BBO-8520, a dual inhibitor of KRASG12C in its GTP- and GDP-bound states, has a novel mechanism of action that enables effector blockade and optimal target coverage, even in growth factor activated states, and a phase I trial is ongoing.
Abstract
Approved inhibitors of KRASG12C prevent oncogenic activation by sequestering the inactive, GDP-bound (OFF) form rather than directly binding and inhibiting the active, GTP-bound (ON) form. This approach provides no direct target coverage of the active protein. Expectedly, adaptive resistance to KRASG12C (OFF)-only inhibitors is observed in association with increased expression and activity of KRASG12C(ON). To provide optimal KRASG12C target coverage, we have developed BBO-8520, a first-in-class, direct dual inhibitor of KRASG12C(ON) and (OFF) forms. BBO-8520 binds in the Switch-II/Helix3 pocket, covalently modifies the target cysteine, and disables effector binding to KRASG12C(ON). BBO-8520 exhibits potent signaling inhibition in growth factor–activated states, in which current (OFF)-only inhibitors demonstrate little measurable activity. In vivo, BBO-8520 demonstrates rapid target engagement and inhibition of signaling, resulting in durable tumor regression in multiple models, including those resistant to KRASG12C(OFF)-only inhibitors. BBO-8520 is in phase 1 clinical trials in patients with KRASG12C non–small cell lung cancer.
Significance: BBO-8520 is a first-in-class direct, small molecule covalent dual inhibitor that engages KRASG12C in the active (ON) and inactive (OFF) conformations. BBO-8520 represents a novel mechanism of action that allows for optimal target coverage and delays the emergence of adaptive resistance seen with (OFF)-only inhibitors in the clinic.
Introduction
RAS is the most frequently mutated oncogene in human cancer. RAS mutations occur in approximately 260,000 new cancer cases per year in the United States and 3.4 million per year worldwide (1). RAS genes encode small GTPase proteins that cycle between active GTP-bound (ON) and inactive GDP-bound (OFF) states. In the (ON) state, RAS activates multiple downstream signaling pathways, including the MAPK and phosphoinositide 3-kinase (PI3K) pathway, promoting proliferation, migration, and survival (2). KRASG12C is an oncogenic mutation that leads to insensitivity to GTPase activating protein (GAP)–mediated hydrolysis, which significantly increases the proportion of KRASG12C in the (ON) state and promotes tumor cell growth (2). KRASG12C mutations are found in approximately 14% of non–small cell lung cancers (NSCLC), 3% of colorectal cancers, and 1% of pancreatic cancers (3).
Sotorasib (AMG-510, LUMAKRAS) and adagrasib (MRTX-849, KRAZATI) are allele-specific KRASG12C(OFF) covalent inhibitors that have been approved for patients with KRASG12C locally advanced or metastatic NSCLC (4–6). Although these inhibitors have improved the treatment paradigm for patients with tumors harboring KRASG12C mutations, they are limited with regards to the depth and duration of response, which are suboptimal compared with other driver-targeted therapies in NSCLC, like osimertinib and alectinib (7, 8). This suboptimal efficacy from (OFF)-only inhibitors is likely driven by cancer cell adaptation through increasing the amount of drug-insensitive KRASG12C(ON; refs. 9–11). Patient responses may be improved with a compound that can inhibit KRASG12C(ON). In fact, clinical data from RMC-6291 (12), a KRASG12C(ON)-only inhibitor that depends on a tri-complex formation with cyclophilin A, has recently reported an objective response rate of 50% in patients with NSCLC undergoing recent prior KRASG12C(OFF)-only inhibitor treatment (13).
Here, we introduce BBO-8520, a potent, selective, orally bioavailable, direct covalent inhibitor of KRASG12C, with dual activity against both the (OFF) and (ON) states of KRASG12C. BBO-8520 is predicted to be more efficacious than sotorasib and adagrasib because of optimal target coverage of both the (ON) and (OFF) states, increased target engagement, and the potential to overcome sotorasib and adagrasib resistance mechanisms, including KRASG12C amplification and activation of receptor tyrosine kinases (RTK).
Results
Identification of BBO-8520
While exploring the druggable space in the Switch-II/Helix3 pocket of KRAS, we identified compound 1, with a quinazoline scaffold, which displayed a dissociation constant (KD) of 0.009 µmol/L with GDP-bound KRASG12D and submicromolar affinity (KD of 0.52 µmol/L) binding to KRASG12D in the active conformation bound to nonhydrolyzable GTP analog GppNHp (Fig. 1A). We hypothesized that binding of compound 1 to KRASG12D(ON) may cause conformational changes in the protein leading to disruption of RAS/effector interaction. To evaluate the effectiveness of compound 1 in disrupting KRASG12D and RAF1 binding, we used a biochemical protein–protein interaction (PPI) Homogeneous Time–Resolved Fluorescence (HTRF) assay. Compound 1 disrupted KRASG12D binding to the RAS-binding domain (RBD) of RAF1 with an IC50 of 1.4 µmol/L.
Figure 1.
Discovery of BBO-8520. A, The compound progression chart shows the development of BBO-8520. Noncovalent KRASG12D inhibitor, compound 1, showed binding activity (SPR) to GppNHp-bound KRASG12D, and disruption of KRASG12D/RAF1(RBD) interaction in PPI assay. Compound 1 was equipped with a covalent warhead to generate compound 2. Optimization of cell potency and ADME properties gave rise to compound 3. Further improvement of the GTP-bound KRASG12C activity by replacing the aminobenzothiazole of compound 3 with a cyano-amino benzothiophene at quinazoline 7-position gave rise to BBO-8520. BBO-8520 shows covalent labeling of C12 in the active, GppNHp-, or GTP-bound KRASG12C, as well as disruption of KRASG12C/RAF1(RBD) binding. B, The table summarizes the ability of compounds to modify C12 of GDP-, GppNHp-, and GTP-bound KRASG12C as measured by MALDI-TOF mass spectrometry; and their ability to disrupt GppNHp- or GTP-bound KRASG12C binding to RAF1(RBD) in PPI assay. C, kinact/Ki measurements of sotorasib, adagrasib, and BBO-8520 in the GDP-bound (OFF) and GTP-bound (ON) conformations of KRASG12C.
This measurable noncovalent activity against KRASG12D(ON) inspired the development of dual inhibitors that could bind covalently to cysteine 12 in both (ON) and (OFF) states of KRASG12C. Adding an acrylamide warhead to compound 1 gave rise to compound 2. Using a matrix-assisted laser desorption/ionization—time-of-flight (MALDI-TOF) mass spectrometry–based assay, we evaluated target engagement in KRASG12C proteins in both (ON) and (OFF) conformations. Compound 2 showed covalent modification of C12 in the (ON) state of KRASG12C although to a lesser degree than in the (OFF) state (Fig. 1B). Interestingly, the ability to modify KRASG12C (ON) translated to strong activity in the KRASG12C/RAF1(RBD) PPI disruption assay with an IC50 of 140 nmol/L (Fig. 1B). Further modification of the quinazoline 2- and 4-position to improve cell potency and ADME properties (clearance and oral bioavailability) gave rise to compound 3. Although compound 3 showed equal potency against the GDP- and GppNHp-bound proteins in the MALDI-TOF assay (>90% modification), it lost significant potency when the natural ligand (GTP) was used instead of the nonhydrolyzable analog GppNHp (38% modified). The loss of potency against the GTP-bound protein was also observed in the PPI assay, in which the 120-nmol/L IC50 for GppNHp-bound protein shifted 30-fold with the GTP-bound protein (Fig. 1B). This discrepancy could be attributed to an artificial increase in state 1 conformation of KRASG12C protein resulting from the use of the nucleotide mimetic (14).
Further optimization using structure-based drug design to improve binding to KRASG12C identified BBO-8520 (Fig. 1A), a potent, selective, and direct dual inhibitor of KRASG12C in both (ON) and (OFF) states. BBO-8520 engaged the target cysteine (C12) rapidly, regardless of nucleotide status, including GTP, as evidenced by MALDI-TOF mass spectrometry measurements (Fig. 1B). BBO-8520 also potently disrupted KRASG12C/RAF1(RBD) interaction with an IC50 of <100 nmol/L whether KRASG12C was bound to GppNHp or GTP (Fig. 1B). As expected, both sotorasib and adagrasib showed no activity in this assay.
To confirm the improved potency of BBO-8520 in comparison to sotorasib and adagrasib, we determined the kinact/Ki of sotorasib, adagrasib, and BBO-8520 using a mass spectrometry–based method as well as measuring the inhibition of pERK in NCI-H358 cells. BBO-8520 had a kinact/Ki of 20,000 M−1s−1 in GTP-bound (ON) and 2,743,000 M−1s−1 in GDP-bound (OFF) KRASG12C, compared with sotorasib and adagrasib which had no activity against (ON), and 11,000 and 180,000 M−1s−1 against (OFF) conformations of the protein, respectively (Fig. 1C; Supplementary Fig. S1A–S1D). In the heterozygous KRASG12C cell line NCI-H358, BBO-8520 had a kinact/Ki of 43,000 M−1s−1 compared with 1,064 M−1s−1 for adagrasib and 776 M−1s−1 for sotorasib for 40- and 55-fold increases, respectively (Supplementary Fig. S1E and S1F). BBO-8520 consistently showed stronger activity against the KRASG12C(OFF) protein than the KRASG12C (ON) state in all assays.
BBO-8520 Binds to Both (ON) and (OFF) KRASG12C as Revealed by Crystal Structures
To gain structural insights into the binding modes of BBO-8520 to KRASG12C, we tethered the compound to GDP- and GppNHp-bound proteins and solved their crystal structures at 1.67 and 2.10 Å resolution, respectively (Supplementary Table S1). Both crystal structures showed a high degree of similarity, with BBO-8520 binding in the pocket between Switch-II and alpha Helix 3 (Fig. 2A). The Switch-I region responsible for binding effectors adopts the open conformation in both structures. Although Switch-I does not interact with BBO-8520 directly, its C-terminal end is affected by the conformation that Switch-II adopts to accommodate the ligand. Importantly, in the GppNHp-bound structure, Switch-I moves away from the nucleotide, adopting a state 1 conformation that is not compatible with effector binding. This conformation, also detected by 31P NMR as described below, is characterized by the loss of direct coordination between the hydroxyl group of T35 and the Mg2+ ion.
Figure 2.
Binding mode of BBO-8520 to KRASG12C (ON) and (OFF) conformation. A, GDP-bound (OFF; top, PDB 8V3A) and GppNHp-bound (ON; bottom, PDB 8V39) forms in surface and ribbon representations. The protein is colored light orange (OFF) or light blue (ON), with Switch-I, Switch-II, and Helix 3 highlighted in pink, cyan, and orange, respectively. BBO-8520 (light green at the top, pink at the bottom), nucleotide, C12, and T35 are shown as sticks, and Mg2+ (green) and the coordinating waters (red) are shown as spheres. B and C, Enlarged view of the binding pocket in the GDP-bound form (PDB 8V3A), focusing on the regions around (B) Switch-II and Helix 3, and (C) C12 and GDP. The protein and BBO-8520 are colored light orange and light green, respectively. H-bonds are indicated by dashed lines. D and E, Enlarged view of the binding pocket in the GppNHp-bound form (PDB 8V39), focusing on the regions around (D) Switch-II and Helix 3, and (E) C12 and GppNHp. The protein and BBO-8520 are colored light blue and light pink, respectively. F and G, Overlay of the GDP-bound and GppNHp-bound structures, focusing on the regions around (F) Switch-II and Helix 3, and (G) C12 and the nucleotide.
In the interaction of BBO-8520 with KRASG12C(OFF), the quinazoline core is sandwiched between E62 and Y96, and the nitrogen at the N1 position makes a H-bond with the H95 side chain (Fig. 2B). At the C2 position, the pyrrolizidine group rests between H95 and D92, and its positively charged nitrogen makes a salt bridge with E62. The CF3 at the C6 position and the aminobenzothiophene at the C7 position make extensive van der Waals contacts with residues in the C-terminal halves of Switch-II and Helix 3. Furthermore, the amino group of the aminobenzothiophene forms H-bonds with the D69 side chain and the backbone carbonyl of E63. The cyano group substituted at the 3-position of the aminobenzothiophene makes an H-bond with the backbone NH of E63. Alternatively, it could engage in a water-mediated interaction with R68. At the C4 position, the dimethylpiperazine adopts a chair conformation with the 2-methyl pointing toward A59 of Switch-II and the 5-methyl sandwiched between the C12 sulfur and the quinazoline N3 nitrogen (Fig. 2C). The acrylamide vinyl forms a covalent bond with the sulfur of C12, and the acrylamide carbonyl makes an H-bond with the K16 side chain.
In the (ON) structure, the interactions between BBO-8520 and KRASG12C remained largely the same, with the quinazoline core, pyrrolizidine, CF3, and aminobenzothiophene making the same H-bond and salt bridge contacts with E62, E63, R68, D69, and H95 (Fig. 2D). However, at the C4 position, the dimethylpiperazine rotates by ca. 180°, such that the 5-methyl points toward Switch-II and makes van der Waals contacts with both A59 and G60, whereas the 2-methyl sits next to the quinazoline N3 nitrogen (Fig. 2E). This rotation is triggered by the repositioning of the acrylamide, necessary because the carbonyl is now too close (2 Å) to the gamma-phosphate of the GppNHp (Fig. 2F and G). Consequently, this rotation results in the loss of the H-bond between the acrylamide carbonyl and the K16 side chain, which is partially compensated by the van der Waals contact with the G60 backbone (Fig. 2E). Overall, the BBO-8520 binding pose is very similar in both structures, especially for the aminobenzothiophene component, whereas the slight tilt of the core is well accommodated by the protein (Fig. 2F). These properties allow BBO-8520 to bind tightly to both (ON) and (OFF) states of KRASG12C, although its affinity for KRASG12C(ON) is lower likely because of the loss of the H-bond to K16. The electron density maps of BBO-8520 bound to KRASG12C in both (ON) and (OFF) states are shown in Supplementary Fig. S2A–S2D.
BBO-8520 Displays a Dual (ON/OFF) Mechanism of Action and Differential Selectivity for KRASG12C
In solution, GTP-bound (ON) RAS exists in two interconverting conformational states, state 1 (signaling incompetent) and state 2 (signaling competent). This can be observed as two separate γ phosphate (γP) peaks (γ1 and γ2, respectively) in 31P NMR spectroscopy. When in complex with effector RAF1(RBD), the state 2 conformation of RAS is stabilized.
We investigated the perturbation of the KRASG12C(ON) state 1–state 2 conformational equilibrium upon BBO-8520 binding using 31P NMR. As depicted in Fig. 3A, BBO-8520 binding significantly alters the protein conformational equilibrium resulting in the emergence of a new predominant γ1Protein–Ligand (γ1PL) peak at ∼ −4.5 ppm, and a reduced γ1 peak (<10% of peak intensity relative to γ1PL). The γ1PL peak represents a “state 1–like” (signaling incompetent) inactive conformation. This perturbation profile strongly resembles that reported recently for KRASG12C(ON) binding of a close analog of BBO-8520 (14). A considerable perturbation is also noted for the β peak (shifted by −1.4 ppm relative to β2), however, to a lesser degree than noted for the γ peak, whereas the α peak is mostly unaffected. Importantly, the signaling-competent conformer (γ2) is not apparent in the spectrum. The downfield shift of γ1PL from γ1 in the protein–ligand (PL) complex is largely caused by the induced displacement of the Switch-I region away from the nucleotide and suggests that the strong inhibitory effect of BBO-8520 is caused by forcing KRASG12C into a signaling incompetent (inactive) conformation.
Figure 3.
Mechanism of action and selectivity of BBO-8520. A, BBO-8520 [ligand (L)] binding to KRASG12C-GTP [protein (P)] shifts the state 1–state 2 equilibrium of protein to the inactive, state 1-like conformation (induced γ1PL peak located most downfield) in the protein–ligand (PL) binary complex spectrum. Peak β1PL represents L binding induced inactive conformation of β GTP. Chemical shifts corresponding to peaks α1, β1, and γ1 belong to state 1 (inactive, effector binding–deficient) conformation, whereas α2, β2, and γ2 to the state 2 (active, effector binding–enabled) conformation. RAF1 RBD loading is unable to induce γ2 (active conformation) population (see P + L + RBD spectrum). Shown on top and bottom are the control spectra (in the presence of DMSO) of P + RBD and P, respectively. The γ1PL peak emergence is only noted in the presence of an inhibitor. The control spectra of KRASG12C-GTP alone or in the presence of RBD do not show this peak. B, BBO-8520 inhibits SOS-mediated nucleotide exchange of GDP with BODIPY-GDP KRAS. Avi-KRAS mutants indicated and Avi-NRAS WT were loaded with BODIPY-GDP, then BBO-8520 was added in a 2-fold dilution series starting at 30 nmol/L. The assay was started by the addition of SOS1 (aa564-1048) and GDP, then analyzed after 4 and 24 hours of incubation. KRASG12C shows the highest inhibition of nucleotide exchange with BBO-8520. NRAS WT was used as a control. C, pERK inhibitory activity of BBO-8520, sotorasib, and adagrasib against a panel of mouse embryonic fibroblasts (MEF) driven by KRAS mutants, HRAS, NRAS, and BRAFV600E. BBO-8520 demonstrates the best pERK inhibitory activity in KRASG12C-driven MEF cells, compared with MEFs driven by other KRAS mutants or WT KRAS. BBO-8520 shows no pERK inhibitory activity in MEFs driven by HRAS, NRAS, or BRAFV600E.
The addition of equimolar RAF1(RBD) to KRASG12C(ON) in the PL binary sample induces a conformational redistribution by shifting nearly half of the γ1PL peak to γ1 only, representing effector binding–deficient state 1, and does not shift back to the signaling-competent state 2 (Fig. 3A). Spoerner and colleagues (15) noted that RAS mutants were nononcogenic if the presence of effector protein was unable to shift the state 1 conformation to state 2. The inability of RAF1(RBD) to induce state 2 in the KRASG12C(ON)–BBO-8520 complex indicates that the compound forces the GTP-bound (ON) protein into the inactive, effector binding–deficient conformation. This NMR-based mechanistic data, together with evidence provided in Fig. 3B that BBO-8520 potently inhibits SOS-mediated nucleotide exchange, demonstrate a dual mechanism of action encompassing inducing the state 1 (effector binding deficient) conformation in KRASG12C (ON) as well as locking KRASG12C(OFF) in its inactive state.
Next, we used mouse embryonic fibroblasts (MEFs) expressing unique RAS isoforms and KRAS mutants to test the potency and selectivity of BBO-8520 (16). Treatment of MEFs with BBO-8520 showed no activity against HRAS, NRAS, and BRAFV600E (IC50 > 10 μmol/L) confirming exquisite selectivity for the KRAS protein. As expected, BBO-8520 displayed the highest activity against KRASG12C with single-digit-nmol/L IC50 on pERK inhibition (>10× better than sotorasib and adagrasib; Fig. 3C). Interestingly, the improved noncovalent interactions within the binding pocket allowed BBO-8520 to gain potency against other KRAS mutant isoforms or WT protein. This was also observed in the nucleotide exchange data in Fig. 3B. BBO-8520 showed potency against KRASG13D and WT KRAS4b (albeit at 5–10× lower than KRASG12C). Activity against other KRAS mutant isoforms was measurable but right shifted >100-fold from KRASG12C activity and may not be biologically significant in the in vivo or clinical settings.
BBO-8520-Induced Conformational Changes within Switch-I/II Region Translate to a Rapid and Potent Inhibition of Oncogenic Signaling
BBO-8520 showed rapid (within 30 minutes) engagement of Cys-12 in the KRASG12C mutant MIA PaCa-2 and SW1463 cell lines, leading to KRASG12C covalent modification and strong pERK signal suppression, consistent with its dual (ON/OFF) mechanism of action (Fig. 4A). This early effect, only achievable by engaging KRASG12C (ON), was absent from sotorasib and adagrasib even at 5× higher concentrations (100 nmol/L; Fig. 4A). Peak downregulation of the MAPK signaling pathway was observed within the first 2 hours and lasted for at least 24 hours in both cell lines. A time course of pERK inhibition using HTRF, as a complementary method, in both MIA PaCa-2 and SW1463 cells confirmed the rapid and sustained inhibition of ERK phosphorylation by BBO-8520 (Fig. 4B).
Figure 4.
BBO-8520 potently inhibits KRASG12C signaling in tumor cells. A, Target engagement and ERK phosphorylation time course in the KRASG12C cancer cell lines MIA PaCa-2 and SW1473. BBO-8520 at 20 nmol/L displays rapid pERK inhibition at 30 minutes which is sustained for up to 24 hours and is compared with 20 or 100-nmol/L sotorasib and adagrasib, which take longer and show less inhibition of pERK. B, HTRF analysis of phosphorylated ERK demonstrates time- and dose-dependent inhibition in response to BBO-8520 in MIA PaCa-2 and SW1463 cells, which is sustained for up to 34 hours posttreatment. C to E, Potent effects of BBO-8520 on 2-hour pERK inhibition, pAKT inhibition, and 7-day 3D viability measurements in KRASG12C cell lines as compared with sotorasib, adagrasib, and RMC-6291. The activity of BBO-8520 in KRASG12C cell lines is compared against a cell line panel comprising KRAS wild type along with G12C/D/V and S, G13D, and BRAFV600Emutants. IC50 (nmol/L) values for each cell line are captured in Supplementary Table S2.
To better understand the potency and selectivity of BBO-8520 in malignant cells, we then profiled the compound in a panel of ∼50 cancer cell lines harboring either wild type or mutant KRAS (G12C, G12D, G12S, G12V, and G13D) or a BRAFV600E mutation. Cells were assayed for pERK after treatment with compounds for 2 hours, pAKT after treatment for 4 hours, or in a 7-day spheroid three-dimensional (3D) viability assay (Fig. 4C–E). BBO-8520 compared favorably against sotorasib and adagrasib in both the ERK phosphorylation inhibition and 3D viability assays displaying better than 10-fold gain in potency (Supplementary Table S2). Comparison with RMC-6291 in six KRASG12C cell lines showed similar potency for both compounds. BBO-8520 demonstrated selectivity for KRASG12C over other KRAS codon 12 mutations (50- to 500-fold selectivity for pERK and 500- to 30,000-fold for 3D viability), G13D mutations (18-fold for pERK and 390-fold for 3D viability), and wild-type KRAS (>1,600-fold for pERK and >450-fold for 3D viability) and had no demonstrable activity in the BRAFV600E mutant cell line A375 (>10,000-nmol/L IC50 for pERK and 3D viability; Fig. 4C and E). BBO-8520 also demonstrated potent inhibition of pAKT signaling in some KRASG12C cell lines with IC50 below 10 nmol/L (Fig. 4D).
BBO-8520 Is Potent against GTP-Bound KRASG12C in Malignant Cells
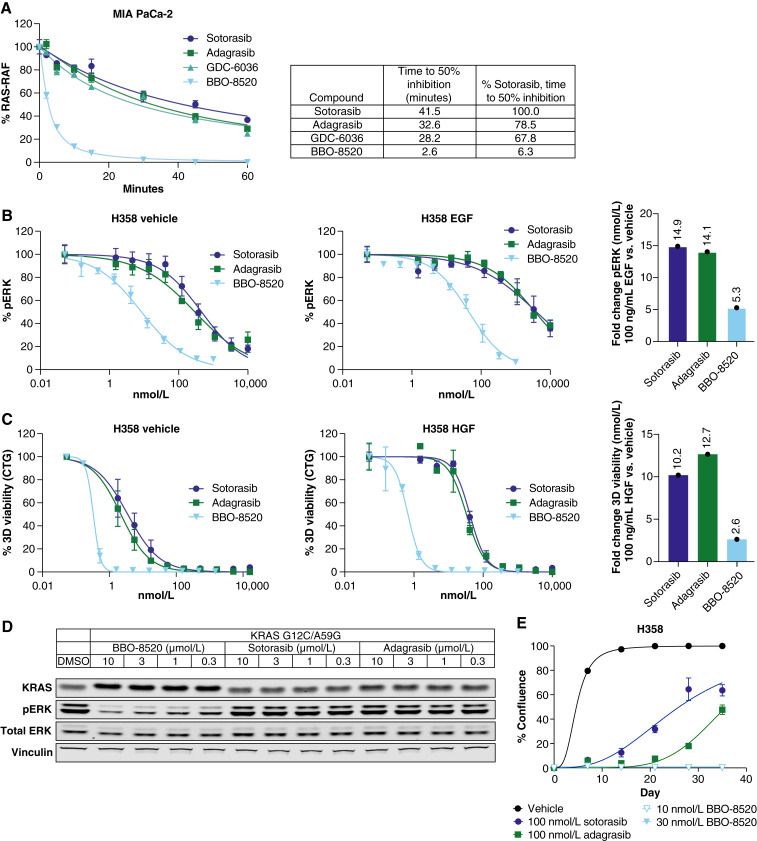
Evidence supporting the engagement of BBO-8520 with KRASG12C(ON) in malignant cells was first acquired using a RAS:RAF ELISA assay. This assay measures KRASG12C(ON) by capturing it with the RBD of RAF1. As shown in Fig. 5A, BBO-8520 started to inhibit the RAF1(RBD):KRASG12C(ON) interaction within the first 2 minutes of treatment. In striking contrast to (OFF)-only inhibitors, BBO-8520 achieved complete inhibition of KRASG12C(ON) within 15 minutes. At this early time point, (OFF)-only inhibitors, sotorasib, adagrasib, and GDC-6036 only reached 20% inhibition while requiring more than 60 minutes to achieve maximal activity. Further analysis of the KRASG12C(ON) inhibitory activity of BBO-8520 was performed by artificially increasing the amount of cellular GTP-bound KRASG12C through growth factor stimulation. We serum-starved NCI-H358 cells and stimulated them either with EGF in the presence or absence of (OFF)-only inhibitors or BBO-8520. Measurement of pERK inhibition showed that BBO-8520 retained potency in the presence of EGF while (OFF)-only inhibitors displayed a large loss of potency (Fig. 5B). In a parallel experiment, NCI-H358 cells were stimulated with hepatocyte growth factor (HGF) in the presence or absence of compounds for a viability endpoint. As observed in the EGF stimulation experiment, (OFF)-only inhibitors lost potency to a much greater extent than BBO-8520 in the presence of HGF (Fig. 5C). Final validation of BBO-8520’s activity against the (ON) state was gathered using a HeLa cell inducible model, in which doxycycline treatment was used to express a KRASG12C/A59G double mutant. A59G is a transition state mutant that abrogates GTPase activity and locks KRAS in the (ON) conformation (4). In this system, the inhibitory effect of sotorasib or adagrasib on MAPK signaling was significantly attenuated. In contrast, BBO-8520 retained potent activity (Fig. 5D).
Figure 5.
BBO-8520 maintains potency in the active state of KRASG12C. A, RAS:RAF ELISA assay in MIA PaCa-2 cells shows rapid dissociation of KRASG12C (ON) with RAF1 by BBO-8520 compared with the (OFF)-only KRASG12C inhibitors sotorasib, adagrasib, or GDC-6036. B and C, NCI-H358 cells were serum starved and then treated with 100 ng/mL of EGF, compound, and assayed for pERK HTRF 20 minutes after compound addition (B) or treated with 100 ng/mL of HGF and compound and assayed for a 5-day viability assay (C). Average potency shifts are shown to the right demonstrating the fold changes of IC50 following EGF or HGF stimulation compared with vehicle. D, HeLa cells were engineered to express a KRASG12C/A59G double mutant known for attenuated GTP hydrolysis and assayed for pERK inhibition following treatment with 0.3, 1, 3, and 10 µmol/L of BBO-8520, sotorasib, or adagrasib. BBO-8520 demonstrated a potent inhibition of the pERK signal compared with the (OFF)-only inhibitors, which showed no activity. E, A long-term clonogenic assay using IC90 concentrations (2-hour pERK) of BBO- 8520, sotorasib, and adagrasib, shows that only BBO-8520 can drive complete growth suppression for up to 35 days in culture compared with sotorasib or adagrasib.
We hypothesized that the dual activity of BBO-8520 would be beneficial to address the development of resistance. First, we tested the activity of BBO-8520 on previously described acquired resistance mutations. We generated Ba/F3 cells with a series of KRASG12C mutations that have altered states of GTP hydrolysis (9, 10). As demonstrated in Supplementary Fig. S3A and S3B, BBO-8520 demonstrated activity on all mutants tested in the Ba/F3 cell system. Second, we analyzed whether BBO-8520’s dual activity could delay the onset of resistance. Using concentrations equivalent to the IC90 of each compound by a 2 hours pERK HTRF assay, we treated NCI-H358 cells in a 2D viability clonogenic assay and observed that BBO-8520 caused complete growth suppression up to 35 days in culture, as compared with (OFF)-only inhibitors that allowed resistance to emerge between days 18 and 20 (Fig. 5E). This demonstrates that the dual activity of BBO-8520 could drive deeper tumor responses and delay the development of resistance.
Cysteine-Proteome Profiling and Transcriptional Regulation Changes Confirm Selectivity for KRASG12C
A risk associated with the use of covalent inhibitors is the potential for nonspecific, off-target reactivity with cysteine residues in the proteome. To determine the selectivity of the BBO-8520 covalent interaction with KRASG12C, and to identify potential off-target liabilities, cysteine-proteome profiling by mass spectrometry was performed as previously described (17). After 2 hours of treatment with DMSO or 20-nmol/L BBO-8520, the cysteine proteome was enriched, and peptides were identified. The Cys12 peptide from KRASG12C was the most significantly engaged cysteine (P-value = 7.1e−5; 98.9% decrease) in the analyzed proteome (Supplementary Fig. S4A). DUS4 (P = 3.3e−4; 62.9% decrease) also demonstrated reduction at two separate cysteines upon BBO-8520 treatment, suggesting downregulation of the protein rather than selective binding with BBO-8520.
Head-to-head RNA-seq studies with sotorasib (1 µmol/L) and adagrasib (300 nmol/L) were performed to compare the global transcriptional regulation driven by BBO-8520 in MIA PaCa-2 cells. Analysis revealed the strongest repression of genes involved in DNA replication and cell cycle progression (Supplementary Fig. S4B). A comparison with the gene set identified using adagrasib in NCI-H1373 and NCI-H358 cells (6) showed similar strong repression of MAPK signaling (DUSP4/6, ETV4/5, SPRY2/4) but at a 10-fold lower concentration of BBO-8520 (30 nmol/L compared with 300 nmol/L of adagrasib). Gene set enrichment analysis of the 50 hallmark signatures was performed on the differentially regulated genes from the MIA PaCa-2 RNA-seq study. A heatmap of the gene set enrichment analysis demonstrates a significant overlap in the signatures with all three compounds strongly supporting a common mechanism of action (Supplementary Fig. S4C). To determine if any kinases were hit by BBO-8520 leading to potential off-target effects, we performed a kinomescan on 468 kinases (Supplementary Fig. S4D). We found that 1 µmol/L BBO-8520 had binding activity on only three kinases (93% inhibition of CDK8, 87% inhibition of CDK11, and 86% inhibition of HIPK1). Follow-up biochemical dose response data for these three kinases showed Kd (nmol/L) of 150, 4,600, and 10,000 for CDK11, CDK8, and HIPK1, respectively. These studies demonstrate that BBO-8520 is selective for KRASG12C, induces global transcriptional regulation changes highly similar to other KRASG12C inhibitors, and carries a low likelihood of off-target activity.
BBO-8520 Exhibits Robust In Vivo Efficacy
The in vivo potency of BBO-8520 was first measured in a single-dose PK/PD Matrigel plug assay using MIA PaCa-2 cells. Following an oral dose of 3, 10, or 30 mg/kg of BBO-8520, a statistically significant reduction in pERK was observed in all groups with 46%, 73%, and 92% pERK inhibition, respectively (Fig. 6A). The pERK IC50 was 85 nmol/L. Similar potency was observed using MIA PaCa-2 tumors, with a pERK IC50 of 61 nmol/L (Supplementary Fig. S5A). The in vivo EC50 was consistent with the free fraction adjusted in vitro IC50 of 55 nmol/L. Evaluation of time-dependent pERK inhibition at 2, 6, 24, 48, and 72 hours following a single dose of BBO-8520 (30 mg/kg) resulted in 40%, 92%, 83%, 57%, and 41% pERK inhibition, respectively (Fig. 6B). In the MIA PaCa-2 PK/PD tumor model, pERK inhibition correlated well (R2 = 0.84) with tumor target engagement as assessed by a band shift assay on KRASG12C by Western blot analysis (Supplementary Fig. S5A and S5B).
Figure 6.
BBO-8520 demonstrates dose- and time-dependent inhibition of pERK and strong efficacy in KRASG12C models. A, BBO-8520 shows dose-responsive inhibition of pERK at 6 hours following a dose of 3, 10, and 30 mg/kg in a MIA PaCa-2 Matrigel plug PD assay (*, P < 0.01; **, P < 0.0001). B, Suppression of pERK was observed up to 72 hours following treatment with 30 mg/kg of BBO-8520 in the MIA PaCa-2 Matrigel plug PD (*, P < 0.01; **, P < 0.0001). C, In the corresponding MIA PaCa-2 CDX model, BBO-8520 showed significant antitumor activity at 0.1, 0.3, 1, 3, and 10 mg/kg following 28 days of treatment (*, P < 0.0001). D, In the NCI-H358 CDX model, BBO-8520 demonstrated significant and robust efficacy at 0.3, 1, 3, and 10 mg/kg following 28 days of treatment (*, P < 0.0001). E, In the KCP NSCLC GEMM, BBO- 8520 demonstrated significant and robust efficacy at 10 mg/kg (*, P < 0.0001). F, In the CT26-KRASG12C-luciferase syngeneic liver tumor model, BBO-8520 extended the median survival as a monotherapy or in combination with anti-PD-1.
The efficacy of BBO-8520 was first assessed in the MIA PaCa-2 CDX tumor model. Following daily oral dosing of BBO-8520 for 28 days at 0.1, 0.3, 1, 3, or 10 mg/kg, we observed statistically significant tumor growth inhibition of 21%, 49%, 69%, 99%, and 90% mean tumor volume regression, respectively (Fig. 6C). The ED50 was 0.3 mg/kg, and ED90 was 1.8 mg/kg. All treatments were well tolerated (Supplementary Fig. S5C). Efficacy was also studied in the NSCLC heterozygous KRASG12C model NCI-H358. Orally administered BBO-8520, once daily for 28 days at 0.3, 1, 3, or 10 mg/kg resulted in tumor volume reductions of 20% and 71%, and mean tumor regressions of 19% and 100%, respectively. The ED50 was 0.6 mg/kg, and ED90 was 1.6 mg/kg (Fig. 6D). Tumors from 1/10 and 10/10 mice in the 3- and 10-mg/kg BBO-8520 groups, respectively, had complete regressions.
The efficacy of BBO-8520 was also tested in two syngeneic, orthotopic, and immunocompetent models to assess the role of the organ microenvironment and the immune system. Our first model was the KrasG12C; Trp53R270H (KCP) NSCLC genetically engineered mouse model (18). Lung tumor–bearing KCP mice were orally administered vehicle or 10-mg/kg BBO-8520 once daily for 42 days. Statistically significant antitumor activity was observed following daily oral treatments with BBO-8520, with 56% mean tumor regression and no signs of resistance developing over the course of the study (Fig. 6E). These results were similar to those reported with the combination of a KRASG12C (OFF) inhibitor and SHP2 inhibitor in this model (19). Our second model was a CT-26-KRASG12C liver tumor model. Liver tumor–bearing mice were orally administered vehicle or 10-mg/kg BBO-8520 once daily until day 28, 10-mg/kg anti-PD-1 intraperitoneally twice weekly until day 21, or the combination of BBO-8520 and anti-PD-1. Notably, 40% (4/10) of the mice treated with BBO-8520 survived until the end of the study and were confirmed to have complete tumor regressions by IVIS imaging and necropsy. The combination of BBO-8520 and anti-PD-1 was also highly efficacious, with 60% (6/10) of the mice surviving confirmed cured. The median survival was 34 days in the vehicle group. The median survival was significantly increased with BBO-8520 treatment to 59 days (P = 0.0240) and increased to 48 days with anti-PD-1 treatment, but this increase was not significant (P = 0.3952; Fig. 6F). Although there was a statistically significant survival benefit between the anti-PD-1 and combination groups (P = 0.0033), there was only a trend toward a survival benefit between the BBO-8520 and combination groups (P = 0.1932) because of the strong effect of BBO-8520 monotherapy. All treatments in syngeneic models were well tolerated alone or in combination with an anti-PD-1 antibody.
BBO-8520 Retains Activity in Models Resistant to Sotorasib
The efficacy of BBO-8520 was next evaluated in the KRASG12C(OFF)-only inhibitor treatment-resistant NSCLC LUN055 PDX model. In addition to bearing a KRASG12C mutation, patient-derived LUN055 cells overexpress the RTK RET. Overexpression of RET is hypothesized to increase the amount of KRASG12C loaded with GTP, which makes this model more resistant to KRASG12C (OFF)-only inhibitors (11). LUN055 tumor–bearing mice treated daily with 30-mg/kg BBO-8520 for 35 days displayed statistically significant and robust antitumor activity of 23% mean tumor regression (Fig. 7A). Regressions were achieved with a free drug concentration of AUC0–24 hours of 79 hours × ng/mL. The TGI was only 71% following daily oral treatments with 100-mg/kg sotorasib, showing BBO-8520 has significantly greater antitumor activity in this model (P = 0.0007 for 100-mg/kg sotorasib vs. 30-mg/kg BBO-8520). In addition, the free drug concentration was 20-fold higher with sotorasib (free AUC0–24 hours of 1,563 hours × ng/mL with 100-mg/kg sotorasib) compared with BBO-8520.
Figure 7.
BBO-8520 is more efficacious than sotorasib and shows activity in sotorasib-resistant tumors. A, Efficacy of BBO-8520 and sotorasib in the RET-amplified LUN055 PDX model. Both BBO-8520 and sotorasib demonstrate antitumor activity (*, P < 0.0001), with BBO-8520 showing significant tumor regressions (23%) vs. 71% TGI for sotorasib. B, MIA PaCa-2 xenografts were grown under the presence of 10 mg/kg of sotorasib until tumors became resistant under treatment (day 35). On day 35, a cohort of eight mice were switched from sotorasib (10 mg/kg) to 30 mg/kg of BBO-8520. These mice showed strong responses with tumor volume regression. C, Analysis of sotorasib-resistant tumors showing a high proportion of KRASG12C amplification by ddPCR. D, Focused view of mice continuing on 10-mg/kg sotorasib or switched to 30-mg/kg BBO-8520 starting on day 35 (200 mm3). All mice treated with BBO-8520 had a statistically significant (P < 0.01) reduction in tumor volume compared with those continued on sotorasib. This included three mice with complete tumor regressions.
Lastly, the antitumor efficacy of BBO-8520 in mice bearing sotorasib-resistant CDX tumors was evaluated. MIA PaCa-2 tumors which developed resistance to 10-mg/kg sotorasib starting on day 35 and had tripled their average volume by day 75 of dosing (Fig. 7B) were analyzed for KRASG12C amplification by ddPCR (Fig. 7C). Six out of eight sotorasib-resistant tumors revealed significant amplification with 6 to 46 copies of KRASG12C (Fig. 7C). The efficacy of BBO-8520 after sotorasib resistance was assessed in a group of eight mice that were started on 30 mg/kg of BBO-8520 on day 35 post-sotorasib. All mice treated daily with 30 mg/kg of BBO-8520 had a statistically significant (P < 0.01) reduction in tumor volume and deep responses (Fig. 7D). Forty percent of these mice (3/8) had complete tumor regressions within 3 weeks of BBO-8520 dosing and had not regrown by the end of the study on day 92 (Fig. 7D). Treatment for >50 days with 30 mg/kg of BBO-8520, even after 35 days of sotorasib was well tolerated (Supplementary Fig. S5D). A similar experiment in the NCI-H358 model confirmed these results (Supplementary Fig. S6). The results from this study show that a dual inhibitor like BBO-8520 could achieve efficacy in patients who have progressed on an (OFF)-only inhibitor like sotorasib or adagrasib.
Discussion
The evolution of precision oncology therapeutics starts with suboptimal first-generation molecules and ends with exquisitely potent, best-in-class medicines that provide optimal target coverage and clinical benefit. Second- and third- generation inhibitors also differentiate themselves from predecessors by not only targeting the native oncogene but also inhibiting resistant variants that drive tumor progression. In the KRASG12C landscape, sotorasib and adagrasib represent first-generation medicines that inhibit KRASG12C by sequestering the GDP-bound (OFF) form while lacking activity against the GTP-bound (ON) form. The approval of sotorasib and adagrasib has changed clinical practice for patients with KRASG12C-positive NSCLC but their clinical benefit is suboptimal compared with best-in-class driver oncogene inhibitors in other oncogene-driven NSCLC disease settings. Recent data from a new set of potent KRASG12C inhibitors like divarasib, D3S-001, and RMC-6291 have provided clinical evidence that better efficacy is possible with better target coverage or by direct inhibition of the (ON) form of KRASG12C. A phase 1 trial including 60 patients with NSCLC treated with divarasib reported a confirmed ORR of 53% and PFS of 13 months (20). This significant gain in clinical benefit over sotorasib and adagrasib (ORR: ∼35% and PFS ∼6 months) by an (OFF)-only inhibitor with no differentiated mechanism of action highlights the suboptimal nature of first-generation KRASG12C inhibitors. Similarly, clinical data from RMC-6291, a tri-complex inhibitor of KRASG12C (ON) that requires cyclophilin A, has shown an ORR of 50% in KRASG12C (OFF) inhibitor-experienced patients, highlighting the importance of inhibiting the (ON) state (13). Until now, achieving both exquisite potency and coverage of the (OFF) and (ON) form of KRASG12C with a direct, covalent small molecule targeting the Switch-II/helix3 pocket has been deemed technically unattainable. An inhibitor that could accommodate the GTP-bound state of KRASG12C, drive exquisite potency, and directly inhibit the (ON) form of KRASG12C would undoubtedly conform to a novel class of KRASG12C inhibitors with excellent potential to achieve new levels of clinical benefit in patients with NSCLC. In fact, in a recently published study the combination of the (ON) inhibitor RMC-4998 with the (OFF) inhibitor sotorasib exerted superior therapeutic efficacy than either alone providing preclinical evidence supporting the superiority of the “dual” mechanism hypothesis (21).
We have developed BBO-8520, a direct, covalent small molecule inhibitor that engages KRASG12C in both the (ON) and (OFF) conformations with subnanomolar potency. This dual mode of action enables rapid, true full-target engagement and inhibition of KRASG12C. The unique ability to accommodate GTP-bound conformation results in forcing KRASG12C (ON) into state 1, which is unable to bind effectors, providing a novel mechanism of action and differentiating from KRASG12C (OFF)-only inhibitors that are exclusively dependent on GTP-GDP cycling (5, 6, 20). We show that adding blockade of effector binding as a mechanism leads to exquisite potency, and optimal target coverage and delays the emergence of adaptive resistance often seen with (OFF)-only inhibitors in the clinic.
Increased KRASG12C(ON), through amplification of the mutant allele or by growth factor–activated RTKs, seems to be the prominent mechanism of resistance (20, 22–24). Newly synthesized KRAS most likely is GTP-bound because of a 10-fold higher concentration of GTP than GDP in the cell (25). Therefore, transcriptional upregulation of the mutant protein provides an easy escape route for cells to overcome (OFF)-only inhibitors. Similarly, RTK activation can effectively overcome (OFF)-only inhibitors by maintaining a high population of KRASG12C(ON; ref. 26). Our observations show that the presence of EGF and HGF have a profound negative effect on the potency of (OFF)-only inhibitors in vitro, whereas amplification of KRASG12C is also detrimental in vivo. In contrast, BBO-8520 maintains exquisite potency in the presence of growth factors, mutant allele amplification, and even when mutations are engineered to maintain KRASG12C in the (ON) form. This superior profile should result in clinical benefits that would hopefully better resemble current best-in-class targeted agents in NSCLC.
BBO-8520 has entered phase 1 clinical trials in patients with KRASG12C NSCLC that are either KRASG12C inhibitor- or have experienced first-generation (OFF)-only inhibitors (NCT06343402). Our data supports the hypothesis that targeting both the (ON) and (OFF) forms of KRASG12C results in greater potency, deeper responses, and slowed development of resistance leading to significant benefits over approved, (OFF)-only KRASG12C inhibitors in NSCLC.
Methods
Reagents and Cell Cultures
BBO-8520 was synthesized as described in WO 2023/004102, Example 313 (details provided in Supplementary Materials and Methods S1), and stored at room temperature protected from light in a powder form. BBO-8520 was dissolved in 100% DMSO and aliquoted for long-term storage at −20°C.
All cells were purchased from ATCC or JCRB and cultured according to the protocols provided by the supplier. All cell lines were maintained at 37°C in a humidified incubator at 5% CO2 and were periodically checked for Mycoplasma. Cell lines used for in vivo studies were confirmed pathogen and Mycoplasma-free by IMPACT 1 assessment (IDEXX BioAnalytics) prior to implant. Cell lines were carried out for no more than 15 cell passages in this work.
Generation of Recombinant Proteins: Cloning, Expression, and Purification
All protein reagents were produced in-house by Protein Expression Laboratory, FNLCR. Entry clones containing E. coli optimized DNA sequences (ATUM) with an upstream tobacco etch virus protease cleavage site were used to generate expression clones in pDest-566 (Addgene #11517) using the protocols outlined by Esposito and colleagues (27). E. coli BL21 (DE3) STAR (rne131) containing rare tRNAs (pRar) was transformed with the expression plasmids as described (27, 28), and expression and purification were carried out as described (29, 30).
Nucleotide Exchange
KRAS proteins were loaded with GppNHp or GTP as described previously (31).
Mass Spectrometry–Based Covalent Engagement Assay
A 1-µmol/L solution of GTP, GppNHp, and GDP-loaded KRAS4bG12C/C118S (amino acids 1–169) protein was prepared and dispensed onto plates, and 30 nL of tested compounds from 1 mmol/L DMSO stocks were then added to the appropriate wells. At 15 minutes, 2 µL of each reaction mixture was pipetted into 15-µL MALDI matrix solution deposited onto plates. The resulting solution was mixed by aspiration, centrifuged at 2,000 g for 1 minute, and 1.5-µL aliquots were then dispensed on pretreated MALDI target. MALDI-TOF measurements were performed on Bruker Daltonics rapifleX Tissuetyper TOF-TOF mass spectrometer using linear mode and mass range from 18.6 to 21.6 kDa. Percent modification was calculated as a ratio of the peak height for the protein modified by compound to the sum of the peak height of the remaining protein plus the peak height for the protein modified by compound. For the detailed protocol see (32).
PPI Assay for RAS-RAF Disruption
A PPI HTRF assay was used to determine the effectiveness of compounds in disrupting KRAS protein and effector (RAF1) binding. Avi-KRASG12C (amino acids 2–169) GTP or GppNHp and RAF1 RBD-3xFLAG (amino acids 51–131) were used. Compounds were dispensed in an assay plate (384-well, Grenier Bio-One) using Echo (model 555) with dose response settings: 200-nL final volume, titration from 30 µmol/L as a 10-point dilution series. Proteins and HTRF reagents were mixed and dispensed onto plates, 20 µL per well, and then incubated for 1 hour at room temperature, with 700-rpm shaking, and the data were collected and analyzed as described previously (33).
Crystallization and Structure Determination
Protein samples for crystallography were prepared by tethering BBO-8520 to KRAS4b(1-169)G12C/C118S bound to GppNHp or GDP. GDP or GppNHp-loaded protein (20.5 mg) was diluted to 100 µmol/L in a buffer containing 20-mmol/L HEPES, 150-mmol/L NaCl, and 2-mmol/L MgCl2 at pH 7.3. A three-fold molar excess of a 10-mmol/L solution of BBO-8520 in DMSO was added, followed by brief mixing at room temperature for 2 minutes. Modification completion was confirmed by MALDI-TOF MS. The reaction mixture was purified on a low-pressure chromatographic system (NGC, Bio-Rad) using five in-line connected 5-mL Sephadex G-25 HiTrap desalting columns (Cytiva), the same buffer used for the reaction at 4-mL/minute flow rate and monitoring eluent at 280 nm. Protein-containing fractions were collected, and modified protein quality was confirmed by MALDI-TOF MS.
Crystallization screenings were set up using the sitting drop vapor diffusion method, as described previously (34). Fifteen mg/mL of the GppNHp-bound tethered KRASG12C or 40 mg/mL of the GDP-bound tethered KRASG12C was mixed with an equal volume (200 nL) of the reservoir solution. Crystals of GppNHp-bound KRASG12C were obtained from a reservoir solution consisting of 56-mmol/L NaH2PO4 and 1,343-mmol/L K2HPO4. Crystals of GDP-bound KRASG12C were obtained from a reservoir solution consisting of 0.1-mol/L Na3 citrate pH 5.5, 20% PEG 4000, and 10% isopropanol. Crystals were cryo-protected with 30% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected from Argonne National Laboratory Advanced Photon Source beamline 24-ID-C at 100 K and wavelength 0.979 Å. Data were integrated and scaled using XDS (35). Structure solution was obtained with molecular replacement using Phaser (36) as implemented in the Phenix programs suite, with the MRTX849-bound KRASG12C (PDB: 6UT0; ref. 37) as the search model. Iterative model building and refinement were performed with COOT and Phenix refine (38). Crystal parameters, data collection, and refinement statistics are summarized in Supplementary Table S1. The GppNHp dataset was affected by lattice-translocation disorder in the crystal, but its impact on chain A was minimal. Supplementary Fig. S2 shows the omit map around BBO-8520, nucleotide, Mg2+ ion, and C12 in chain A of both structures. Crystallographic and structural analysis software support is provided by the SBGrid consortium (39). The atomic coordinates and structure factors of the GDP-bound and GppNHp-bound KRASG12C tethered with BBO-8520 have been deposited in the Protein Data Bank and are available under accession numbers 8VCA and 8VC9, respectively.
31P NMR
All NMR samples included 800 mmol/L of KRASG12C-GTP protein in 93% H2O/7% D2O solvent composition in a buffer comprising 20-mmol/L HEPES (pH 7.3), 150-mmol/L NaCl, 2-mmol/L MgCl2, and 500-µmol/L 2,2-dimethyl-2-silapentanesulfonic acid (DSS) as internal standard, as described previously (14). Binary and ternary complexes of KRASG12C-GTP-BBO-8520 and KRASG12C-GTP-BBO-8520-RAF1 RBD were prepared in 1:2 and 1:2:1 stoichiometric ratios, respectively. All data were collected on a Bruker 500 MHz spectrometer at 278K. See Supplementary Materials and Methods S1 for details.
Western Blotting
Western blot experiments were performed according to the standard protocols. Primary antibodies were obtained from Cell Signaling Technology and were used at a concentration of 1:1,000: p-Akt S473 (#9271), AKT (#9272), p-p44/42 MAPK T202/204 (#9101), ERK1/2 (#9102), vinculin (#13901), and GAPDH (#2118). KRAS4b antibody was purchased from Proteintech (12063-1-AP).
HTRF Assay for pERK
Phospho-ERK levels were analyzed as described previously (40).
3D Viability Assay
Cells were seeded at 1,000 cells per well in ultralow attachment 96-well plates. Two days after plating, cells were treated with a 9-point dose titration of BBO-8520 starting at 10 µmol/L in 1:3 dilution increments using a Tecan D300e. Cells were incubated with BBO-8520 for a period of 4 to 7 days. After treatment, viability was assessed using a 3D CTG reagent (Promega). Luminescence was read on a Clariostar plate reader, and the data was imported into GraphPad Prism 9, log-transformed, and normalized to DMSO as 100% and media only as 0%. Following normalization, nonlinear regression was performed on a log (inhibitor) versus normalized response curve fit to generate an IC50.
k inact/Ki Measurements
Second-order rate constant kinact/Ki of covalent inhibition of KRASG12C was determined as described (41) with modifications. Details are provided in Supplementary Materials and Methods S1.
RNA-seq
Sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Homo sapiens GRCh38 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b to generate BAM files. Unique gene hit counts were calculated using featureCounts from the Subread package v.1.5.2. The hit counts were summarized and reported using the gene_id feature in the annotation file. Only unique reads that fell within exon regions were counted. If a strand-specific library preparation was performed, the reads were strand-specifically counted. After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the customer-defined groups of samples was performed. The Wald test was used to generate P-values and log2 fold changes. Genes with an adjusted P-value of <0.05 and absolute log2 fold change >1 were called differentially expressed genes for each comparison.
RAS-RAF ELISA Assay
MIA Paca-2 cells were seeded at 280,000 cells/well in 1 mL of DMEM with 10% FBS in a 12-well plate and placed in a 37°C incubator and allowed to adhere overnight. Cells were treated with 1 µmol/L of BBO-8520, sotorasib, adagrasib, or GDC-6036 using the Tecan D300e for 2, 5, 10, 15, 30, and 60 minutes. Following treatment, the cells were processed according to the manufacturer’s protocol (Abcam, Cat# 134640). Luminescence was read on a BMG Labtech Clariostar plate reader, and data was exported to GraphPad Prism 9, in which it was normalized to dimethyl sulfoxide-treated cells (DMSO) as 100% and blank as 0%. Following normalization, nonlinear regression was performed on an inhibitor versus normalized response curve fit to generate a time for 50% inhibition for each cell line treated with compound.
KRASG12C/A59G Experiments
HeLa cells were engineered to express KRASG12C/A59G under the control of the doxycycline-induced promoter. Cells were transduced with lentivirus and selected with 1-µg/mL puromycin for several passages. Cells were plated at 1.25e6 cells in a 10-cm dish into media containing 200-ng/mL doxycycline, allowed to attach for 24 hours, and then treated for 2 hours with various doses of compound. Following treatment, cell lysates were collected and processed for Western blot using phospho-ERK (Thr202/204), total ERK, KRAS, and vinculin according to the Western blotting methods above.
In Vivo Studies
All in vivo procedures were reviewed and approved by the Institutional Animal Care and Use Committee prior to execution and performed in accordance with the regulations and guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care.
In vivo studies were performed at Charles River Accelerator and Development Lab, Crown Biosciences, Inc., GenenDesign Co., Ltd., and NYU Langone Health in accordance with protocols and Institutional Animal Care and Use Committee guidelines. The vehicle used was the BBO-8520 and sotorasib formulation buffer for all studies (10% v/v N-methyl-pyrrolidone (Sigma-Aldrich, catalog 328634), 20% w/v solutol (Sigma-Aldrich, catalog 42966), and 30% v/v polyethylene glycol 300 (Sigma-Aldrich, catalog 8074841000) in 50-mmol/L citrate buffer pH 4 to 5). Plasma compound concentration levels were measured following protein precipitation using LC-MS/MS at Cytoscient, LLC or BioDuro-Sundia, Inc.
For Matrigel plug PK/PD studies, each athymic nude mouse was inoculated subcutaneously with 5 × 106 MIA PaCa-2 tumor cells suspended in growth factor reduced Matrigel (Sigma ECM, Sigma-Aldrich, catalog E6909), and treatments were administered the following day. Plugs were processed into lysates, and pERK tumor levels were measured using MSD (MSD, catalog N45107B-1) according to the manufacturer’s instructions. pERK tumor levels normalized to vehicle tumor levels [% pERK (normalized to vehicle) = [experimental (pERK/ERK)]/[vehicle (pERK/ERK)] × 100] were reported.
Standard protocols were followed to establish all subcutaneous CDX and PDX models. When CDX or PDX tumors reached a mean size of 175 to 210 mm3, mice were randomized into treatment groups (n = 10 per group) and orally dosed daily for 28 to 35 days. For the sotorasib-resistant efficacy study, MIA PaCa-2 tumor–bearing BALB/c nude mice were treated orally once daily with vehicle for 54 days or with 10-mg/kg sotorasib until resistance developed on day 35 (tumor volume reached 188 mm3). These mice were then randomized (10 animals/group) and treated daily with 10-mg/kg sotorasib from days 37 to 85 or with 30-mg/kg BBO-8520 from days 37 to 97. Tumor volumes were shown until one mouse in each group had to be euthanized because of large tumor volume. Standard methods were used to extract genomic DNA from tumors and measure levels of KRAS amplification using predesigned ddPCR copy number assay probes for human KRAS (Bio-Rad, catalog 10031240) and the reference gene RPP30 (Bio-Rad, catalog 10031241) according to the manufacturer’s instructions.
For the NSCLC genetically engineered mouse model efficacy study, KrasG12C;Trp53R270H mice (mixed background; ref. 18) were monitored by MRI for tumor development after intranasal induction with adeno-Cre (2.5 × 106 PFU). When lung tumors reached a mean size of 84 mm3, mice were randomized into treatment groups (n = 10 per group) and orally dosed daily with vehicle or 10-mg/kg BBO-8520 for 42 days. Lung tumor volume was monitored by MRI every 2 weeks.
For the syngeneic liver tumor model efficacy study, the murine colorectal cancer KRASG12D CT26-g-GFP/luciferase cell line (Creative Biogene, catalog CSC-RR0238) was engineered to introduce the KRASG12C mutation. Each anesthetized BALB/c mouse was injected intrahepatically with 5 × 105 CT26-KRASG12C-luciferase tumor cells suspended in PBS. Standard methods were used to measure the bioluminescence signal with an IVIS Spectrum (Perkin Elmer). When liver tumor bioluminescence signal reached a mean of 3.6 × 106 photons/second, mice were randomized into treatment groups (n = 10 per group) and dosed orally with vehicle or BBO-8520 until day 28 or intraperitoneally anti-PD-1 (clone RMP1-14, Bio X Cell, catalog BE0146) biweekly for 3 weeks. Mice were euthanized when the bioluminescence signal was >8.9 × 108 photons/second or when they showed defined signs of a large tumor.
Data analyses were performed using Microsoft Excel or GraphPad Prism software (version 9). For statistical analyses comparing pERK Matrigel plug levels in the vehicle group to all other groups, one-way ANOVA of all group means followed by posthoc Dunnett’s multiple comparisons were performed using GraphPad Prism software. For the statistical analyses comparing the vehicle group to all other groups in the efficacy studies, two-way repeated measures or mixed-effects ANOVA followed by posthoc Dunnett’s multiple comparisons test of the means was applied over the indicated number of days using GraphPad Prism software.
Data Availability
The data generated in these analyses are available within the article and its Supplementary Materials. The RNA-seq data generated in this study are publicly available in Gene Expression Omnibus at (GSE278656). The atomic coordinates and structure factors of the GDP-bound and GppNHp-bound KRAS-G12C tethered with BBO-8520 have been deposited in the Protein Data Bank and are available under accession numbers 8VCA and 8VC9, respectively.
Supplementary Material
Supplementary Material and Methods
Supplementary Table S1 summarizes the crystallography data for BBO-8520 bound to GMPPNP- and GDP-bound KRAS G12C
Supplementary Table S2 summarizes the potency of BBO-8520, sotorasib, adagrasib and RMC-6291 on pERK and viability in a panel of KRAS mutant, wild type and BRAF V600E mutant cell lines
Supplementary Figure S1 shows the Kinact/KI values for BBO-8520, sotorasib and adagrasib in biochemical and cell-based assays
Supplementary Figure S2 shows the electron density map for BBO-8520 bound to KRAS G12C in the GDP and GppNHp states
Supplementary Figure S3 shows the activity of BBO-8520, sotorasib, and adagrasib on Ba/F3 cells with KRAS G12C mutations with altered states of GTP hydrolysis
Supplementary Figure S4 summarizes cysteine profiling, RNA-seq and kinomescan studies demonstrating that BBO-8520 is selective for KRAS G12C and MAPK inhibition.
Supplementary Figure S5 shows that BBO-8520 demonstrates in vivo pERK inhibition, KRAS G12C target engagement and is well tolerated
Supplementary Figure S6 shows that BBO-8520 shows anti-tumor activity in NCI-H358 CDX sotorasib-resistant tumors
Acknowledgments
We thank William Burgan, Katie Powell, Scott Eury, Lauryl Scott, Nick Wright, Vanessa Wall, Bill Gillette, Simon Messing, Mukul Sherekar, Jennifer Mehalko, Shelley Perkins, Peter Frank, Troy Taylor, Nitya Ramakrishnan, and Brianna Higgins of the Protein Expression Laboratory (Frederick National Laboratory for Cancer Research) for their help in generating cell lines and recombinant proteins. We thank Monalisa Swain for her help with SPR experiments. We thank Mariam Ashraf, Derek Bratcher, and Sathiya Dharmaiah for their help in protein crystallization. We thank Kyle Sullivan, Nadage Gitego, Cindy Feng, and Carlos Stahlhut for non-KRASG12C cell line IC50 measures. X-ray diffraction data were collected at Northeastern Collaborative Access Team beamlines (24-ID-C), funded by the NIH (NIGMS P30 GM124165). The Pilatus 6M detector on the 24-ID-C beamline is supported by an NIH-ORIP HEI grant (S10 RR029205). This research utilized resources at the Advanced Photon Source, operated by Argonne National Laboratory for the US Department of Energy Office of Science (Contract DE-AC02-06CH11357). Part of this work was supported by Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344 (Release: LLNL-JRNL-864652). We thank the computing support from Lawrence Livermore National Laboratory institutional grand challenge program. This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
All BridgeBio Oncology Therapeutics (BBOT) authors are employees and stockholders of BBOT, a private company. B. Wang, R. Xiu, E. Wallace, Z. Zhang, and Y. Yang report a patent for PCT/US2022/037992(WO2023004102A2) pending. A.E. Maciag reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study, as well as a patent for PCT/US2022/037992(WO2023004102A2) pending, licensed, and with royalties paid from TheRas/BridgeBio. A.K. Sharma reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. A.H. Chan reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study, as well as a patent for PCT/US2022/037992 (WO2023004102A2) pending, licensed, and with royalties paid from TheRas/BridgeBio. M. Dyba reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study and owning stock in BridgeBio. B.P. Smith reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study, as well as owning stock in BridgeBio Pharma, Inc. D. Rabara reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. E.K. Larsen reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. J.-P. Denson reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. P.A. Alexander reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. M. Abreu Blanco reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. D.M. Turner reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study, as well as a patent for PCT/US2022/037992 (WO2023004102A2) pending, licensed, and with royalties paid from TheRas/BridgeBio. F.C. Lightstone reports a patent for PCT/US2022/037992(WO2023004102A2) pending and licensed to BBOT. K.-K. Wong reports grants from BridgeBio and Mirati during the conduct of the study, as well as other support from Cogent, Iambic, Pfizer, and Janssen outside the submitted work. A.G. Stephen reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. D.K. Simanshu reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study, as well as a patent for PCT/US2022/037992 (WO2023004102A2) pending, licensed, and with royalties paid from TheRas/BridgeBio. D.V. Nissley reports support from a Collaborative Research and Development Agreement with TheRas/BridgeBio and from NCI contract 75N91019D00024 during the conduct of the study. F. McCormick reports personal fees from BBOT and Leidos Biomedical during the conduct of the study, as well as personal fees from BridgeBio, Quanta, and Amgen outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A.E. Maciag: Conceptualization, supervision, investigation, writing–original draft, writing–review and editing. J.P. Stice: Conceptualization, formal analysis, supervision, validation, methodology, writing–original draft, writing–review and editing. B. Wang: Conceptualization, supervision, validation, methodology, writing–original draft, writing–review and editing. A.K. Sharma: Investigation, writing–original draft. A.H. Chan: Investigation, writing–original draft. K. Lin: Supervision, validation, visualization, writing–original draft. D. Singh: Investigation, visualization. M. Dyba: Investigation, methodology. Y. Yang: Conceptualization, software, formal analysis, investigation, methodology. S. Setoodeh: Investigation, visualization. B.P. Smith: Investigation. J.H. Ju: Data curation, validation, visualization. S. Jeknic: Data curation, validation, visualization. D. Rabara: Investigation. Z. Zhang: Conceptualization. E.K. Larsen: Investigation. D. Esposito: Investigation. J.-P. Denson: Investigation. M. Ranieri: Investigation. M. Meynardie: Investigation. S. Mehdizadeh: Investigation, visualization. P.A. Alexander: Investigation. M. Abreu Blanco: Investigation. D.M. Turner: Investigation. R. Xu: Conceptualization. F.C. Lightstone: Conceptualization, supervision. K.-K. Wong: Supervision, methodology. A.G. Stephen: Supervision, investigation. K. Wang: Conceptualization, supervision. D.K. Simanshu: Supervision, investigation, writing–original draft. K.W. Sinkevicius: Conceptualization, formal analysis, supervision, validation, methodology, writing–original draft, writing–review and editing. D.V. Nissley: Supervision, investigation, writing–original draft. E. Wallace: Conceptualization, methodology, writing–review and editing. F. McCormick: Funding acquisition, writing–review and editing. P.J. Beltran: Conceptualization, formal analysis, supervision, validation, writing–original draft, writing–review and editing.
References
- 1. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19:533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRASG12C somatic mutations across race, sex, and cancer type. N Engl J Med 2021;384:185–7. [DOI] [PubMed] [Google Scholar]
- 4. Lito P, Solomon M, Li L-S, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016;351:604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217–23. [DOI] [PubMed] [Google Scholar]
- 6. Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 8. Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829–38. [DOI] [PubMed] [Google Scholar]
- 9. Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired resistance to KRASG12C inhibition in cancer. N Engl J Med 2021;384:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tanaka N, Lin JJ, Li C, Ryan MB, Zhang J, Kiedrowski LA, et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov 2021;11:1913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lietman CD, Johnson ML, McCormick F, Lindsay CR. More to the RAS story: KRASG12C inhibition, resistance mechanisms, and moving beyond KRASG12C. Am Soc Clin Oncol Educ Book 2022;42:1–13. [DOI] [PubMed] [Google Scholar]
- 12. Schulze CJ, Seamon KJ, Zhao Y, Yang YC, Cregg J, Kim D, et al. Chemical remodeling of a cellular chaperone to target the active state of mutant KRAS. Science 2023;381:794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jänne PA, Bigot F, Papadopoulos K, Eberst L, Sommerhalder D, Lebellec L, et al. Abstract PR014: preliminary safety and anti-tumor activity of RMC-6291, a first-in-class, tri-complex KRASG12C(ON) inhibitor, in patients with or without prior KRASG12C(OFF) inhibitor treatment. Mol Cancer Ther 2023;22(Suppl 12):PR014. [Google Scholar]
- 14. Sharma AK, Pei J, Yang Y, Dyba M, Smith B, Rabara D, et al. Revealing the mechanism of action of a first-in-class covalent inhibitor of KRASG12C (ON) and other functional properties of oncogenic KRAS by 31P NMR. J Biol Chem 2024;300:105650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spoerner M, Hozsa C, Poetzl JA, Reiss K, Ganser P, Geyer M, et al. Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J Biol Chem 2010;285:39768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esposito D, Stephen AG, Turbyville TJ, Holderfield M. New weapons to penetrate the armor: novel reagents and assays developed at the NCI RAS initiative to enable discovery of RAS therapeutics. Semin Cancer Biol 2019;54:174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 2016;6:316–29. [DOI] [PubMed] [Google Scholar]
- 18. Li S, Liu S, Deng J, Akbay EA, Hai J, Ambrogio C, et al. Assessing therapeutic efficacy of MEK inhibition in a KRASG12C-driven mouse model of lung cancer. Clin Cancer Res 2018;24:4854–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fedele C, Li S, Teng KW, Foster CJR, Peng D, Ran H, et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J Exp Med 2021;218:e20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sacher A, LoRusso P, Patel MR, Miller WH Jr, Garralda E, Forster MD, et al. Single-agent divarasib (GDC-6036) in solid tumors with a KRAS G12C mutation. N Engl J Med 2023;389:710–21. [DOI] [PubMed] [Google Scholar]
- 21. Nokin M-J, Mira A, Patrucco E, Ricciuti B, Cousin S, Soubeyran I, et al. RAS-ON inhibition overcomes clinical resistance to KRAS G12C-OFF covalent blockade. Nat Commun 2024;15:7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ryan MB, Coker O, Sorokin A, Fella K, Barnes H, Wong E, et al. KRASG12C-independent feedback activation of wild-type RAS constrains KRASG12C inhibitor efficacy. Cell Rep 2022;39:110993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, et al. Vertical pathway inhibition overcomes adaptive feedback resistance to KRASG12C inhibition. Clin Cancer Res 2020;26:1633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akhave NS, Biter AB, Hong DS. Mechanisms of resistance to KRASG12C-targeted therapy. Cancer Discov 2021;11:1345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Esposito D, Garvey LA, Chakiath CS. Gateway cloning for protein expression. Methods Mol Biol 2009;498:31–54. [DOI] [PubMed] [Google Scholar]
- 28. Wall VE, Garvey LA, Mehalko JL, Procter LV, Esposito D. Combinatorial assembly of clone libraries using site-specific recombination. Methods Mol Biol 2014;1116:193–208. [DOI] [PubMed] [Google Scholar]
- 29. Taylor T, Gillette W. Production of isotopically labeled KRAS4b. Methods Mol Biol 2024;2797:23–34. [DOI] [PubMed] [Google Scholar]
- 30. Kopra K, Vuorinen E, Abreu-Blanco M, Wang Q, Eskonen V, Gillette W, et al. Homogeneous dual-parametric-coupled assay for simultaneous nucleotide exchange and KRAS/RAF-RBD interaction monitoring. Anal Chem 2020;92:4971–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waybright T, Stephen AG. Nucleotide exchange on RAS proteins using hydrolysable and non-hydrolysable nucleotides. Methods Mol Biol 2024;2797:35–46. [DOI] [PubMed] [Google Scholar]
- 32. Dyba M, Denson J-P, Maciag AE. MALDI-TOF mass spectrometry-based assay for measuring covalent target engagement of KRAS G12C inhibitors. Methods Mol Biol 2024;2797:145–57. [DOI] [PubMed] [Google Scholar]
- 33. Larsen EK, Abreu-Blanco M, Rabara D, Stephen AG. KRAS4b:RAF-1 homogenous time-resolved fluorescence resonance energy transfer assay for drug discovery. Methods Mol Biol 2024;2797:159–75. [DOI] [PubMed] [Google Scholar]
- 34. Chan AH, Simanshu DK. Crystallographic studies of KRAS in complex with small molecules and RAS-binding proteins. Methods Mol Biol 2024;2797:47–65. [DOI] [PubMed] [Google Scholar]
- 35. Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 2010;66:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007;40:658–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fell JB, Fischer JP, Baer BR, Blake JF, Bouhana K, Briere DM, et al. Identification of the clinical development candidate MRTX849, a covalent KRASG12C inhibitor for the treatment of cancer. J Med Chem 2020;63:6679–93. [DOI] [PubMed] [Google Scholar]
- 38. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66:213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, et al. Collaboration gets the most out of software. Elife 2013;2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smith BP, Rigby M, Ma R, Maciag AE. High-throughput cell-based screening of small molecule KRAS signaling inhibitors using a homogeneous time-resolved fluorescence (HTRF) assay. Methods Mol Biol 2024;2797:271–85. [DOI] [PubMed] [Google Scholar]
- 41. Li KS, Quinn JG, Saabye MJ, Guerrero JFS, Nonomiya J, Lian Q, et al. High-throughput kinetic characterization of irreversible covalent inhibitors of KRASG12C by intact protein MS and targeted MRM. Anal Chem 2022;94:1230–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material and Methods
Supplementary Table S1 summarizes the crystallography data for BBO-8520 bound to GMPPNP- and GDP-bound KRAS G12C
Supplementary Table S2 summarizes the potency of BBO-8520, sotorasib, adagrasib and RMC-6291 on pERK and viability in a panel of KRAS mutant, wild type and BRAF V600E mutant cell lines
Supplementary Figure S1 shows the Kinact/KI values for BBO-8520, sotorasib and adagrasib in biochemical and cell-based assays
Supplementary Figure S2 shows the electron density map for BBO-8520 bound to KRAS G12C in the GDP and GppNHp states
Supplementary Figure S3 shows the activity of BBO-8520, sotorasib, and adagrasib on Ba/F3 cells with KRAS G12C mutations with altered states of GTP hydrolysis
Supplementary Figure S4 summarizes cysteine profiling, RNA-seq and kinomescan studies demonstrating that BBO-8520 is selective for KRAS G12C and MAPK inhibition.
Supplementary Figure S5 shows that BBO-8520 demonstrates in vivo pERK inhibition, KRAS G12C target engagement and is well tolerated
Supplementary Figure S6 shows that BBO-8520 shows anti-tumor activity in NCI-H358 CDX sotorasib-resistant tumors
Data Availability Statement
The data generated in these analyses are available within the article and its Supplementary Materials. The RNA-seq data generated in this study are publicly available in Gene Expression Omnibus at (GSE278656). The atomic coordinates and structure factors of the GDP-bound and GppNHp-bound KRAS-G12C tethered with BBO-8520 have been deposited in the Protein Data Bank and are available under accession numbers 8VCA and 8VC9, respectively.