH4–04.
Analysis of case 2 by clone specific PCR was not possible because of a lack of sufficient material.
DISCUSSION
An increasing amount of evidence has accumulated to show that B cells with rearranged Ig genes can modify their Ig receptor specificity by replacing their rearranged VH segments.6,7,15,16 Depending on the developmental stage of the B cells analysed, different terms and purposes have been assigned to this phenomenon. For immature B cells of the bone marrow it is thought that this mechanism is used to change the reactivity of the autoreactive Ig receptor (a phenomenon known as receptor editing),10,17–19 whereas for peripheral B cells it has been proposed that this modification—which probably occurs in the germinal centre reaction—is a further way of enhancing the affinity and/or specificity of the Ig receptor (known as receptor revision), in addition to the elimination of autoreactive or unproductive antibodies.21,22,29
The revised receptors found in mature B cells consist mainly of so called “hybrid” VH segments, which are composed of parts derived from two different VH segments. It was proposed that cryptic recombination signal sites, present in the germline genes, enable the recombination between a recipient and a donor VH segment, leading to a partial VH replacement in an existing, functional VDJ rearrangement.6,7 The detection of revised Ig sequences that were clonally related, as demonstrated by shared somatic mutations in the common VH recipient part, showed that the recombination events took place in the germinal centre during ongoing somatic hypermutation.15 Because this process serves to enhance antibody affinity, this finding indicates that receptor revision might be a further mechanism, in addition to somatic mutation, that shapes the antigen selected B cell repertoire.
“In both cases, the tumour cell immunoglobulins share the same CDR3 and JH portions but differ in their VH configuration”
Unexpectedly, despite the availability of many Ig sequences derived from the clonal Ig rearrangements of a large variety of B cell tumours, no convincing evidence of receptor revision has yet been described for human lymphomas. This raises the question of whether these Ig receptor modifications are absent from malignant lymphomas or are present, but have been overlooked or misinterpreted as artefacts in previous studies.
Here, we describe the presence of receptor revision in two cases of human MALT B cell lymphoma. In both cases, the tumour cell Igs share the same CDR3 and JH portions but differ in their VH configuration. In case 1, most of the sequences displayed a VH3–23 rearrangement, thus reflecting the predominant tumour cell population. In addition, two further rearrangements involving VH3–30 and VH3–07 germline segments, but with the same CDR3/JH portion as the VH3–23 rearrangement, were found (fig 1 ▶). In case 2, the VH4–59 segment was rearranged in most clonal sequences, but VH4–61 and VH4–04 were also detectable in three sequences and in one further sequence, all with concordant CDR3/JH regions (fig 3 ▶).
In agreement with the findings in mature germinal centre derived B cells, only a part of the VH segment was replaced during receptor revision in the MALT lymphomas investigated in our study.15 In both cases, the 3′-VH part consistently belonged to the initial rearrangement, whereas the 5′-VH portion was replaced by part of a segment that originated either from a more 5′ located VH germline segment or from a VH germline segment located on the other allele.
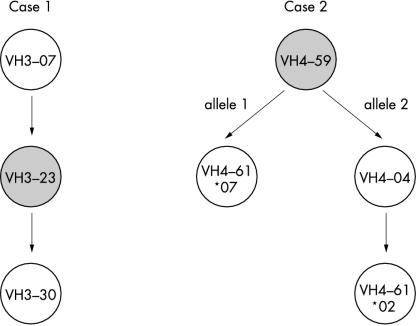
Based on the genomic arrangement of the VH segments on chromosome 14,30 the initial rearrangement of case 1 involved a VH3–07 segment, which was (probably during the course of a germinal centre reaction) replaced by VH3–23 (fig 5 ▶). The fact that the rearrangement involving VH3–23 was found in most sequences indicates that this receptor revision took place early in the germinal centre reaction. This idea is supported by the mutational pattern of the initial and revised rearrangements, both of which display the same number of mutations in their IgH sequences (figs 1 and 2 ▶ ▶). In contrast, the second receptor revision involving the germline segment VH3–30 (located 5′ of VH3–23 on chromosome 14) seems to have appeared at a much later stage in the germinal centre reaction. This hypothesis is supported by the low number of mutations (two) in the 5′-VH3–30 part of group 3.2, the complete absence of somatic mutations in the 5′-VH3–30 part of group 3.1, and the maintenance of the mutational pattern of the remaining 3′-VH part found in this rearrangement.
Figure 5.
Order of revision events in the tumour cells of two cases of human mucosa associated lymphoid tissue lymphoma. Presumable sequence of events leading to the presence of revised immunoglobulin receptors in cases 1 and 2. The circles represent the related clones of each case, labelled with the respective recipient (upper circles) and donor VH segments used for the VH replacements. Dominant tumour clone rearrangements are marked by grey circles. Arrows denote receptor revision events.
To establish the order of the revision events in case 2, two observations had to be considered in addition to the chromosomal arrangement of the germline segments, namely: (1) one mutation in the common 3′-VH portion of the sequences was only present in 53% of the VH4–59 rearrangements and in the VH4–61*02 and the VH4–04 rearrangements (position 240); and (2) the VH4–61 germline segment was rearranged on both alleles (fig 3 and 4 ▶ ▶). Taking these facts into account, the most likely sequence of events is that the initial rearrangement used a VH4–59 segment, which then became the dominant tumour clone (fig 5 ▶). In the course of the germinal centre reaction, two clonal lines evolved from this rearrangement. They shared two mutations in the 5′-VH region and one in the 3′-VH part (position 255), but were differentiated by one mutation in the 3′-VH part, which was only present in one clonal line (comprising 53% of the detected VH4–59 rearrangements). Both of these clones from the initial VH4–59 rearrangement became a target for receptor revision. In one clone, 5′-VH4–59 was replaced with the respective portion of the VH4–61*07 gene segment, located further towards the 5′ end on the same allele. The other clone was revised using the VH4–04 germline gene from the second allele. This VH4–04/VH4–59 rearrangement in turn gave rise to a new hybrid using the more 5′ localised VH4–61*02 segment. All the revised rearrangements acquired further somatic mutations.
The observed VH replacements might have been mediated by recombination signal sites that were present near the 3′ ends of the VH sequences (figs 1 and 3 ▶ ▶). In case 2, the cryptic heptamer beginning at C207 was a possible junction between the VH4–59 recipient and the donor VH genes. For case 1, the breakpoint would probably have been at C224, after the last region of sequence homology to the respective donor VH germline genes, but before the common 3′-VH region. However, a heptamer-like sequence can only be detected before (C204) and after (C233) this point.
In early PCR studies for the amplification of antigen receptor rearrangements, the generation of chimaeric molecules as a result of PCR recombination events has been described repeatedly.31–34 However, since then PCR conditions have improved considerably, particularly by the introduction of new types of polymerases and newly designed primer systems. The Ig PCR system used in our study is based on up to date polymerases (TaqGold) and on VH family specific primers, which have been approved by their application to several thousand lymphoproliferative disorders, including extensive single cell studies.27 In none of these cases have artificial chimaeric PCR products been produced. Furthermore, all experiments were performed three times, and concordant results were produced. The derivation of our Ig sequences from individual rearrangements is further underscored by detailed sequence analysis of the PCR products. All rearrangements are in frame and, even more striking, there are common and different somatic mutations in the 3′ portions of the VH segments. These mutation patterns, which fulfill the criteria for ongoing somatic mutations, cannot be generated by coincidental PCR recombinations, but reflect the presence of different individual B cell clones in our cases.
“Our findings of receptor revision in mucosa associated lymphoid tissue lymphomas demonstrate that this process of rearranging is not only active in physiological germinal centre B cells but also in lymphomas with ongoing immunoglobulin mutations”
To confirm the presence of the revised Ig rearrangements by an independent approach we performed additional PCRs with forward primers designed for the CDR2 of each of the four rearrangements of case 1 (group 1, VH3–07; group 2, VH3–23; group 3.1 and 3.2, VH3–30). The reverse primer was located in the common clone specific CDR3 portion. Amplification was carried out separately for each primer pair and PCR products were obtained with all four primer combinations. Sequence analysis of these PCR products demonstrated that the sequences generated by the clone specific PCR matched the corresponding rearrangements amplified by the previous FR1 PCR (data not shown). This unequivocally confirms the existence of revised IgH rearrangements in the tumour cells of case 1. Unfortunately, there was no more suitable material left for the re-analysis of case 2.
Our findings of receptor revision in MALT lymphomas demonstrate that this process of rearranging is not only active in physiological germinal centre B cells but also in lymphomas with ongoing Ig mutations. This might have implications for the interpretation of clonality studies based on Ig rearrangements because the expected clonal appearance might be confused by additional dominant B cell clones, leading to the interpretation of an oligoclonal reactive B cell expansion instead of a clonal malignant B cell population. Finally, the presence of antigen receptor revision might interfere with anti-idiotypic vaccination strategies because the tumour cell clone might possess more than one type of Ig receptor.
Take home messages.
VH replacement was found in the two mucosa associated lymphoid tissue lymphomas studied, and the mutational pattern was in line with the hypothesis that in mature B cells these replacements are associated with active somatic hypermutation
Thus, VH replacements appear to be a further mechanism for reshaping antigen affinity and specificity
These receptor modifications are not restricted to normal and reactive germinal centre B cells, but can occur in close association with the development of malignant B cell lymphomas
This might have implications for the interpretation of clonality studies based on immunoglobulin rearrangements
Acknowledgments
This study was supported by the Deutsche Krebshilfe.
Abbreviations
FR, framework
Ig, immunoglobulin
IgH, immunoglobulin heavy chain
MALT, mucosa associated lymphoid tissue
PCR, polymerase chain reaction
REFERENCES
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature 1983;302:575–81. [DOI] [PubMed] [Google Scholar]
- 2.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D–JH fusions. Proc Natl Acad Sci U S A 1982;79:4118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakano H, Kurosawa Y, Weigert M, et al. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature 1981;290:562–5. [DOI] [PubMed] [Google Scholar]
- 4.Jacob J, Kelsoe G, Rajewsky K, et al. Intraclonal generation of antibody mutants in germinal centres. Nature 1991;354:389–92. [DOI] [PubMed] [Google Scholar]
- 5.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell 1991;67:1121–9. [DOI] [PubMed] [Google Scholar]
- 6.Reth M, Gehrmann P, Petrac E, et al. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature 1986;322:840–2. [DOI] [PubMed] [Google Scholar]
- 7.Kleinfield R, Hardy RR, Tarlinton D, et al. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature 1986;322:843–6. [DOI] [PubMed] [Google Scholar]
- 8.Berinstein N, Levy S, Levy R. Activation of an excluded immunoglobulin allele in a human B lymphoma cell line. Science 1989;244:337–9. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity 1997;6:97–105. [DOI] [PubMed] [Google Scholar]
- 10.Gay D, Saunders T, Camper S, et al. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med 1993;177:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiernholm NB, Berinstein NL. Immunoglobulin somatic variation; studies of receptor editing in a human B cell lymphoma. Leuk Lymphoma 1994;12:333–41. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Nagy Z, Prak EL, et al. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity 1995;3: 747–55. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand FE, Golub R, Wu GE. V(H) gene replacement occurs in the spleen and bone marrow of non-autoimmune quasi-monoclonal mice. Eur J Immunol 1998;28:3362–70. [DOI] [PubMed] [Google Scholar]
- 14.Cascalho M, Ma A, Lee S, et al. A quasi-monoclonal mouse. Science 1996;272:1649–52. [DOI] [PubMed] [Google Scholar]
- 15.Wilson PC, Wilson K, Liu YJ, et al. Receptor revision of immunoglobulin heavy chain variable region genes in normal human B lymphocytes. J Exp Med 2000;191:1881–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K, Meffre E, Albesiano E, et al. Immunoglobulin heavy chain variable region gene replacement as a mechanism for receptor revision in rheumatoid arthritis synovial tissue B lymphocytes. J Exp Med 2000;192:1151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemazee D, Hogquist KA. Antigen receptor selection by editing or downregulation of V(D)J recombination. Curr Opin Immunol 2003;15: 182–9. [DOI] [PubMed] [Google Scholar]
- 18.Radic MZ, Erikson J, Litwin S, et al. B lymphocytes may escape tolerance by revising their antigen receptors. J Exp Med 1993;177:1165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med 1993;177:1009–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Zheng B, Schatz DG, et al. Neoteny in lymphocytes: Rag1 and Rag2 expression in germinal center B cells. Science 1996;274:2094–7. [DOI] [PubMed] [Google Scholar]
- 21.Han S, Dillon SR, Zheng B, et al. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science 1997;278:301–5. [DOI] [PubMed] [Google Scholar]
- 22.Papavasiliou F, Casellas R, Suh H, et al. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science 1997;278:298–301. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organisation. World Health Organisation classification of tumours. Pathology and genetics of tumours of haematopoetic and lymphoid tissues. Lyon: IARC Press, 2001.
- 24.Isaacson PG. Gastrointestinal lymphoma. Hum Pathol 1994;25: 1020–9. [DOI] [PubMed] [Google Scholar]
- 25.Qin Y, Greiner A, Hallas C, et al. Intraclonal offspring expansion of gastric low-grade MALT-type lymphoma: evidence for the role of antigen-driven high-affinity mutation in lymphomagenesis. Lab Invest 1997;76:477–85. [PubMed] [Google Scholar]
- 26.Du M, Diss TC, Xu C, et al. Ongoing mutation in MALT lymphoma immunoglobulin gene suggests that antigen stimulation plays a role in the clonal expansion. Leukemia 1996;10:1190–7. [PubMed] [Google Scholar]
- 27.Marafioti T, Hummel M, Anagnostopoulos I, et al. Origin of nodular lymphocyte-predominant Hodgkin’s disease from a clonal expansion of highly mutated germinal-center B cells. N Engl J Med 1997;337:453–8. [DOI] [PubMed] [Google Scholar]
- 28.Chothia C, Lesk AM, Tramontano A, et al. Conformations of immunoglobulin hypervariable regions. Nature 1989;342:877–83. [DOI] [PubMed] [Google Scholar]
- 29.Klonowski KD, Monestier M. Ig heavy-chain gene revision: leaping towards autoimmunity. Trends Immunol 2001;22:400–5. [DOI] [PubMed] [Google Scholar]
- 30.Cook GP, Tomlinson IM, Walter G, et al. A map of the human immunoglobulin VH locus completed by analysis of the telomeric region of chromosome 14q. Nat Genet 1994;7:162–8. [DOI] [PubMed] [Google Scholar]
- 31.Shuldiner AR, Nirula A, Roth J. Hybrid DNA artifact from PCR of closely related target sequences. Nucleic Acids Res 1989;17:4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deane M, Norton JD. Immunoglobulin heavy chain gene rearrangement involving V–V region recombination. Nucleic Acids Res 1990;18:1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob J, Przylepa J, Miller C, et al. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med 1993;178:1293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ford JE, McHeyzer-Williams MG, Lieber MR. Chimeric molecules created by gene amplification interfere with the analysis of somatic hypermutation of murine immunoglobulin genes. Gene 1994;142: 279–83. [DOI] [PubMed] [Google Scholar]