Abstract
Background: Deletions in chromosome 3 occur frequently in uterine cervical carcinoma (CA-CX). The common consensus regions deleted during CA-CX development are not well defined, and have not been correlated with tumour progression.
Aims: To define specific regions of chromosome 3 deleted during development of CA-CX and to correlate these with clinicopathological data.
Methods: Deletion mapping of chromosome 3 was done in seven cervical intraepithelial neoplasia (CIN) and 43 primary CA-CX samples using 20 highly polymorphic microsatellite markers.
Results: Deletions of chromosome 3 were significantly associated with tumour progression. High frequencies (33–53%) of loss of heterozygosity (LOH) were found in 3p26.1, 3p22.3, 3p21.2, and 3p13, suggesting the location of putative tumour suppressor genes (TSGs) in these regions. Among these four regions, deletions in 3p21.2 were suggested to occur early during CA-CX development. A significant correlation was found between LOH at 3p26.1 and 3p22.3 with tumour progression from stage I/IIB to stage III/IV. No association was found with the highly deleted regions and human papillomavirus positivity, parity, or menopausal status. Microsatellite size alteration was seen in only seven of the samples. However, rare biallelic alterations were seen in and around the highly deleted regions. Loss of normal copy of chromosome 3 and interstitial alterations in chromosome 3p were seen in some samples.
Conclusion: These four regions on chromosome 3p may be differentially deleted during specific stages of CA-CX development. The putative TSGs located in these regions may have a cumulative effect on tumour progression.
Keywords: squamous cell carcinoma of the uterine cervix, chromosome 3 alterations, tumour suppressor gene(s), human papilloma virus infection
Globally, uterine cervical carcinoma (CA-CX) is the fifth most common cancer,1 and each year there are approximately 450 000 new cases of invasive cervical cancer diagnosed (about 12% of all cancers in women), with 300 000 deaths from the disease (World Health Organisation 1996). CA-CX is the most common cancer among Indian women,2 and approximately 1/5 to 1/6 of cervical cancers worldwide occur in India.3
Several possible contributory factors have been implicated in the development of CA-CX, such as the use of oral contraceptives, young age at marriage, multiparity, immunosuppression, low socioeconomic and educational status, multiple sex partners, smoking, and others.4–8 However, no one has linked menopausal status with the incidence of CA-CX, although an association has been found between menopausal status and breast and biliary tract cancer.9,10 Human papillomavirus (HPV) infection has been shown to be one of the most important aetiological factors in the development of CA-CX, particularly infection with high risk HPV types 16, 18, 33, and 42.11–14 Because only a minority of HPV positive lesions progress to invasive cancer after a long latent period, additional somatic events, including the functional inactivation of tumour suppressor genes (TSGs) and activation of oncogenes, are thought to be required for malignant transformation.15,16
“Whereas gain on chromosome 3q may represent the presence of an oncogene at this locus, chromosomal losses might represent the location of putative tumour suppressor genes”
Cytogenetic analysis of CA-CX lesions, although limited, has revealed frequent non-random chromosomal aberrations, including deletions, amplifications, and isochromosome formation.16 Comparative genomic hybridisation of cervical tumours has identified over-representation of chromosome 3q as a recurrent event in invasive tumours and under-representation of chromosomal arms 2q, 3p, 6p, 8p, 11q, and 13q during tumour progression.17,18 Whereas gain on chromosome 3q may represent the presence of an oncogene at this locus, chromosomal losses might represent the location of putative TSGs.
Loss of heterozygosity (LOH) studies of primary CA-CX have shown allelic loss on several chromosomal arms, such as 3p, 6p, 11q, and 18q.19,20 Although the involvement of chromosome 3p deletions in the development of CA-CX has been confirmed by LOH studies using chromosome specific probes, there are several discrepancies between the reports defining the commonly deleted regions and frequencies of LOH. Mullokandov and colleagues20 found an overall frequency of 39% LOH at the 3p21.3, 3p22.1–24.1, and 3p25.1–25.3 regions, whereas Larson and colleagues21 detected an overall frequency of 70% LOH in CA-CX, with the most frequent deletion being at the 3p14 region. However, the highest frequencies of LOH detected by Mitra et al were 40% and 32% at the 3p21.33–22.1 and 3q28–29 regions, respectively, in CA-CX samples from Indian patients using restriction fragment length polymorphism (RFLP) markers.19 Some investigators reported a gradual increase in the deletion frequency of chromosome 3p alleles along with tumour progression using samples at different stages from the same individual, but no statistical correlations were made with the associated regions, probably because of the low sample number.22,23 In addition to LOH, microsatellite size alteration (MA) is another genetic change that has been associated with several human cancers.24 The mechanisms underlying MA are currently unknown, but they probably represent a form of genomic instability.24 About 35% of CA-CX samples of Western origin have been shown to exhibit MA.22 Hampton and colleagues25 have demonstrated an association between ethnicity and aetiological factors and the chromosomal deletions seen in CA-CX development; the frequency of chromosome 4p deletion differed between CA-CX samples of Indian origin and those from patients of white/Hispanic/American black origin using the same RFLP probes. Until now, few attempts have been made to carry out deletion mapping of chromosome 3 in CA-CX samples from Indian patients using highly polymorphic microsatellite markers to evaluate the deletion pattern, in addition to localising the candidate TSGs associated with the development of this tumour.
Thus, in our present study, we have attempted to map the frequently deleted regions on chromosome 3 in 50 cervical lesions of Indian patients using 20 highly polymorphic microsatellite markers. The deletion pattern and MA were correlated with different clinicopathological parameters, such as clinical stage, HPV infection, and menopausal status, to investigate whether there is an association between any of these factors and chromosomal alterations.
MATERIALS AND METHODS
Sample collection and clinical data
Fifty freshly operated uterine cervical lesions from previously untreated and unrelated individuals with corresponding normal tissue or peripheral blood leucocytes were used for our analysis (table 1 ▶). The tissues were obtained from the patients treated at the hospital section of Chittaranjan National Cancer Institute, India after appropriate informed consent by the patients and approval of the protocol by the hospital authorities. The samples were frozen immediately after collection and stored at −80°C until use.
Table 1.
Clinicopathological features and FAL index of uterine cervical carcinoma samples
| Tumour | Age/Religion | Clinical stage | Histology | Years married | Menopausal status | Parity | HPV status | FAL index | Mean FAL index |
| 310 | 40/H | CIN-I | Mild Dys | 15 | Pre | 0+0 | 16 | 0.14 | |
| 4736 | 48/M | CIN-II | Mod Dys | 15 | Post | 4+0 | 18 | 0 | |
| 4737 | 55/H | CIN-II | Mod Dys | 18 | Post | 2+0 | A | 0 | |
| 2740 | 47/H | CIN-II | Mod Dys | 20 | Post | 5+0 | 16 | 0.05 | 0.107 |
| 2661 | 42/H | CIN-III | Sev Dys | 19 | Post | 3+0 | 16 | 0 | |
| 224 | 48/H | CIN-III | Sev Dys | 13 | Post | 3+0 | 16 | 0.06 | |
| 930 | 40/H | CIN-III | Sev Dys | 16 | Post | 3+0 | 16 | 0.5 | |
| 4725 | 50/H | I | WDSCC | 16 | Post | 2+0 | A | 0 | |
| 3574 | 30/H | I | WDSCC | 13 | Pre | 6+0 | A | 0.06 | |
| 2890 | 52/H | I | MDSCC | 13 | Post | 5+0 | 16 | 0.06 | |
| 567 | 42/H | I | WDSCC | 16 | Pre | 3+2 | 18 | 0.14 | |
| 1441 | 42/H | I | WDSCC | 16 | Pre | 6+0 | 16 | 0.21 | |
| 3036 | 46/H | I | WDSCC | 10 | Pre | 1+0 | 16 | 0.27 | |
| 3776 | 48/H | I | WDSCC | 12 | Post | 5+0 | 16 | 0.35 | |
| 1684 | 46/H | I | WDSCC | 22 | Post | 3+0 | 16 | 0.47 | |
| 1319 | 40/H | I | WDSCC | 15 | Post | 5+0 | 16 | 1 | |
| 5210 | 60/H | IB | WDSCC | 10 | Post | 10+0 | 16 | 0.19 | |
| 506 | 56/H | IB | WDSCC | 17 | Post | 5+0 | 18 | 0 | |
| 1466 | 40/H | IB | MDSCC | 10 | Pre | 4+0 | 18 | 0.07 | |
| 4251 | 65/H | IB | WDSCC | 20 | Post | 4+0 | A | 0.08 | 0.196 |
| 3120 | 45/H | IB | MDSCC | 18 | Pre | 3+0 | 16 | 0.14 | |
| 2753 | 48/H | IB | MDSCC | 18 | Post | 8+0 | 16 | 0.17 | |
| 2884 | 60/H | IB | CIS | 15 | Post | 3+0 | 16 | 0.29 | |
| 1118 | 43/H | IB | WDSCC | 21 | Pre | 3+0 | 16 | 0.38 | |
| 2961 | 40/H | II | WDSCC | 18 | Pre | 4+0 | A | 0.07 | |
| 4840 | 47/H | II | WDSCC | 11 | Post | 4+0 | 18 | 0.08 | |
| 113 | 46/H | II | MDSCC | 17 | Pre | 3+0 | 16 | 0.18 | |
| 4900 | 25/H | IIB | WDSCC | 10 | Pre | 1+0 | 16 | 0 | |
| 5091 | 40/H | IIB | PDSCC | 14 | Pre | 5+0 | 16 | 0.07 | |
| 5169 | 56/H | IIB | MDSCC | 13 | Post | 6+0 | A | 0.13 | |
| 4849 | 40/H | IIB | WDSCC | 10 | Pre | 6+2 | 16 | 0.25 | |
| 3493 | 40/M | IIB | WDSCC | 25 | Pre | 2+0 | 16 | 0.25 | |
| 1145 | 48/H | III | WDSCC | 18 | Pre | 5+0 | 16 | 0.07 | |
| 2756 | 45/H | III | MDSCC | 19 | Pre | 4+0 | 16 | 0.28 | |
| 3221 | 38/H | III | WDSCC | 24 | Pre | 2+0 | 18 | 1 | |
| 4320 | 30/H | IIIB | PDSCC | 11 | Pre | 2+0 | 16 | 0.08 | |
| 3609 | 65/H | IIIB | MDSCC | 20 | Post | 2+0 | 16 | 0.15 | |
| 2760 | 38/H | IIIB | WDSCC | 17 | Pre | 3+0 | 18 | 0.2 | |
| 1178 | 60/H | IIIB | PDSCC | 15 | Post | 3+0 | 16 | 0.27 | |
| 568 | 50/H | IIIB | MDSCC | 14 | Post | 3+0 | 16 | 0.29 | |
| 54 | 47/H | IIIB | MDSCC | 15 | Post | 5+0 | 16 | 0.31 | 0.343 |
| 1636 | 60/H | IIIB | WDSCC | 13 | Post | 7+0 | 16 | 0.43 | |
| 1058 | 42/H | IIIB | PDSCC | 19 | Pre | 1+0 | 16 | 0.53 | |
| 2060 | 65/H | IIIB | WDSCC | 20 | Post | 6+0 | 16 | 0.61 | |
| 574 | 65/H | IV | WDSCC | 13 | Post | 8+0 | 16 | 0.08 | |
| 4171 | 55/H | IV | PDSCC | 13 | Post | 2+0 | 16 | 0.13 | |
| 3297 | 43/H | IV | WDSCC | 20 | Pre | 4+0 | 16 | 0.13 | |
| 4839 | 38/M | IV | PDSCC | 10 | Pre | 8+0 | 18 | 0.38 | |
| 666 | 50/M | IV | WDSCC | 11 | Post | 7+0 | 16 | 0.53 | |
| 1879 | 40/H | IV | WDSCC | 10 | Pre | 6+0 | 16 | 0.71 |
Religion: H, hindu; M, muslim; Histology: Mod, moderate; Sev, severe; Dys, dysplasia; WDSCC, well differentiated squamous cell carcinoma; MDSCC, moderately differentiated squamous cell carcinoma; PDSCC, poorly differentiated squamous cell carcinoma; Menopausal status: Pre, premenopausal; Post, postmenopausal; HPV status: A, HPV absent; 16, HPV-16 present; 18, HPV-18 present.
CIN, cervical intraepithelial neoplasia; FAL, fractional allele loss.
Table 1 ▶ provides a detail clinical history of the patients. Clinically, the tumours were graded and staged according to the FIGO classification. The average age of the patients was recorded as 44 years, with a mean length of time married of 16 years and parity ranging from one to 10. Twenty three of the patients were premenopausal, whereas the remainder (27) were postmenopausal.
Microdissection and DNA extraction
The normal cells present as contaminants in the primary uterine cervical lesions were removed by microdissection.26 Samples containing < 60% tumour cells were not analysed. High molecular weight genomic DNA was extracted from the tissues according to the standard procedure.26
Selection of microsatellite markers
Twenty highly polymorphic microsatellite markers spanning the entire length of chromosome 3 (fig 1 ▶) were selected for our analysis. Fifteen markers were taken from the chromosome 3p region and five markers from 3q (fig 1 ▶). The SST marker from the chromosome 3q27.3 region was used as a control. All the markers showed > 69% informativity in our samples (data not shown). The localisation and physical distance between the markers were obtained from http://genome.ucsc.edu, except for D3S1289, in which case the information was obtained from the Genome Database.
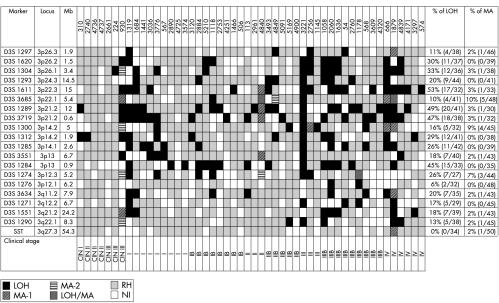
Figure 1.
Allele status of the chromosome 3 markers in the primary uterine cervical lesions. LOH, loss of heterozygosity; MA-1, microsatellite size alteration of one allele; MA-2, microsatellite size alteration of both alleles; LOH/MA, loss of one allele and size alteration of the other; RH, retention of heterozygosity; NI, non-informative; CIN, cervical intraepithelial neoplasia. D3S1297 is 1.9 Mb from chromosome 3p telomere.
Microsatellite analysis and interpretation of LOH and MA
A standard polymerase chain reaction (PCR) analysis containing a [γ-32P] ATP end labelled forward primer was performed in a 20 μl reaction volume, as described by Dasgupta et al.26 The DNA samples were amplified for 30 cycles in a thermal cycler (Techne, Cambridge, UK), comprising one minute at 95°C, one minute at the appropriate annealing temperature (50–60°C), and two minutes at 72°C, followed by a final extension step at 72°C for seven minutes. The analysis of PCR products on denaturing polyacrylamide sequencing gels was performed as described by Dasgupta et al.26 LOH was determined by densitometric scanning (CS-900; Shimadzu, Kyoto, Japan) of the autoradiographs. For informative cases, allelic loss was scored if there was complete loss of one allele or if the relative band intensity of one allele was decreased at least 50% in the tumour, compared with the same allele in the corresponding normal control. The value was calculated as the ratio of the band intensities of the larger to the smaller alleles in the tumour DNA divided by the same ratio in the corresponding normal DNA sample. An LOH index of > 1.5 (loss of the smaller allele) or < 0.67 (loss of the larger allele) corresponded to at least a 50% reduction in relative band intensities.27 MA was scored as present if one or both alleles at a given locus showed size variation; that is, either expansion or contraction, in comparison with the same allele in normal control DNA.28 When calculating LOH, samples showing homozygosity and MA only were not considered, whereas when calculating MA, the samples showing LOH only were not considered.29 Those samples showing both LOH and MA at the same locus was considered for calculating both LOH and MA. All samples showing LOH and/or MA were subjected to repeat analysis after a second independent amplification for confirmation, and typed with additional RFLP markers (NAT-KpnI, Hb-HinfI, Hb-HincIII, CYPIAI-MspI, and ALAD-RsaI) to verify correct pairing of the samples.30 In all cases, the results were in concordance with our findings (data not shown). This LOH/MA analysis procedure could detect LOH when 50% tumour DNA was present and MA when only 10–30% tumour DNA was present.26
Detection of HPV-16 and HPV-18
The presence of HPV in the cervical lesions was detected by performing PCR using primers (MY09 and MY11) from the consensus L1 region.31 Typing of HPV-16/18 in the L1 positive samples was done by means of PCR using specific primers from the E6 region of HPV-1632 and the E7 region of HPV-18.33 The PCR products were electrophoresed in 2% agarose gel, stained with ethidium bromide, visualised under ultraviolet light, and photographed. For final confirmation of the HPV types, after gel electrophoresis the PCR products were transferred on to a nylon membrane for Southern hybridisation with [32P] labelled HPV type specific probes.34 DNA from the SiHa (for HPV-16) and HeLa (for HPV-18) cell lines and the HPV type specific plasmids were used as positive controls.
Statistical analysis of the clinical data
We used multiple polymorphic markers (n = 20) to investigate LOH at different regions of chromosome 3. For data analysis, we used two approaches: (1) for individual samples we determined the frequencies of LOH at individual chromosomal regions; (2) to determine whether the deletions in chromosome 3 were associated with the progression of the tumour, we determined frequencies of loss of individual markers using a fractional allele loss (FAL) index, as defined below by Wistuba and colleagues28:
FAL index = total number of chromosome 3 markers with LOH/total number of informative chromosome 3 markers
To correlate regional loss with the progression of the tumour, we calculated the mean FAL index for each histological category (table 1 ▶). The t statistic was used for comparison of the mean FAL index of these histological groups.
We performed χ2 analysis to determine the association between LOH at highly deleted regions and different clinicopathological features, such as parity, the length of time married, menopausal status, HPV infection (table 2 ▶), and tumour stage (fig 2 ▶). A p value < 0.05 with Yates correction was considered to be significant.
Table 2.
Clinicopathological correlation of the high LOH regions
| 3p26.1 region | 3p22.3 region | 3p21.2 region | 3p13 region | |||||||||
| Characteristics | LOH+ | LOH− | p Value | LOH+ | LOH− | p Value | LOH+ | LOH− | p Value | LOH+ | LOH− | p Value |
| Parity (0–3) | 4 | 18 | 0.602 | 7 | 15 | 0.772 | 17 | 5 | 0.048 | 7 | 15 | 0.803 |
| Parity (4–10) | 8 | 20 | 10 | 18 | 14 | 14 | 8 | 20 | ||||
| HPV+ | 12 | 32 | <1 | 17 | 27 | <1 | 29 | 15 | 0.274 | 14 | 30 | 0.775 |
| HPV− | 0 | 6 | 0 | 6 | 2 | 4 | 1 | 5 | ||||
| Years married (10–17) | 7 | 26 | 0.52 | 11 | 22 | 0.889 | 20 | 13 | 0.777 | 9 | 24 | 0.55 |
| Years married (18–25) | 5 | 12 | 6 | 11 | 11 | 6 | 6 | 11 | ||||
| Premenopause | 5 | 18 | 0.729 | 7 | 16 | 0.623 | 16 | 7 | 0.309 | 8 | 15 | 0.495 |
| Postmenopuase | 7 | 20 | 10 | 17 | 15 | 12 | 7 | 20 | ||||
HPV, human papillomavirus; LOH, loss of heterozygosity.
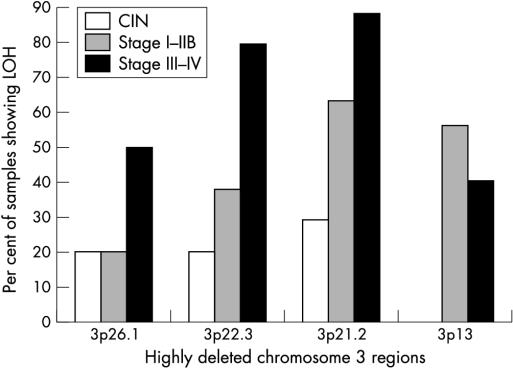
Figure 2.
Pattern of deletions in the highly deleted regions of chromosome 3 during uterine cervical carcinoma progression. CIN, cervical intraepithelial neoplasia; LOH, loss of heterozygosity.
RESULTS
Molecular abnormalities (LOH and MA) on chromosome 3
Deletion mapping of chromosome 3 in the primary cervical lesions revealed that 44 of the 50 samples showed LOH/MA for at least one marker (figs 1, 3 ▶ ▶). All the 44 samples with alterations showed LOH, whereas only seven showed MA, indicating the importance of chromosome 3 deletions in the development of CA-CX. The allelic losses of chromosome 3 gradually increased with progression of the clinical stage, and there was a significant correlation between the mean FAL index (p < 0.05) and progression of the tumour (table 1 ▶).
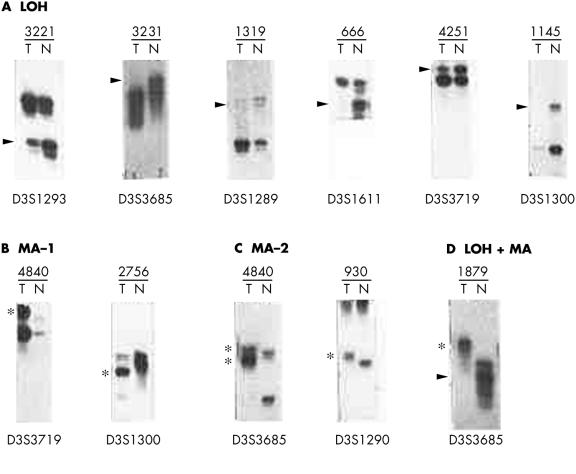
Figure 3.
Representative autoradiograph showing (A) loss of heterozygosity and (B) microsatellite size alteration at different marker loci on chromosome 3 in different samples. T, DNA from dysplastic/tumour cells after microdissection; N, corresponding normal tissue or peripheral blood leucocytes. Sample numbers are the same as in fig 1 ▶. Arrows indicate loss of the corresponding allele and the asterisks indicate size alteration of one or both alleles.
The highest LOH frequencies of 33%, 53%, 48%, and 45% were detected at the chromosomal regions 3p26.1 (D3S1304), 3p22.3 (D3S1611), 3p21.2 (D3S1289/D3S3719), and 3p13 (D3S1284), respectively (fig 1 ▶). However, the highest frequency of MA (10%) was seen at the 3p22.1 (D3S3685) locus (fig 1 ▶). No sample showed high microsatellite instability; that is, ≥ 40% MA in the markers tested.
Four cervical lesions (930, 1319, 3221, and 1879) showed molecular abnormalities (LOH or MA) at all the informative loci, indicating loss of normal copy of chromosome 3 in these lesions (fig 1 ▶). Eight samples (1684, 3776, 4840, 3493, 2756, 1058, 2060, and 666) showed LOH and/or MA in more than two consecutive markers, particularly around the high LOH regions (fig 1 ▶). No homozygous deletion was found in these samples. However, five lesions (930, 4840, 4849, 1178, and 1879) showed rare biallelic alterations (MA-2 or LOH + MA) at four loci (fig 1 ▶) in and around the high LOH regions.
Detection of HPV
Forty four of the 50 cervical lesions were positive for HPV infection (table 1 ▶) using the consensus L1 primers. Among the HPV positive samples, 36 of 44 were positive for HPV-16 and eight of 44 were positive for HPV-18. No correlation was found between HPV infection with other clinicopathological parameters (data not shown).
Clinical correlation of the molecular abnormalities (LOH/MA)
Chi square analysis revealed a significant correlation between LOH at the 3p26.1 and 3p22.3 regions and clinical stage (p = 0.005–0.04) and LOH at 3p21.2 with parity (p = 0.048) (table 2 ▶). No such correlation was found between the four highly deleted regions mentioned earlier and other clinicopathological parameters, such as length of time married, menopausal status, and HPV infection (table 2 ▶). In addition, no association was found between the frequency of MA and different clinicopathological parameters (data not shown).
DISCUSSION
In our present study, we attempted to delineate a detailed deletion map of chromosome 3 in seven CIN lesions and 43 CA-CX lesions at different clinical and histological stages from an Indian patient population. We found that the short arm of chromosome 3 was preferentially deleted during the development of CA-CX. The gradual increases in the mean FAL index from CIN to stage III–IV tumours were indicative of the accumulative nature of the deletions occurring in different chromosome 3 regions during progression of the tumour. Although a similar type of accumulative increase in the frequency of chromosome 3 deletions was observed during the development of CA-CX in a Western patient population,22,23 and in the development of lung carcinoma,28 the analysis procedure used in these studies was different to ours. Those investigators analysed CIN/dysplastic samples and invasive samples from the same individuals, whereas we analysed CIN and invasive samples from different individuals.
Among the four highly deleted regions, deletions in the 3p26.1 and 3p22.3 regions appeared to be associated with the development of stage III–IV CA-CX tumours because of a significant increase of LOH frequency compared with stage I–IIB tumours (p = 0.04–0.005) (fig 2 ▶). Although the deletion in these two regions overlapped with the deleted regions seen in CA-CX tumours in other studies,21,23,35 none of these investigators showed a significant association of the deletions with progression of the tumour. The marker D3S1304 on the 3p26.1 region used in our study is approximately 3.3 Mb further towards the telomere than the VHL gene, which is associated with the development of renal cell carcinoma (RCC) and related tumours.36 Thus, VHL or another nearby gene might be involved in the progression of later stages of CA-CX tumours. The D3S1611 marker on the 3p22.1 region is intragenic to the mismatch repair gene MLH1.37 However, we did not see a high frequency of MA in the cervical lesions studied. This indicates that the mutator phenotype might not be prevalent in the development of CA-CX. This observation corroborates well with other studies on this tumour.16 Thus, the high frequency of allelic losses at the MLH1 locus suggests that apart from mismatch repair function, the deregulation of other possible functions of MLH1 (for example, homologous recombination, mediation of the G2 checkpoint, transcription coupled nucleotide excision repair, recognition of DNA damage, and apoptosis) might be associated with progression of the tumour.38 However, the analysis of other genetic/epigenetic alterations (mutation, methylation, etc) of the MLH1 gene in cervical lesions is necessary to know the exact mechanism of inactivation of this gene. Adjacent to the MLH1 locus another TSG, DLC1, was found to be located about 1 Mb towards the centromere,39 but its involvement in the development of CA-CX has not yet been studied.
“Deletions in the 3p26.1 and 3p22.3 regions appeared to be associated with the development of stage III–IV uterine cervical carcinoma tumours”
Deletion in the 3p21.2 region increased gradually from CIN to stage III–IV tumours and this region showed the highest percentage of deletion of all the four highly deleted regions at every stage of disease (fig 2 ▶). Thus, it seems that deletion in the 3p21.2 region might be the earliest event among the four highly deleted regions. Similar to our results, Kersemaekers and colleagues40 and Guo and colleagues23 have detected a high frequency of LOH in this region in this tumour type. However, no candidate TSG has been identified in this region, although a cluster of candidate TSGs including FUS1, RASSF1, SEMA3F, ARP, and BAP1 are located about 3.0 Mb away, between the telomere and the D3S1289 locus of the 3p21.2 region.41 Nevertheless, an association of these genes with the development of CA-CX has not yet been documented.
The absence of LOH at 3p13 in CIN lesions but the high incidence of LOH in later stages of carcinoma indicate that deletions in this region might be necessary for progression of the tumour from CIN to stage I–IIB (fig 2 ▶). This region overlaps with the homozygously deleted region observed at chromosome 3p12–13 in a cervical carcinoma cell line.42 Similarly, Wong et al have identified 52% LOH at the 3p13 region in CA-CX tumours.43 The chromosome 3p12–14 region has been shown to suppress the tumorigenicity of an RCC cell line in a microcell hybrid system, suggesting the presence of a candidate TSG in this region.36 Two candidate TSGs, NRC1 and ROBO1/DUTT1, associated with the development of RCC and lung cancer, respectively, are located in the 3p12–14 region.44,45 However, we did not see high LOH in the ROBO1/DUTT1 locus using the adjacent marker D3S1274, although we have seen high LOH in this locus in primary head and neck squamous cell carcinoma.46 The role of these genes in the development of CA-CX has not been analysed.
The low incidence (16%) of LOH in the D3S1300 locus, an intragenic marker of the FHIT gene, indicates that this gene is probably not associated with the development of CA-CX. However, using the same marker, Wistuba et al detected 56% LOH in this tumour type.22 This difference in deletion frequency might result from differences in ethnicity, as reported by Hampton et al.25 The involvement of this gene in the development of CA-CX cannot be ruled out completely without analysis of the mutation, methylation, and aberrant transcription profiles of this gene.
The presence of rare biallelic alterations in and around the high LOH regions indicates that the LOH/MA in one allele might impose some selective pressure on the other allele for deletion or size alteration, which would result in a growth advantage for the tumour. A similar type of phenomenon has also been seen in the allelotyping of chromosome 3 in primary head and neck squamous cell carcinoma samples in Indian patients.26 The occurrence of interstitial alterations and loss of normal copy of chromosome 3 in some samples suggest that these chromosomal alterations are needed for progression of the tumour. These types of chromosomal abnormalities in chromosome 3 and 17 have also been reported in primary head and neck squamous cell carcinoma tumours.47,48
The high incidence (88%) of HPV infection in these samples suggests that HPV infection is a prerequisite for the development of cervical lesions. However, the absence of a significant correlation between HPV infection and the highly deleted regions indicates that HPV infection might have a causal association with the onset of CA-CX by affecting normal proliferation, differentiation, and apoptosis, and thus resulting in the accumulation of more mutations in the infected cells.49
Thus, it can be concluded from our analysis that the differential deletions in the four highly deleted regions seen in chromosome 3p in the uterine cervical lesions are necessary for the development of specific stages of CA-CX progression. The loss of function of TSGs located in these regions may have a sequential cumulative effect in the development of this tumour. In addition, Huebner50 has suggested that compound mutations in different TSGs located in different chromosome 3p regions are necessary for the development of lung, kidney, and other cancers. Along with these deletions, other molecular changes in the chromosome, such as MA, biallelic alterations, interstitial alterations, and loss of normal copy of chromosome 3, might play some role in tumour progression by imposing some selective pressure that provides a growth advantage for the tumour.
Take home messages.
Deletions of chromosome 3 were significantly associated with tumour progression
High frequencies of loss of heterozygosity (LOH) were found in 3p26.1, 3p22.3, 3p21.2, and 3p13, suggesting the location of putative tumour suppressor genes in these regions, which may have a cumulative effect on tumour progression
Deletions in 3p21.2 appear to occur early during cervical carcinoma development
There was a significant association between LOH at 3p26.1 and 3p22.3 and tumour progression from stage I/IIB to stage III/IV
These highly deleted regions were not associated with human papillomavirus positivity, parity, or menopausal status
Microsatellite size alteration was seen in only seven of the samples
Acknowledgments
We are thankful to the Director, Chittaranjan National Cancer Institute (CNCI), Calcutta-700 026, India and Drs PS Basu, R Mondal, and S Mondal for their encouragement and help during this work. We are also grateful to Professor HZ Hausen and Dr EM de Villiers for their generous gift of HPV-16/18 plasmids. Financial support for this work was provided by grant number 27 (0111)/00/EMR-II from CSIR, Government of India.
Abbreviations
CA-CX, uterine cervical carcinoma
CIN, cervical intraepithelial neoplasia
FAL, fractional allele loss
HPV, human papillomavirus
LOH, loss of heterozygosity
MA, microsatellite size alteration
PCR, polymerase chain reaction
RCC, renal cell carcinoma
RFLP, restriction fragment length polymorphism
TSG, tumour suppressor gene
REFERENCES
- 1.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer 1993;54:594–606. [DOI] [PubMed] [Google Scholar]
- 2.Sankaranarayanan R, Nair MK, Jayprakash PG, et al. Cervical cancer in Kerala: a hospital registry-based study on survival and prognostic factors. Br J Cancer 1995;72:1039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murthy NS, Mathew A. Screening for cancer of the uterine cervix and approaches adopted in India. Indian J Cancer 1999;36:154–62. [PubMed] [Google Scholar]
- 4.Brinton LA, Fraumeni JF, Jr. Epidemiology of uterine cervical cancer. J Chronic Dis 1986;39:1051–65. [DOI] [PubMed] [Google Scholar]
- 5.Ponten J, Adami HO, Bergstrom R, et al. Strategies for global control of cervical cancer. Int J Cancer 1995;60:1–26. [DOI] [PubMed] [Google Scholar]
- 6.Schiffmann MH, Brinton AB. The epidemiology of cervical cancer. Cancer 1995;76:1888–901. [DOI] [PubMed] [Google Scholar]
- 7.Franco EL. Epidemiology in the study of cancer. Encyclopedia of Cancer 1997;1:621–41. [Google Scholar]
- 8.Yliotalo N, Sorensen P, Josefesson A, et al. Smoking and oral contraceptives as risk factors for cervical carcinoma in situ. Int J Cancer 1999;81:365–75. [DOI] [PubMed] [Google Scholar]
- 9.Azzena A, Zen T, Ferrara A, et al. Risk factor for breast cancer. Case–control study results. Eur J Gynaecol Oncol 1994;15:386–92. [PubMed] [Google Scholar]
- 10.Tavani A, Negri E, La Vecchia C. Menstrual and reproductive factors and biliary tract cancers. Eur J Cancer Prev 1996;5:241–7. [DOI] [PubMed] [Google Scholar]
- 11.Zur Hausen. Human papilloma virus. Annu Rev Microbiol 1994;48:427–47. [DOI] [PubMed] [Google Scholar]
- 12.Howley PM. Papillomavirinae. The viruses and their replication. In: Fields BN, Knipe DM, Howley PM, eds. Virology, Vol. 2. Boston: Harvard University, 1995:2045–76.
- 13.Shah KV, Howley PM. Papilloma viruses. In: Fields BN, Knipe DM, Howley PM, eds. Virology, Vol. 2. Boston: Harvard University, 1995:2077–109.
- 14.Zur Hausen. Papilloma virus causing cancer: evasion from host cell control in early events in carcinogenesis. J Natl Cancer Inst 2000;92:690–8. [DOI] [PubMed] [Google Scholar]
- 15.Larson AA, Kern S, Sommers RL, et al. Analysis of replication error phenotypes in cervical carcinoma. Cancer Res 1996;56:1426–31. [PubMed] [Google Scholar]
- 16.Lazo PA. The molecular genetics of cervical carcinoma. Br J Cancer 1999;80:2008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heselmeyer K, Schrock E, du Manoir S, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A 1996;93:479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen DG, White DJ, Hutchins AM, et al. Progressive genetic aberrations detected by comparative genomic hybridisation in squamous cell cervical cancer. Br J Cancer 2000;83:1659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra AB, Murty V, Li RG, et al. Allelotype analysis of cervical carcinoma. Cancer Res 1994;54:4481–7. [PubMed] [Google Scholar]
- 20.Mullokandov MR, Kholodilov NG, Atkin NB, et al. Genomic alteration in cervical carcinoma: losses of chromosome heterozygosity and human papilloma virus tumor status. Cancer Res 1996;56:197–205. [PubMed] [Google Scholar]
- 21.Larson AA, Kern S, Curtiss S, et al. High resolution analysis of chromosome 3p alterations in cervical cancer. Cancer Res 1997;57:4082–90. [PubMed] [Google Scholar]
- 22.Wistuba II, Montellao FD, Milchgrub S, et al. Deletions of chromosome 3p are frequent and early events in the pathogenesis of uterine cervical carcinoma. Cancer Res 1997;57:3154–8. [PubMed] [Google Scholar]
- 23.Guo Z, Hu X, Afink G, et al. Comparisons of chromosome 3p deletions between cervical precancer synchronous with and without invasive cancer. Int J Cancer 2000;86:518–23. [DOI] [PubMed] [Google Scholar]
- 24.Fishel R. Genomic instability, mutators, and the development of cancer: is there a role for p53? J Natl Cancer Inst 1996;88:1608–9. [DOI] [PubMed] [Google Scholar]
- 25.Hampton GM, Larson AA, Baergen RN, et al. Simultaneous assessment of loss of heterozygosity at multiple microsatellite loci using semi-automated fluorescence-based detection: subregional mapping of chromosome 4 in cervical carcinoma. Proc Natl Acad Sci U S A 1996;93:6704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dasgupta S, Mukherjee N, Roy S, et al. Mapping of the candidate tumor suppressor genes’ loci on human chromosome 3 in head and neck squamous cell carcinoma of Indian patient population. Oral Oncol 2002;38:6–15. [DOI] [PubMed] [Google Scholar]
- 27.Melamed J, Einhorn JM, Ittamann MM. Allelic loss on chromosome 13q in human prostate carcinoma. Clin Cancer Res 1997;3:1867–72. [PubMed] [Google Scholar]
- 28.Wistuba II, Behrens C, Milchgrub S, et al. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene 1999;18:643–50. [DOI] [PubMed] [Google Scholar]
- 29.Dillon EK, Boer WB, De Papadimitriou JM, et al. Microsatellite instability and loss of heterozygosity in mammary carcinoma and its probable precursors. Br J Cancer 1997;76:156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishwad CS, Ferrel RE, Rossie KM, et al. Microsatellite instability in oral cancer. Int J Cancer 1995;64:332–5. [DOI] [PubMed] [Google Scholar]
- 31.D’Costa J, Saranath D, Dedhia P, et al. Detection of HPV-16 genome in human oral cancers and potentially malignant lesions from India. Oral Oncol 1998;34:413–20. [DOI] [PubMed] [Google Scholar]
- 32.Park JS, Dong SM, Kim HS, et al. Detection of p16 gene alteration in cervical cancer using tissue microdissection and LOH study. Cancer Lett 1999;136:101–8. [DOI] [PubMed] [Google Scholar]
- 33.Balaram P, Nalinakumari KR, Abraham E, et al. Human papilloma viruses in 91 oral cancers from Indian betel quid chewers—high prevalence and multiplicity of infections. Int J Cancer 1995;61:450–4. [DOI] [PubMed] [Google Scholar]
- 34.Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papilloma virus and a subset of head and neck cancers. J Natl Cancer Inst 2000;92:709–20. [DOI] [PubMed] [Google Scholar]
- 35.Chung GT, Huang DP, Lo KW, et al. Genetic lesions in the carcinogenesis of cervical cancer. Anticancer Res 1992;12:1485–90. [PubMed] [Google Scholar]
- 36.Kok K, Naylor SL, Buys CHCM. Deletion of the short arm of chromosome 3 in solid tumors and the search for tumor suppressor genes. Adv Cancer Res 1997;71:27–92. [DOI] [PubMed] [Google Scholar]
- 37.Papadopoulos N, Necolaids NC, Wei YF, et al. Mutation of a mutL homologue in hereditary colon cancer. Science 1994;263:1625–9. [DOI] [PubMed] [Google Scholar]
- 38.Benachenhou N, Guiral S, Gorska-Flipot I, et al. Frequent loss of heterozygosity at the DNA mismatch repair loci hMLH1 and hMSH3 in sporadic breast cancer. Br J Cancer 1909;79:1012–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daigo Y, Nishiwaki T, Kawasoe T, et al. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res 1999;59:1966–72. [PubMed] [Google Scholar]
- 40.Kersemaekers AM, Hermans J, Fleuren GJ, et al. Loss of heterozygosity for defined regions on chromosome 3, 11 and 17 in carcinomas of the uterine cervix. Br J Cancer 1998;77:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene 2002;21:6915–35. [DOI] [PubMed] [Google Scholar]
- 42.Aburatani H, Wang Y, Shibata H, et al. Identification of a region of homozygous deletion in cervical carcinoma [abstract]. Am J Hum Genet 1994;55:A51. [Google Scholar]
- 43.Wong WF, Chung TK, Cheung TH, et al. Frequent loss of heterozygosity of chromosome 3 short arm detected by PCR based microsatellite polymorphism in cervical squamous cell carcinoma. Cancer Lett 1997;115:161–4. [DOI] [PubMed] [Google Scholar]
- 44.Sundaresan V, Chung G, Heppel-Parton A, et al. Homozygous deletion at 3p12 in breast and lung cancer. Oncogene 1998;17:1723–9. [DOI] [PubMed] [Google Scholar]
- 45.Lovell M, Lott ST, Wong P, et al. The genetic locus NRC-1 within chromosome 3p12 mediates tumor suppression in renal cell carcinoma independently of histological type, tumor microenvironment, and VHL mutation. Cancer Res 1999;59:2182–9. [PubMed] [Google Scholar]
- 46.Chakraborty SB, Dasgupta S, Roy A, et al. Differential deletions in chromosome 3p are associated with the development of head and neck squamous cell carcinoma from Indian patients. Cancer Genet and Cytogenet [In press.] [DOI] [PubMed]
- 47.Scholes AGM, Woolger JA, Boyle MA, et al. Synchronous oral carcinomas: independent or common clonal origin? Cancer Res 1998;58:2003–6. [PubMed] [Google Scholar]
- 48.Adamson R, Jones AS, Field JK. Loss of heterozygosity studies on chromosome 17 in head and neck cancer using microsatellite markers. Oncogene 1994;9:2077–83. [PubMed] [Google Scholar]
- 49.Munger K, Heselmeyer K. The molecular pathogenesis of cervical cancer; the role of human papilloma virus. In: Gazdar A, Srivastava S, Henson DE, et al, eds. Molecular pathology of early cancer. Amsterdam: IOS Press, 1999:97–111.
- 50.Huebner K. Tumor suppressor on 3p: a neoclassic quartet. Proc Natl Acad Sci U S A 2001;98:14763–5. [DOI] [PMC free article] [PubMed] [Google Scholar]