Abstract
Background: X linked hyper-IgM (XHIM) is a primary immunodeficiency caused by mutations in the tumour necrosis factor superfamily 5 gene, TNFSF5, also known as the CD40 ligand (CD40L) gene. Patients often present with recurrent infections, and confirmation of a diagnosis of XHIM enables appropriate therapeutic interventions, including replacement immunoglobulin, antibiotics, and bone marrow transplantation.
Aim: To review and optimise the institution’s diagnostic strategy for XHIM.
Method: Samples from 65 boys were referred to this centre for further investigation of suspected XHIM. The results, which included a flow cytometric whole blood assay for CD40L expression followed by mutation analysis in selected patients, were reviewed.
Results: Twenty one patients failed to express CD40L and TNFSF5 mutations were found in 20 of these patients. In contrast, no TNFSF5 mutations were found in 16 patients with weak expression of CD40L. Interestingly, one quarter of patients with confirmed XHIM who had TNFSF5 mutations had low concentrations of IgG, IgA, and IgM. Most of the remaining patients with XHIM had the classic pattern of normal or raised IgM with low concentrations of IgA and IgG.
Conclusions: This study demonstrates the usefulness of the whole blood staining method as a rapid screen to select patients for subsequent TNFSF5 mutation analysis, and shows the benefits of a unified protein/genetic diagnostic strategy.
Keywords: CD40 ligand, X linked hyper-IgM, immunodeficiency, diagnostic strategy, TNFSF5
X linked hyper-IgM (XHIM) syndrome is a primary immunodeficiency caused by mutations in the tumour necrosis factor superfamily 5 gene, TNFSF5. Patients with XHIM syndrome usually present in the first years of life with recurrent infections, including respiratory tract infections and Pneumocystis carinii pneumonia (PCP). Typically, laboratory investigations reveal hypogammaglobulinaemia of the A and G isotypes, with a normal or raised IgM value. Proportions of T, B, and natural killer cells are normal. Over time, patients may be neutropenic, may contract cryptosporidium causing diarrhoea and/or liver disease, and may develop malignancy. Currently, the only curative treatment is bone marrow transplantation, although some patients remain reasonably well on replacement immunoglobulin and prophylactic antibiotics (reviewed by Notorangelo and Hayward1).
The TNFSF5 gene responsible for the XHIM syndrome encodes the CD40 ligand (CD40L; also known as CD154), a type II membrane glycoprotein expressed predominately on activated CD4+ T cells.2,3 CD40, the receptor for CD40L, is expressed on antigen presenting cells including B cells, dendritic cells, and macrophages. CD40–CD40L interactions provide a costimulatory signal for T cells and lead to T cell activation (reviewed by van Kooten and Banchereau4). The engagement of CD40 by CD40L on B cells leads to B cell proliferation and immunoglobulin class switching (reviewed by Durie and colleagues5). The defects observed in affected patients may result from a lack of B and T cell activation and communication.
“The TNFSF5 gene responsible for the X linked hyper-IgM syndrome encodes the CD40 ligand, a type II membrane glycoprotein expressed predominately on activated CD4+ T cells”
Several laboratories have developed immunostaining techniques using CD40L monoclonal antibodies or a CD40–Ig fusion protein,6,7 and the failure of expression of CD40L on activated CD4 cells has been included in the diagnostic criteria for the XHIM syndrome.8,9 Previous studies have primarily been performed by immunostaining of separated peripheral blood mononuclear cells.6,7 Several different staining patterns have been described, with partial CD40L expression being documented in patients with a milder form of XHIM.7 In addition, patients may express a non-functional form of CD40L and thus, although normal by protein expression analysis, have XHIM syndrome.10 Like most primary immunodeficiencies, no genotype–phenotype correlation has been described.11,12
We have reviewed results obtained using a whole blood CD40L expression assay and describe the correlation between protein expression and mutation detection. Furthermore, we suggest a stratified protein/genetic diagnostic approach for XHIM.
MATERIALS AND METHODS
Study population
Consultant immunologists referred 65 patients for XHIM testing based on clinical suspicion. All patients were screened by immunostaining for CD40L expression. Patients whose cells failed to express CD40L were screened for mutations in their TNFSF5 gene. Patients whose cells showed weak expression of CD40L had their TNFSF5 gene analysed if there was a strong clinical suspicion of XHIM syndrome. Table 1 ▶ summarises the patients’ data. Informed consent was given for CD40L and TNFSF5 analysis and blood was collected under institutional guidelines.
Table 1.
Details of patients whose CD40 ligand (CD40L) expression was analysed by immunostaining
| Patient | CD40L expression | Age | IgM | IgA | IgG | Mutation | Evidence of mutation being disease causing |
| 1 | Weak | 11 years | H | H | H | Not done | |
| 2 | Weak | 11 years | L | L | No sample | ||
| 3 | Weak | 2 months | N | N | N | Not done | |
| 4 | Weak | 3 months | N | N | N | Not done | |
| 5 | Weak | 3 years | N | H | Not done | ||
| 6 | Weak | 15 years | L | L | None found | ||
| 7 | Weak | 11 years | L | L | L | None found | |
| 8 | Weak | 13 years | L | L | L | Not done | |
| 9 | Weak | 18 years | N | N | N | Not done | |
| 10 | Weak | 45 years | N | H | R | Not done | |
| 11 | Weak | 5 months | N | N | M | None found | |
| 12 | Weak | 1 years | None found | ||||
| 13 | Weak | 1 years | L | Not done | |||
| 14 | Weak | 32 years | N | L | H | No sample | |
| 15 | Weak | 1 month | None found | ||||
| 16 | Weak | 3 months | None found | ||||
| 17 | Absent | 9 years | L | L | L | W140G (c.418t→g) Exon 5 | Mutation previously described15 |
| AC: 4M0002313 C007014 | Disrupts conserved a.a. in TNF homology domain | ||||||
| 18 | Absent | 18 years | H | L | L | c.444-448gagca→c Exon 5 | Patient previously described16 |
| AC: 4M0003613 C002914 | Disrupts reading frame | ||||||
| 19 | Absent | 13 years | L | L | R | G38R(c.112g→c) Exon 1 | Patient previously described16 |
| AC: 4M00018 & 4M0001913 C002814 | Disrupts transmembrane domain | ||||||
| 20 | Absent | 3 months | H | H | N | No sample | |
| 21 | Absent | 1 years | N | L | L | c.302-303insA Exon 3 | Reading frame shift |
| 22 | Absent | 4 months | H | L | L | IVS3+1g→t | May affect splicing |
| 23 | Absent | 15 years | N | L | R | IVS3-915a→t | May affect splicing |
| 24 | Absent | 3 years | L | L | L | c.158-161delTAGA Exon 2 | Mutation previously described14 |
| 25 | Absent | 20 years | H | L | R | c.158-161 delTAGA Exon 2 | Mutation previously described14 |
| 26 | Absent | 29 years | c.539delA Exon 5 | Reading frame shift | |||
| 27 | Absent | 29 years | H | L | L | c.508-516delTATATCTAT Exon 5 | 3 a.a. deletion from TNF homology domain |
| 28 | Absent | 3 years | N | L | L | None found | |
| 29 | Absent | 1 years | c.521-522delAA Exon 5 | Reading frame shift | |||
| 30 | Absent | 2 months | N | L | M | Q174R (c.521a→g) Exon 5 | Relative previously described.16 Arisen de novo |
| AC: 4M0002513 C003814* | Great grandmother did not carry the mutation | ||||||
| 31 | Absent | 11 years | H | L | T254M (c.761c→t) Exon 5 | Mutation previously described16 | |
| AC: 4M00021 & 4M0002213 | Not seen in 100 normal chromosomes | ||||||
| 32 | Absent | 2 months | c.242-243insT Exon 2 | Relative previously described16 | |||
| AC: 4M0004713 C000814 | |||||||
| 33 | Absent | 10 years | H | L | del ex 5 Exon 5 | Mutation previously described16 | |
| AC: 4M0003513 | |||||||
| 34 | Absent | 16 years | L | L | R | G257S (c. 769g→a) Exon 5 | Patient previously described14,16 |
| AC: 4M0002013 C003014 | Disrupts conserved a.a. in TNF homology domain | ||||||
| 35 | Absent | 29 years | T254M (c.761c→t) Exon 5 | Mutation previously described14 | |||
| Not found in 100 normal X chromosomes | |||||||
| 36 | Absent | 4 months | N | L | L | c.511-512delAT Exon 5 | Reading frame shift |
| 37 | Absent | 13 years | H | L | L155P (c.464t→c) Exon 5 | Mutation previously described14 | |
| Not found in 100 normal X chromosomes | |||||||
| 3 | Present | 4 months | N | N | M | ||
| 38 | Present | 7 years | L | L | L | ||
| 39 | Present | 8 years | L | L | N | ||
| 40 | Present | 6 years | L | L | L | ||
| 41 | Present | 5 years | L | L | L | ||
| 42 | Present | 11 years | N | N | N | ||
| 43 | Present | 49 years | H | N | N | ||
| 44 | Present | 20 years | L | L | L | ||
| 45 | Present | 4 years | N | N | N | ||
| 46 | Present | 4 years | N | N | N | ||
| 47 | Present | 9 months | N | L | N | ||
| 48 | Present | 10 years | N | L | L | ||
| 49 | Present | 4 months | H | H | N | ||
| 50 | Present | 19 years | N | N | L | ||
| 51 | Present | 1 years | |||||
| 52 | Present | 4 years | N | N | N | ||
| 53 | Present | 23 years | |||||
| 54 | Present | 14 years | N | L | N | ||
| 55 | Present | 7 years | |||||
| 56 | Present | 7 years | N | N | N | ||
| 57 | Present | 11 months | N | N | L | ||
| 58 | Present | 23 years | N | N | L | ||
| 59 | Present | 12 years | L | L | L | ||
| 60 | Present | 23 years | N | H | L | ||
| 61 | Present | 45 years | L | L | L | ||
| 62 | Present | 23 years | N | N | N | ||
| 63 | Present | 41 years | H | N | N | ||
| 64 | Present | 2 months | H | N | H | ||
| 65 | Present | 8 years | N | N | L |
CD40L expression is described as present, absent, or weak. The age listed is the age of the patient when immunostaining was performed. Immunoglobulin concentrations are listed as high (H), normal (N), low/absent (L), maternal (M), or replacement (R) using age appropriate normal ranges. Mutation data are only presented for TNFSF5 gene analysis—the exon/intron in which the mutation resides is given, together with the reference if previously reported, and CD40Lbase accession number (AC). Mutation nomenclature is according to the nomenclature working group.17 *Accession number for a relative. a.a., amino acid.
Cell preparation and culture
A 1 ml aliquot of blood was collected in lithium heparin from the patient. Patient samples were analysed in parallel with a normal control. The blood was mixed with an equal volume of RPMI (Invitrogen, Paisley, UK) containing 10% fetal calf serum (Labtech, Ringmer, UK). Half was incubated as an unstimulated sample at 37oC overnight. To the remainder, 3 ng of phytohaemagglutinin (Murex Diagnostics, Dartford, UK) and 2 pg of phorbol myristate acetate (Sigma, Poole, Dorset, UK) were added and the cells incubated at 37oC overnight.
Immunostaining
A 100 μl aliquot of the cell suspension was stained with anti-CD45 PerCP antibody (Becton Dickinson, Oxford, UK) along with a fluorescein isothiocyanate (FITC) isotype control (Becton Dickinson), anti-CD25 FITC (Becton Dickinson), anti-CD4 PE (Becton Dickinson), anti-CD69 FITC (Becton Dickinson), or anti-CD154 FITC (Alexis, Nottingham, UK). Cells were lysed, washed, and fixed according to Becton Dickinson protocols. Two thousand and five hundred CD45+ lymphocyte events were acquired on a FacsCalibur (Becton Dickinson). Events were analysed using CellQuest (Becton Dickinson). CD45+ lymphocytes were gated based upon side scatter and CD45 expression. Stimulated and unstimulated isotype controls were analysed by histogram overlays. CD154 staining in unstimulated cells was analysed by overlaid histograms with CD69 staining, CD25 staining, and CD154 staining in stimulated cells. The control samples were compared with the patient samples. Samples with suboptimal expression of CD69 and CD25 were reported as poorly activated and the test was repeated on another sample.
Genetic analysis
Polymerase chain reaction primers for exons 1–5 of the CD40L gene were: exon 1 forward (f): 5′-CTC ATG CTG CCT CTG CCA C and reverse (r): 5′-CCT ATG AAT TAG TAA GGA CCC; exon 2 f: 5′-CAT TAG CTG TAT TCT CCT TCC and r: 5′-GAC ACT TTT ACC AGT AAT TAA GC; exon 3 f: 5′-CAA CAG AGT AAT GAC AGA TGC and r: 5′-TTC ATA GAA ATT AGC AAA TAG CAA; exon 4 f: 5′-TGT AGA ACT GGA CCA GAT AG and r: 5′-GGT AAC ATG ACT TCG GCA TC; exon 5 f: 5′-CTC TGC TTC ACC TCA CCA C and reverse.16
Amplification conditions for the exons were 94°C for three minutes, then 29 cycles of 94°C for 30 seconds, 55°C for one minute, and 72°C for 30 seconds, followed by a final extension of 72°C for 10 minutes. Mutation analysis was either by fluorescent cycle sequencing using dRhodamine/BigDye™ kits (PE Applied Biosystems, Foster City, California, USA) or by single strand conformation polymorphism (SSCP) analysis, as described previously,18 followed by sequencing of shifted fragments. Before SSCP gel electrophoresis, the products (20 μl) of exons 1 and 5 were digested with 10 units of restriction enzymes HaeIII and AlwNI/MboI, respectively, according to the manufacturer’s instructions (New England Biolabs, Hitchin, UK). Mutations were named according to the nomenclature guidelines of the nomenclature working group.17 Mutations identified have been submitted to CD40Lbase14 and accession numbers are given in table 1 ▶.
Immunoglobulin quantification
Immunoglobulins were measured using a Dade Behring (Milton Keynes, UK) nephelometer (BNII), according to the manufacturer’s guidelines. Immunoglobulin concentrations were compared with age related normal ranges.
RESULTS
CD40L expression patterns in whole blood
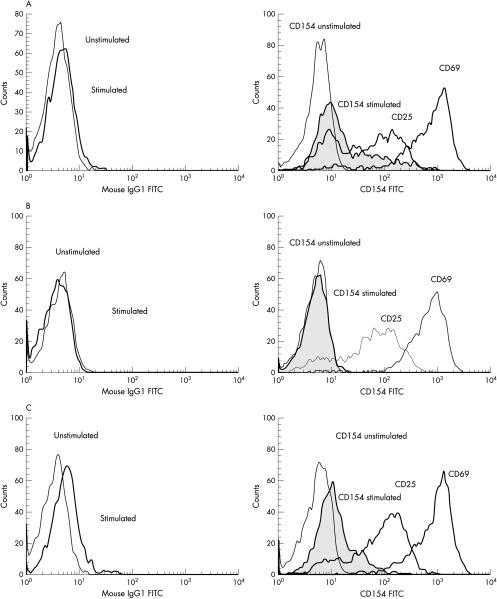
Figure 1A ▶ shows the staining pattern seen in most of the referred patients (45%) who had normal CD40L expression. The 21 patients who failed to express CD40L were easily distinguished (fig 1B ▶). It is important to note that the isotype control shifts upon stimulation (1A and B, left hand panels), and this shift must not be misinterpreted as weak expression. In addition, CD40L expression was examined in 11 healthy children (6 months to 14 years); all expressed normal amounts of CD40L (data not shown).
Figure 1.
CD40 ligand (CD40L) expression as detected by immunostaining. All figures show fluorescence activated cell sorter histograms with fluorescein isothiocyanate (FITC) staining in a log scale along the x axis and counts (number of events) along the y axis. (A) An example of normal CD40L expression. The left hand panel shows the isotype controls in stimulated and unstimulated cells. The right hand panel shows CD40L expression in unstimulated cells (CD154 unstimulated) and CD40L expression in stimulated cells (CD154 stimulated) overlayed on CD25 and CD69 expression. (B) An example of absent CD40L expression. Panels and lines are the same as in (A). (C) An example of weak CD40L expression. Panels and lines are the same as in (A).
We identified a group of patients (16) with weak expression of CD40L (fig 1C ▶). These patients have normally activated CD4 positive T cells, as measured by their CD25 and CD69 expression (fig 1C ▶, right panel), but they fail to express normal amounts of CD40L. Some of these patients were seriously ill (for example, they had sepsis or lymphoproliferative disease), whereas others were under 6 months of age. Based on the CD40L staining pattern and clinical details, TNFSF5 mutation analysis was performed in a subset of these patients and the results are shown in table 1 ▶.
Correlation between CD40L expression and TNFSF5 mutation analysis
Lack of CD40L expression
Mutation analysis was undertaken in 20 of 21 patients lacking CD40L expression (a DNA sample was not available from the remaining patient). By a combination of SSCP screening and sequencing, mutations were found in 19 of these 20 patients (table 1 ▶), confirming the diagnosis of XHIM. Of the mutations identified, seven have not been reported previously in the literature.
As with previous reports, the mutations identified included insertions, deletions, splice site mutations, and missense mutations.19,20 Most of the deletion, insertion, and splice site mutations are predicted to cause a reading frame shift, and are probably pathogenic. The one in frame deletion (c.508–516delTATATCTAT) is predicted to remove three amino acids from a highly conserved part of the tumour necrosis factor homology domain, and is also likely to be pathogenic. One of the deletions (c.158–161delTAGA) was found in two apparently unrelated individuals. This mutation has also been reported in two other individuals on the CD40L database.14
Weak CD40L expression
TNFSF5 mutation analysis was also carried out on eight of the 16 patients who had weak expression of CD40L and no mutations were identified, indicating that these patients are unlikely to have XHIM and that the weak CD40L expression seen was the result of other causes.
The remaining nine patients with weak CD40L expression did not have mutation analysis undertaken for the following reasons: no DNA available (n = 2), clinician declined (n = 2), another diagnosis made (n = 4), and normal CD40L on repeat (n = 1).
Normal CD40L expression
None of the 29 patients with normal CD40L expression was screened for mutations.8,9 Many had originally been referred because they appeared to have either common variable immunodeficiency (CVID) or an undefined combined immunodeficiency (a cellular immunodeficiency with or without a humoral component) and a diagnosis of XHIM was being excluded. This group of patients serves as an important control group for the assay because some of them were ultimately diagnosed with other disorders, including X linked lymphoproliferative disease (XLP; n = 2), lymphoma (n = 2), and Wiskott-Aldrich syndrome (WAS; n = 1), whereas 17 of them fit the European Society for Immunodeficiency diagnostic criteria for CVID.9
Immunoglobulin production and CD40L
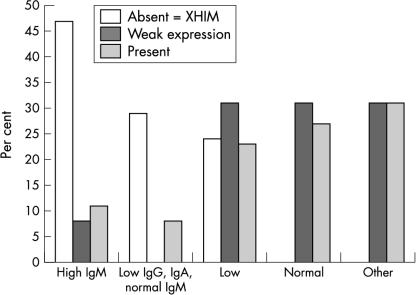
Immunoglobulin measurements were available on 56 patients (fig 2 ▶). Eight of the 17 patients with confirmed XHIM had raised serum concentrations of IgM, whereas only one of the 13 patients in the weak expression group had raised serum IgM concentrations. Five of the 17 patients with confirmed XHIM presented with normal concentrations of IgM and low IgG and IgA, whereas none of the patients with weak expression and only two of the 26 patients with normal staining had this pattern of immunoglobulin production. In all three groups, up to a third of patients had low concentrations of IgG, IgA, and IgM. None of the patients with genetically defined XHIM had normal immunoglobulin concentrations at the time that their CD40L was analysed.
Figure 2.
Patients with X linked hyper-IgM (XHIM) syndrome have abnormal immunoglobulin concentrations. IgM, IgA, and IgG concentrations were measured by nephelometry and are presented graphically here. Patients were grouped by CD40 ligand expression (absent, weak, or normal expression) and by their pattern of immunoglobulin production. The “high IgM” group denotes the immunoglobulin pattern seen in patients with raised concentrations of IgM and normal or low concentrations of IgG and IgA. “Low IgG, IgA, normal IgM” represents those patients with low concentrations of IgG and IgA and normal concentrations of IgM. “Low” represents those patients with low concentrations of IgG, IgA, and IgM. “Other” includes those patients with raised concentrations of IgG or IgA and patients with low IgG, IgA, or IgM, but with the other two classes of immunoglobulin present at normal concentrations.
Infants under 6 months of age may fail to express normal amounts of CD40L
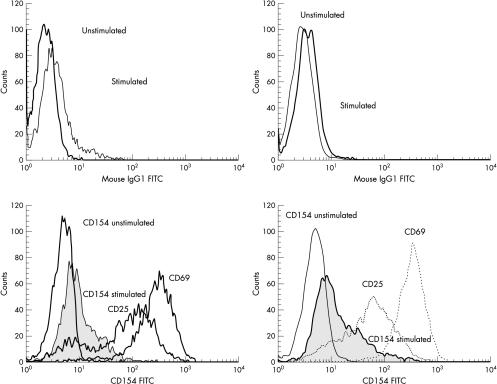
CD40L expression was analysed on peripheral blood cells from 13 patients less than 6 months of age. Three of these patients expressed normal amounts of CD40L, whereas five showed weak expression and five had absent expression (table 1 ▶). Genetic analysis identified mutations in four patients with absent CD40L expression (no DNA sample was available from the fifth). Of the five patients with weak expression, no mutation was found in the three children screened. In addition, one of the patients with abnormal expression had their CD40L immunostaining repeated two months later, and CD40L expression had normalised (fig 3 ▶).
Figure 3.
CD40 ligand (CD40L) expression in infants increases with age. The left hand panels show CD40L expression at 2 months in a representative patient. The right hand panels shows the same patient’s CD40L expression at 4 months. The top panel in each set shows the isotype controls in stimulated and unstimulated cells. The bottom panel in each set shows CD40L expression in unstimulated cells (CD154 unstimulated) and CD40L expression in stimulated cells (CD154 stimulated), alongside CD25 and CD69 expression.
DISCUSSION
Using a whole blood method to assay CD40L expression, we describe the immunostaining analysis of 65 patients suspected of having XHIM syndrome and the results of TNFSF5 mutation analysis and immunoglobulin measurements in a subset of these patients. Our data indicate that immunostaining of whole blood using an anti-CD40L monoclonal antibody is a rapid, efficient, and sensitive method of screening for XHIM syndrome in children older than 6 months. A high mutation detection rate (20 of 21) was found for patients lacking CD40L expression, whereas no mutations were identified in those with weak expression of CD40L. This is in contrast to previously reported studies,7 and may be attributable to different antibodies and methodological differences. In addition, in our study, one quarter of the patients with genetically confirmed XHIM had panhypogammaglobulinaemia (low IgG, IgA, and IgM) at the time of testing.
The whole blood CD40L assay used in our study is a rapid technique and uses only a small blood volume. Accurate interpretation of the assay results requires: (1) inclusion of a normal sample in every assay run to validate the technique; (2) isotype controls for the stimulated and unstimulated samples must be run and the shift in the isotype control after stimulation must be accounted for when examining the expression of CD40L; (3) consideration of the clinical status of the patient because extremely ill patients may express CD40L suboptimally; and (4) knowledge of the patient’s age because infants under 6 months of age may fail to express normal amounts of CD40L (results of our study).21
Our data suggest that weak expression of CD40L is less likely to be the result of mutations in the TNFSF5 gene, but more likely to be a result of other factors, such as patient age and clinical condition.
“The absence of a high IgM should not preclude testing for CD40 ligand expression”
CD40L screening should be performed in the context of appropriate clinical history and baseline immunological investigations. Patients most likely to have XHIM syndrome are those with normal or raised serum IgM and low or absent serum IgG, opportunistic infections such as PCP and cryptosporidium, and an X linked family history.1,8,9,12,14 The high IgM concentrations have been assumed to reflect chronic antigenic stimulation because values may normalise on appropriate treatment.12 As shown in fig 2 ▶, patients with normal immunoglobulin concentrations are extremely unlikely to have XHIM syndrome. We have shown that a large proportion of patients (one quarter) with low IgM also have low/absent IgA and IgG. Thus, the absence of a high IgM should not preclude testing for CD40L expression.
Take home messages.
TNFSF5 mutations were found in 20 of the 21 patients who failed to express CD40 ligand (CD40L), whereas no TNFSF5 mutations were found in 16 patients with weak expression of CD40L
A subset (one quarter) of patients with confirmed X linked hyper-IgM had low concentrations of IgG, IgA, and IgM, whereas most of the remainder had the classic pattern of normal or raised IgM with low concentrations of IgA and IgG
The whole blood staining method is a useful rapid screen to select patients for subsequent TNFSF5 mutation analysis to confirm the diagnosis
A unified protein/genetic diagnostic strategy is a useful approach to identify patients with various immunodeficiencies
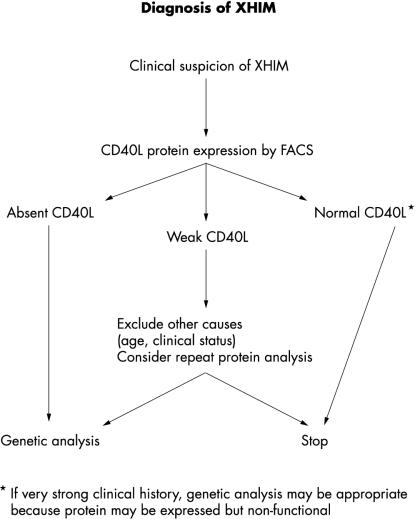
Consistent with previous reports,19 mutation analysis of those patients with no CD40L expression demonstrates that SSCP is a sensitive method for detecting mutations in the TNFSF5 gene, with mutations detected in 20 of the 21 patients. Most (11 of 16) mutations identified involved exon 5. This exon contains approximately half of the coding sequence of the gene, and starting mutation screening with exon 5 may be a time and cost effective strategy. We have shown that lack of CD40L expression is an excellent predictor for the presence of an XHIM mutation. It is possible that a non-functional protein may be expressed, so, if clinical suspicion is high, genetic analysis should be undertaken. We propose that the flow diagram in fig 4 ▶ is a time and cost efficient pathway for the exclusion or diagnosis of XHIM syndrome.
Figure 4.
Flow diagram for the diagnosis of X linked hyper-IgM (XHIM) syndrome. FACS, fluorescence activated cell sorter; CD40L, CD40 ligand.
In conclusion, the use of the fluorescence activated cell sorter based assay described in our study enables the rapid identification of patients who are likely to have XHIM syndrome. The diagnosis of XHIM syndrome can then be confirmed by TNFSF5 mutation analysis, enabling accurate carrier testing and prenatal diagnosis. This approach, of screening by protein expression followed by mutation analysis is being routinely used in our centre to identify patients with several different immunodeficient conditions including X linked severe combined immunodeficiency, WAS, and XLP,18,22–24 and has the advantage of being rapid and cost effective, with the potential to be applied to other genetic conditions.
Acknowledgments
Thanks to T Cranston and Dr F Katz for their contributions to the genetic analysis, to the staff of the clinical immunology laboratory at Great Ormond Street Hospital for their technical help with immunological assays, and to the clinicians caring for the patients reported.
Abbreviations
CD40L, CD40 ligand
CVID, common variable immunodeficiency
f, forward
FITC, fluorescein isothiocyanate
PCP, Pneumocystis carinii pneumonia
r, reverse
SSCP, single strand conformation polymorphism
WAS, Wiskott-Aldrich syndrome
XHIM, X linked hyper-IgM
XLP, X linked lymphoproliferative disease
REFERENCES
- 1.Notarangelo LD, Hayward AR. X-linked immunodeficiency with hyper-IgM (XHIM). Clin Exp Immunol 2000;120:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graf D, Korthauer U, Mages HW, et al. Cloning of TRAP, a ligand for CD40 on human T cells. Eur J Immunol 1992;22:3191–4. [DOI] [PubMed] [Google Scholar]
- 3.Hollenbaugh D, Grossmaire LS, Kulles CD, et al. The human T cell antigen gp39, a member of the TNF gene family, is a ligand for the CD40 receptor: expression of a soluble form of gp39 with B cell co-stimulatory activity. EMBO J 1992;11:4313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Kooten C, Banchereau J. Functions of CD40 on B cells, dendritic cells and other cells. Curr Opin Immunol 1997;9:330–7. [DOI] [PubMed] [Google Scholar]
- 5.Durie FH, Fay TM, Mastris SR, et al. The role of CD40 in the regulation of humoral and cell-mediated immunity. Immunol Today 1994;15:406–11. [DOI] [PubMed] [Google Scholar]
- 6.Callard RE, Smith SH, Herbert J, et al. CD40 ligand (CD40L) expression and B cell function in agammaglobulinemia with normal or elevated levels of IgM (HIM). J Immunol 1994;22:3295–306. [PubMed] [Google Scholar]
- 7.Seyama K, Nonoyama S, Gangsaas I, et al. Mutations in CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper IgM syndrome. Blood 1998;92:2421–34. [PubMed] [Google Scholar]
- 8.Rosen FS, Eibl M, Roifman C, et al. Primary immunodeficiency diseases, report of an IUIS scientific committee. Clin Exp Immunol 1999;118(suppl 1):1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.European Society for Immunodeficiency. Diagnostic criteria for primary immunodeficiency. www.esid/13xhim.html.
- 10.Su L, Garber EA, Hsu YM. CD154 variant lacking tumor necrosis factor homologous domain inhibits cell surface expression of wild-type protein. J Biol Chem 2001;276:1673–6. [DOI] [PubMed] [Google Scholar]
- 11.de Vries E, Noordzij J, Davies EG, et al. The 782C_T (T254M) mutation: lack of a tight phenotype–genotype relationship. Blood 1999;94:1488–9. [PubMed] [Google Scholar]
- 12.Levy J, Espanol-Boren T, Thomas C, et al. Clinical spectrum of X-linked hyper-IgM syndrome. J Pediatr 1997;131:47–54. [DOI] [PubMed] [Google Scholar]
- 13.Original CD40L base (now closed to submission): www.expasy.ch/cd40lbase.
- 14.CD40L base: http://www.uta.fi/imt.bioinfo/CD40Lbase.
- 15.Kroczek RA, Graf D, Brungnoni D, et al. Defective expression of CD40 ligand on T cells causes X-linked immunodeficiency with hyper-IgM (HIGM1). Immunol Rev 1994;183:39–59. [DOI] [PubMed] [Google Scholar]
- 16.Katz F, Hinshelwood S, Rutland P, et al. Mutation analysis in CD40 ligand deficiency leading to X-linked hypogammaglobulinaemia with hyper IgM syndrome. Hum Mutat 1996;8:223–8. [DOI] [PubMed] [Google Scholar]
- 17.Antonarakis SE, and the Nomenclature Working Group. Recommendations for a nomenclature system for human gene mutations. Hum Mutat 1998;11:1–3. [DOI] [PubMed] [Google Scholar]
- 18.Gilmour KC, Cranston T, Loughlin S, et al. Rapid protein-based assays for the diagnosis of T-B+ severe combined immunodeficiency. Br J Haematol 2001;112:671–6. [DOI] [PubMed] [Google Scholar]
- 19.Lin Q, Rohrer J, Cutler Allen R, et al. A single stranded conformation polymorphism study of CD40 ligand: efficient mutation analysis and carrier detection for X-linked hyper IgM syndrome. J Clin Invest 1996;97:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notarangelo LD, Peitsch MC. CD40Lbase: a database of CD40L gene mutations causing X-linked Hyper-IgM syndrome. Immunol Today 1996;17:511–16. [DOI] [PubMed] [Google Scholar]
- 21.Nonoyama S, Penix LA, Edwards CP, et al. Diminished expression of CD40 ligand by activated neonatal cells. J Clin Invest 1995;95:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genevier HC, Hinshelwood S, Gaspar HB, et al. Expression of Bruton’s tyrosine kinase within the B cell lineage. Eur J Immunol 1994;24:3100–5. [DOI] [PubMed] [Google Scholar]
- 23.Qasim W, Gilmour KC, Heath S, et al. Protein assays for diagnosis of Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Br J Haematol 2001;112:861–5. [DOI] [PubMed] [Google Scholar]
- 24.Gilmour KC, Cranston T, Jones A, et al. Diagnosis of X-linked lymphoproliferative disease by analysis of SLAM-associated protein expression. Eur J Immunol 2000;30:1691–7. [DOI] [PubMed] [Google Scholar]