Abstract
Despite the exponential increase in sequencing information driven by massively parallel DNA sequencing technologies, universal and succinct genomic fingerprints for each organism are still missing. Identifying the shortest species-specific nucleotide sequences offers insights into species evolution and holds potential practical applications in agriculture, wildlife conservation, and healthcare. We propose a new method for sequence analysis termed nucleic “quasi-primes,” the shortest occurring sequences in each of 45,076 organismal reference genomes, present in one genome and absent from every other examined genome. In the human genome, we find that the genomic loci of nucleic quasi-primes are most enriched for genes associated with brain development and cognitive function. In a single-cell case study focusing on the human primary motor cortex, nucleic quasi-prime genes account for a significantly larger proportion of the variation based on average gene expression. Nonneuronal cell types, including astrocytes, endothelial cells, microglia perivascular-macrophages, oligodendrocytes, and vascular and leptomeningeal cells, exhibit significant activation of quasi-prime-containing gene associations related to cancer, whereas simultaneously suppressing quasi-prime-containing genes are associated with cognitive, mental, and developmental disorders. We also show that human disease–causing variants, eQTLs, mQTLs, and sQTLs are 4.43-fold, 4.34-fold, 4.29-fold, and 4.21-fold enriched at human quasi-prime loci, respectively. These findings indicate that nucleic quasi-primes are genomic loci linked to the evolution of species-specific traits, and in humans, they provide insights in the development of cognitive traits and human diseases, including neurodevelopmental disorders.
Over the past two decades, the cost of sequencing the complete genome of an organism has rapidly declined. Advances in parallel DNA sequencing technologies have enabled the generation of reference genomes for thousands of biological organisms, across viral, archaeal, bacterial, and eukaryotic species (O'Leary et al. 2016; Schoch et al. 2020). The availability of multiple, diverse organismal genomes has enabled advances in bioinformatic analyses and accelerated scientific discovery. Subsequently, our understanding of evolution, encompassing the mechanisms of horizontal and vertical gene transfer, selection pressures, and the emergence of new species traits, has improved. This information has been utilized in various domains such as human health, pathogen surveillance, agriculture, and species conservation, leading to the development of approaches for disease diagnosis, enhancement of food safety, and mitigation of antibiotic resistance, among other applications (Deurenberg et al. 2017; Brandies et al. 2019; Jagadeesan et al. 2019; Maljkovic Berry et al. 2020). The number of reference organismal genomes is expected to continue to increase and encompass a significant proportion of the genetic diversity present in nature (Lewin et al. 2018; Darwin Tree of Life Project Consortium 2022). For example, the Earth BioGenome Project aims to sequence the genomes of all eukaryotic species within the next 10 years (Lewin et al. 2022). This is essential for understanding evolutionary history, discovering the ecological interactions of living organisms, and developing precise techniques to detect genomic differences between species. Nevertheless, the increase in biological data also necessitates algorithmic advances to capture the most useful information.
As species evolve and diverge, they acquire new traits. This leads to the divergence of lineage-specific DNA at varying rates as subsets of the genome evolve more rapidly (Seehausen et al. 2014). As an example, after the divergence of humans from other primates, there was an expansion of cranial capacity, brain size, and cognitive abilities in humans (Florio et al. 2017). Comparative genomics and phylogeny analyses identified regions of organismal genomes that show patterns of accelerated evolution (Ferris et al. 2018; Foley et al. 2023). The usage of mutation rate patterns, species sequence alignments, and the identification of highly conserved regions can provide insights into phenotypic changes (McLean et al. 2011; Hubisz and Pollard 2014). However, available methods rely on genome alignments and mutational analysis, often lacking the ability to identify the units of accelerated evolution at base pair resolution. The identification of the shortest genomic sequences unique to a species can improve our understanding of how genomic regions evolve and can serve as an alternative method for the identification of genomic loci that are changing, at a base pair resolution.
k-mer analysis involves counting and comparing substrings of length k in biological sequences. The distribution of nucleotide k-mers varies substantially across organismal genomes (Chor et al. 2009; Bussi et al. 2021). The presence of individual nucleotide k-mers in a species is dependent on several factors, including the GC content of its genome, the biological roles associated with each particular k-mer, and the mutation patterns associated with that organism (Bussi et al. 2021). The set of k-mers in a species's genome can serve as a signature of its underlying sequence. Comparing these k-mers and their frequencies enables the identification of distinct characteristics among species. For example, extremophile genomes exhibit some of the most distinct and unique k-mer profiles (Bize et al. 2021). Previous efforts have identified sequences that are clustered in specific taxonomies or are conserved; examples include OrthoVenn3, which identifies orthologous clusters and detects conserved and variable genomic structures (Sun et al. 2023) and Telobase, which provides telomere motifs across organismal genomes in the tree of life (Lyčka et al. 2024).
Nullomers are the subset of nucleotide k-mers that are not observed in a genome (Hampikian and Andersen 2007), and their absence has been previously attributed to mutational patterns and negative selection (Georgakopoulos-Soares et al. 2021b; Koulouras and Frith 2021). Additionally, genome primes are the subset of nullomers that are absent from the genomes of all species (Hampikian and Andersen 2007). Applications have included the usage of nullomers in barcoding (Goswami et al. 2013), deriving anticancer peptides (Alileche et al. 2012), developing vaccine adjuvants (Patel et al. 2012), and in detecting cancer (Georgakopoulos-Soares et al. 2021a; Montgomery et al. 2024). Therefore, the development of efficient algorithms and methodologies that derive highly informative k-mers can result in multiple practical applications.
Previously, we defined a concept termed peptide quasi-primes, the shortest peptide sequences that are unique to a reference proteome (Mouratidis et al. 2023). Here, we extend this concept to nucleic quasi-primes, which are the shortest nucleotide k-mers that are unique to a particular species and are nullomers in all other assembled reference genomes (Fig. 1). We also detect and analyze the set of DNA sequences that are absent from every known genome, also known as DNA primes. As proof of concept, we annotate the set of human quasi-primes and their locations in the human genome. This work provides the methodology for the identification of nucleic quasi-primes and exemplifies their utility for detecting genomic loci that are associated with human-specific traits and that could be potentially important targets in understanding human brain evolution and brain-associated diseases.
Figure 1.
Identification of nucleic quasi-prime sequences in individual species. Schematic displaying the identification of a four-mer quasi-prime sequence in a species. All nucleic k-mers of a specific length are identified for each species, across all the reference genomes. k-mers that are shared between multiple species are removed, and only k-mers appearing in a single reference genome are kept, constituting the set of quasi-primes for that species for that particular length. As an example, we show a short toy sequence that is found in the human reference genome but is absent from all other species in our database, therefore constituting a human nucleic quasi-prime k-mer. Quasi-primes are associated with the evolution of human specific traits. For humans, quasi-prime-containing genes are enriched in the cortex and are associated with brain development and diseases. Human traits and pathogenic variants are significantly enriched in human quasi-prime sequences. Variants including expression quantitative trait loci (eQTLs), methylation QTL (mQTLs), splicing QTL (sQTLs), genome-wide association studies (GWAS) variants, and disease variants are more likely to be found in human quasi-prime sites.
Results
k-mer distribution across taxonomic subdivisions and species
We performed a comprehensive analysis of 45,076 reference genomes spanning the three domains of life and viruses. Our aim was to explore the diversity and uniqueness of nucleotide sequences across different species and identify k-mers that may serve as molecular fingerprints. We first investigated the number of different k-mers across each sequenced reference genome as a function of k-mer length ranging from 1–17 bp. For k-mer lengths of 16 bp, we found that the median number of observed k-mers per genome was 3,796,626. We also examined the number of 16 bp long k-mers detected per genome across taxonomic subdivisions for viral, archaeal, bacterial, and eukaryotic genomes and observed a median of 35,744, 4,912,057, 8,380,257, and 30,028,396 k-mers, respectively, representing 0.000832%, 0.1189%, 0.1976%, and 1.866% of the k-mer space (Supplemental Fig. 1; Fig. 2A). These findings were consistent for k-mer lengths of 15 and 17 bp (Supplemental Fig. 1; Fig. 2A), indicating that the majority of possible k-mers are absent from a given reference genome at these k-mer lengths. Therefore, we proceeded to investigate the extent to why certain k-mers are absent across multiple species and, as an extension, determine if there exist k-mers that are unique to a single species.
Figure 2.
k-mer content as a function of genome length across organisms. (A) Number of different k-mers observed as a function of genome size for k-mer lengths of 15 bp, 16 bp, and 17 bp. (B) Number of expected versus the number of observed k-mers for each reference genome for k-mer lengths of 15 bp, 16 bp, and 17 bp. Viral, archaeal, bacterial, and eukaryotic genomes are colored pink, blue, yellow, and green, respectively, across the A and B figure panels. (C) GC content percentage of nucleic prime sequences. (D) Average number of GpC and CpG occurrences per prime. Error bars in D represent SD. (E) Number of quasi-primes detected in each reference genome.
There are more nullomers than expected across genomes in each taxonomy
In a previous study, we demonstrated that the human genome exhibits a higher prevalence of nullomers than expected by chance (Georgakopoulos-Soares et al. 2021b). However, these findings have not been extended to other species, and it is currently unknown if this pattern is consistent across the different organismal genomes present in the taxonomic subdivisions. To address this gap, we performed a comprehensive analysis of the number of expected and observed frequencies of oligonucleotide k-mers for each species by generating simulated genomes that account for dinucleotide content (see Methods). We then investigated whether there were significant deviations between the two sets of frequencies across different k-mer lengths and organisms.
Our results reveal a consistent pattern of nullomer enrichment across species and taxonomic groups. Specifically, for k-mer lengths of 15 bp, we found that 99.99%, 100%, 99.98%, and 97.95% of viral, archaea, bacteria, and eukaryotic species, respectively, had significantly fewer observed oligonucleotide k-mers than expected by chance (chi-square test with Bonferroni-corrected P-value, P-value < 0.05), whereas only between 0% and 0.02% cases in each of the taxonomies had higher enrichment of observed k-mers per species than expected by chance. Similar results were also obtained for 16 bp and 17 bp k-mer lengths (Fig. 2B). We infer that the nullomer sequences are more frequent than expected by chance across taxonomies, and this is a universal rule across all organismal genomes likely driven by negative selection and genome repetitiveness.
Detection of genome primes across 45,076 reference genomes
In addition to the k-mer diversity and uniqueness across different taxa, we were also interested in identifying oligonucleotides that were absent from all reference genomes, which are termed as genome primes. These sequences are likely to be under selective pressure (Hampikian and Andersen 2007; Georgakopoulos-Soares et al. 2021b) and have biological significance. Additionally, genome primes could serve practical purposes, such as PCR primers, highly specific platforms in CRISPR construct designs, genomic barcodes, and sample labeling. Previous studies examined a limited number of genomes and derived 60,370 15 bp nucleic prime sequences (Hampikian and Andersen 2007). Therefore, we aimed to determine the minimum k-mer length at which we could identify genome primes with the current number of available reference genomes.
We discovered a total of 5,186,757 genome primes at 16 bp. We characterized the genomic features of these sequences and found that they had a high GC content with average GC content of 54.33% (Fig. 2C). The most prominent feature of genome primes was the presence of GC/CG dinucleotides, which occurred in 99.9997% of genome prime sequences. Only 16 genome primes lacked GC/CG dinucleotides. Furthermore, we observed a significant enrichment of CpG sites over GpC sites. CpG sites had 1.94-fold higher frequency relative to GpC sites within genome primes (binomial test, P-value < ×10−100) (Fig. 2D; Supplemental Fig. 2A). These results suggest that genome primes are highly enriched in CpGs, possibly owing to their higher mutation rate (Sved and Bird 1990; Fryxell and Moon 2005).
Detection of nucleic quasi-primes across 45,076 reference genomes
Next, we examined if there is a certain k-mer length at which we can identify sequences that are unique to a single species and absent from every other species examined, which we have defined as nucleic quasi-primes. We found that up to 15 bp in length, every possible k-mer was found in two or more species among all the genomes examined. Therefore, we concluded that nucleic quasi-primes cannot be found up to 15 bp lengths in any genome. However, at 16 bp, we report 14,678,002 nucleic quasi-primes. We report that the number of nucleic quasi-primes detected varies substantially between species, and the median number of nucleic quasi-prime sequences detected is 16 across the studied organisms, with certain organisms not having any nucleic quasi-prime at that k-mer length (Fig. 2E). We also examined the nucleotide composition on nucleic quasi-primes for the three domains of life and for viruses and found, that across them, “CG” and “TA” dinucleotides were highly enriched (Supplemental Fig. 2B). This was also consistent when we separated eukaryotes in plants, fungi, invertebrates, and vertebrate mammalian groups (Supplemental Fig. 2C), indicating that the dinucleotide composition of quasi-primes is similar across taxa.
We were interested to examine how our findings would change based on the increasing number of organismal genomes available in the future. We sorted the genomes according to size and distributed them into five different bins of equal size to ensure a uniform selection. For each percentage (5%, 10%, 25%, 50%, 75%), we performed the following: first, we selected 5% of the genomes from each of the five bins. Incrementally we added more genomes to reach 10%, 25%, 50%, and finally 75%, ensuring that each larger percentage included all genomes from the previous percentage. This approach provides an estimate of how quasi-primes behave as the number of genomes available increases. We performed three different simulations, repeating the described data selection process. For each percentage in each simulation, we applied our quasi-prime detection algorithm for a sequence length of 16 (Supplemental Fig. 2D). To further understand the behavior of quasi-primes as the data set grows, we fitted an inverse function to the results from our simulations. The idea behind fitting this specific function is based on the fact that the number of quasi-primes decreases as the number of genomes increases, following an inverse relationship. We predict that for a data set of 500,000 genomes (∼1111% of the current 45,076 genomes), the number of quasi-primes would be approximately 4,495,722 (Supplemental Fig. 2D). These results indicate that with the addition of more organismal genomes the number of quasi-primes obtained could decline significantly; however, this could be addressed by increasing the quasi-prime length.
Identification of taxonomic quasi-primes: a case study across 246 mammalian genomes
Next, we investigated if there are k-mer sequences that can be found in one or more species within a taxonomic group that are otherwise absent from all species outside that taxonomic group. We termed those sequences taxonomic quasi-primes and reasoned that these sequences are more likely to reflect loci that have evolved in a taxonomic group. Taxonomic quasi-primes are a superset of quasi-primes; that is, all quasi-primes are by definition also taxonomic quasi-primes. We performed an investigation across all 246 mammalian genomes from the Zoonomia project (Zoonomia Consortium 2020) to identify mammalian taxonomic quasi-primes for a 16 bp k-mer length.
First, we identified nucleic quasi-primes in each of the 246 mammalian organisms. We report that on average there are 5779 16 bp nucleic quasi-primes per mammalian species, which are absent from every other organismal genome (Fig. 3A). Additionally, we identified a total of 13,240,656 mammalian taxonomic quasi-primes shared between one or more mammals and otherwise absent from all other species outside this taxonomic group. We also observed that the number of taxonomic quasi-prime sequences shared between multiple species declines precipitously with the number of species sharing them (Fig. 3B,C). We identified that the mammalian quasi-prime found in the largest number of mammals is found in 148 mammals (60% of mammals examined) and nowhere else outside the taxonomy. This mammalian quasi-prime was identified in the coding region of insulin receptor substrate 2 (IRS2), a gene that has emerged during vertebrate evolution (Al-Salam and Irwin 2017), indicating the ability of taxonomic quasi-primes to identify regions that have evolved in a specific taxonomy. We also investigated if the proportion of mammalian taxonomic quasi-prime sequences shared between species enabled their clustering based on their evolutionary relatedness. We find that closely related species share a larger proportion of mammalian taxonomic quasi-prime sequences, resulting in clusters including those of feline, cavy, and primate species (Fig. 3D,G). Thus, we conclude that we can identify taxonomic quasi-prime sequences and that these enable the clustering of species that share a more recent common ancestor.
Figure 3.
Characterization of mammalian taxonomic quasi-primes. (A) Number of species quasi-primes across the Zoonomia project. The top 50 species are shown. (B) Number of taxonomic quasi-primes shared between the mammalian species studied. (C) Number of human taxonomic quasi-primes shared with mammalian species. (D) Clustering of mammalian taxonomic quasi-primes based on the Jaccard index of shared taxonomic quasi-prime k-mers. Highlighted clusters represent the cluster of primate species (E), the cluster of guinea pigs or cavies (F), and the cluster of felines (G).
Human quasi-primes are preferentially found in promoter and 5′ UTR regions
Our next aim was to identify the presence and characteristics of human quasi-prime nucleotide sequences. We detected a total of 19,226 human nucleic quasi-prime sequences throughout the human genome. Analogous to genomic primes, human quasi-primes exhibited a high GC content and an enrichment of CpGs relative to GpCs (t-test, P-value < 1×10−100) (Supplemental Fig. 3A–C). We also mapped the locations of each human quasi-prime sequence in the genome and identified 11,982 loci that harbor human quasi-primes, of which 4525 were in genic regions. We assessed the abundance of human quasi-primes in the following genomic subcompartments: genic, intronic, coding, and 5′ and 3′ UTRs, as well as 2500 bp upstream of the transcription start site (TSS). We found that the highest frequency of human quasi-primes is observed at 5′ UTR and promoter regions (Fig. 4A). We performed an analysis to examine the distribution of quasi-primes across genomic compartments in 15 organisms, including multiple primate genomes, rodents, other mammals, and invertebrates. We find that 5′ UTR regions are most enriched in nucleic quasi-primes, followed by promoter regions (Supplemental Fig. 4A). These results were also consistent in non-human primates, namely, Gorilla gorilla, Pan paniscus, Pongo abelii, Pan troglodytes, and Macaca mulatta, and similarly found enrichment in 5′ UTR and promoters (Supplemental Fig. 4B). This could be because of those regions evolving faster than other genomic compartments and having fewer sequence constraints, which enable the generation of quasi-primes. We also utilized regulatory elements as categorized by ENCODE (The ENCODE Project Consortium et al. 2020) to examine the distribution of human quasi-primes across cis-regulatory elements. The set of cis-regulatory elements encompassed CTCF-only, CTCF-bound, PLS, DNase-H3K4me3, dELS, PLS, and pELS terms. We report that human quasi-prime loci are most likely to be found in PLS (Fig. 4B). We also examined the overlap between long noncoding RNA genes and human quasi-prime loci and found a total of 1837 quasi-primes overlapping 1459 long noncoding RNAs; however, there was no significant enrichment relative to the quasi-prime controls (binomial test, P-value > 0.05). Thus, we infer that specific cis-regulatory elements, including promoter regions and 5′ UTRs, have the highest frequency of human quasi-prime sequences.
Figure 4.
Taxonomic nucleic quasi-prime sequences. (A) Density of human quasi-primes across genic regions. (B) Density of human quasi-primes across cis-regulatory elements. (C) Gene expression of human quasi-primes across tissues. (D–F) GO term analysis for human quasi-primes for biological processes (D), molecular function (E), and cellular components (F).
Human accelerated regions have been previously characterized as regions that are evolutionarily conserved but have diverged in humans (Pollard et al. 2006). Using the set of human accelerated regions collected from five previous studies (Doan et al. 2016), we examined if there is overlap with human quasi-prime loci. We find only three human quasi-prime loci overlapping human accelerated regions, accounting for 0.0156% of the total human quasi-primes, indicating that human quasi-primes capture distinct genomic loci.
Human quasi-primes are associated with brain development and cognitive function
We analyzed the expression of quasi-prime-containing genes across human tissues. In total, we examined the consensus normalized expression from RNA-seq experiments across 50 tissues (Pontén et al. 2008). We observe that brain regions, including the cerebral cortex, basal ganglia, white matter, and thalamus show the highest expression (Fig. 4C), indicating a preference for quasi-primes for brain-related genes.
Subsequently, we conducted a GO term analysis to examine the biological processes associated with human quasi-prime-containing genes. For molecular and biological function terms, we found terms associated with GTPase activity, cell morphogenesis, and neuron morphogenesis being highly enriched (Fig. 4D,E). We found that when examining cellular component terms, neuronal-associated terms are enriched, including synaptic and postsynaptic membrane, neuron-to-neuron synapse, and dendritic and axonal terms (Fig. 4F). Therefore, these results substantiate the brain-related roles of human quasi-prime-containing genes.
Human quasi-prime-containing genes are involved in neurological diseases
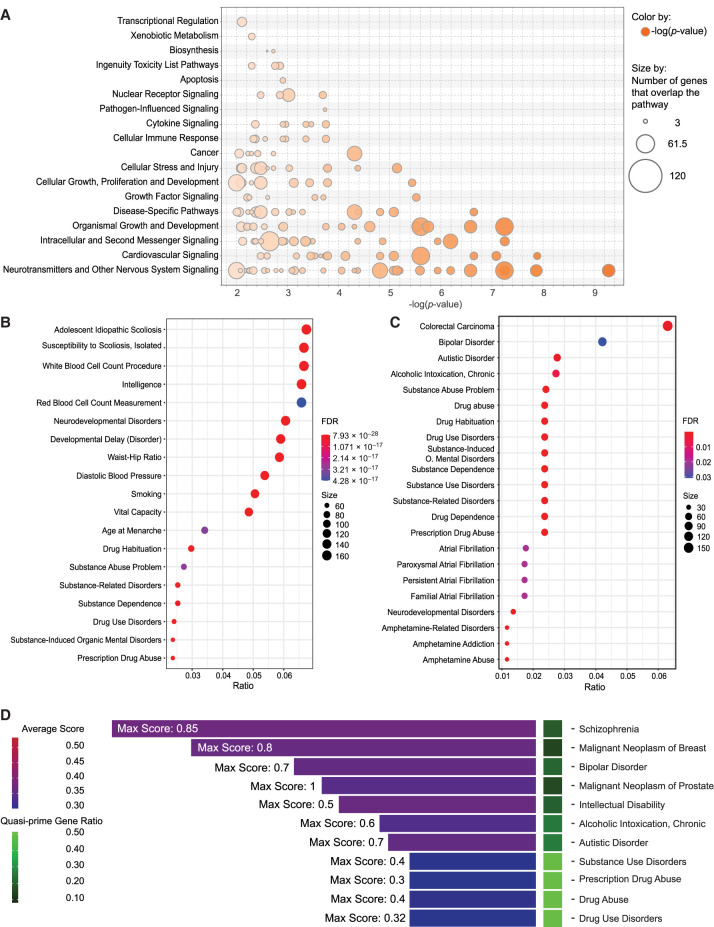
We conducted an ingenuity pathway analysis (IPA) to assess the enrichment of quasi-prime-containing genes in various biological pathways, which we will refer to as quasi-prime genes. We estimated the proportion of genes in each pathway being quasi-prime genes (ratio) and the statistical significance of the enrichment. We found that the most enriched pathways were related to “neurotransmitters and other nervous systems signaling,” such as the opioid signaling pathway (−log P-value = 9.28, ratio = 0.276), circadian rhythm signaling (−log P-value = 7.87, ratio = 0.266), dopamine-DARPP32 feedback in cAMP signaling (−log P-value = 7.25, ratio = 0.289), and axonal guidance signaling (−log P-value = 7.245, ratio = 0.224) (Fig. 5A). Among the highly enriched pathways, the nervous system–related pathways are found among the lowest P-values (Supplemental Fig. 5). Other important pathways are related to cardiovascular signaling (nitric oxide signaling, cellular effects of sildenafil, cardiac hypertrophy signaling, cardiac beta-adrenergic signaling) and endocrine/neuroendocrine functions. Our results suggest that pathways associated with the nervous system are most affected by quasi-prime genes.
Figure 5.
Functional and clinical disease associations of quasi-prime genes. (A) Bubble plot showing the enriched pathway categories from IPA analysis of quasi-prime genes. The size of each bubble represents the proportion of genes in the pathway having human quasi-primes, and the color represents the adjusted P-value of the enrichment. (B) DisGeNET enrichment dot plot showing the top characteristics associated with all quasi-prime genes present in “all” DisGeNET databases. (C) DisGeNET enrichment dot plot showing the top characteristics associated with all quasi-prime genes present in the “CURATED” DisGeNET database. The dots represent the characteristic association score, and the color represents the number of genes in common between the gene set and the disease/characteristic. (D) Gene disease bar plot presenting the count of associated genes across different disease conditions for all diseases with more than six associated genes. The average disease association score for all quasi-prime genes for a given disease within “all” DisGeNET databases is visualized in the color of the bar. The maximum score for all genes within a disease is displayed in text on figure. The number of quasi-prime genes out of the total genes annotated in the database is represented as a ratio shown in the heatmap tile.
We also performed a gene-disease association analysis using DisGeNET (Piñero et al. 2017, 2020). When examining diseases and other traits that are highly enriched for human quasi-prime-containing genes, we find several neurological disorders had the strongest associations, including schizophrenia, bipolar disorder, intellectual disability, drug abuse, and autism (Fig. 5B–D; Supplemental Fig. 6). Specifically, we report that 198 schizophrenia-associated genes and 113 bipolar disorder–associated genes are also quasi-prime-containing genes. We also find that the set of quasi-prime genes associated with these diseases include primarily G-protein-coupled receptors, ion channels, signaling enzymes, and nucleic acid binding proteins (Supplemental Fig. 7A).
From an evolutionary perspective, we aimed to understand whether quasi-primes play a role of highly constrained genes and predicted loss-of-function (pLOF) variants in disease pathogenesis. Therefore, we performed an intersection analysis of our gene set with the pLOF variant Genome Aggregation Database (gnomAD). Our findings revealed a significant enrichment (pLI—odds ratio: 1.66, P-value: 2.65 × 10−45, effect size [Cohen's h]: 0.223; LOEUF—odds ratio: 1.77, P value: 1.23 × 10−57, effect size [Cohen's h]: 0.225) of highly constrained genes being quasi-prime-containing genes (Supplemental Fig. 7B). We observed that highly constrained quasi-prime genes, as indicated by lower LOEUF deciles, were enriched (P-value: 9.31 × 10−8, rho: −0.987) with potentially deleterious pathogenic variants such as frameshift, splice acceptor, splice donor, or stop gained variants (Supplemental Fig. 7C). Our results highlight the potential role of quasi-prime-containing genes in disease pathogenesis. We therefore discover disease associations with human quasi-primes, primarily those relating to cognition, with evidence to support an evolutionary role.
Human quasi-prime genes account for a significant proportion of the variation in expression of primary motor cortex cells
We explored the contribution of quasi-prime genes to the cellular diversity of the human primary motor cortex (M1) using single-cell transcriptomics (Fig. 6A). We leveraged the data from the Allen Institute for Brain Science, which obtained single-cell profiles from postmortem and neurosurgical donor brains with dissected cortical layers (Supplemental Fig. 8; Bakken et al. 2021). We identified 442 quasi-prime genes (22.1%, P-value: 1.613 × 10−173, effect size: 0.165, odds ratio: 2.7) among the top 2000 genes with the highest average expression variability across M1 cell types (Fig. 6A; Supplemental Fig. 9A,B). The most variable quasi-prime genes were RELN, THSD7B, FBXL7, GPC5, and ADARB2. The most variable quasi-prime gene, RELN, encodes for reelin, a protein that regulates neuron signaling during brain development. Our results demonstrate that quasi-prime genes account for a significant proportion of the single-cell expression variation among M1 primary motor cortex cells.
Figure 6.
Differentially expressed genes associated with quasi-prime sequences among cell types found in the human single-cell primary motor cortex brain atlas. (A) Two thousand genes were thresholded by variation to capture the highly variable genes. The resulting highly variate genes were labeled either as genes with quasi-prime loci or without. Four hundred forty-two genes associated with quasi-prime loci (22.1%) capture high levels of variation as shown in the pie chart (Fisher's exact test P-value: 1.613 × 10−173, effect size: 0.165, odds ratio: 2.7). (B) Differential expression of quasi-prime genes thresholded by an absolute log2 fold change > 1 and P adjusted value < 0.05 represented by a dot plot. The top four most significant genes per above cell type ranked by positive log2 fold change are shown. (C) Differentially expressed quasi-prime genes were utilized in gene set enrichment analysis of the Gene Ontology (GO) database representing the top 20 terms per category thresholded by a P-value < 0.01. (D) A cmap network graph shows the clustering of top categories in the GO term GSEA analysis.
Human quasi-prime genes are linked to neuronal support and protection in the primary motor cortex among nonneuronal cells
Differential expression analysis across cell types was conducted to determine cell type–specific expression patterns and function using quasi-prime genes. Our analysis revealed nonneuronal cell types including astrocytes, endothelial cells, microglia perivascular-macrophages, oligodendrocytes, and vascular and leptomeningeal cells exhibited the most distinct expression profiles of quasi-prime genes with cell type–specific markers (Fig. 6B; Supplemental Fig. 10. We performed a Gene Ontology (GO) gene set enrichment analysis (GSEA) of the most significantly differentially expressed quasi-prime-containing genes (log2 fold change > 1 and P-value < 0.05) for each of the 20 cell types. We observed that the quasi-prime gene sets of the five nonneuronal cell types (astrocytes, endothelial cells, microglia perivascular macrophages, oligodendrocytes, and vascular leptomeningeal cells) had common characteristics. These cell types showed upregulation of quasi-prime gene sets involved in metabolic processes, cellular senescence, cell adhesion and proliferation, and immune system pathways (Fig. 6C,D). They also showed downregulation of quasi-prime gene sets related to cell/tissue development, organization, signaling, and transport. Among these cell types, microglia perivascular macrophages exhibited enhanced expression of quasi-prime gene sets associated with immune development, proliferation, adhesion, and activation; reduced expression of quasi-prime gene sets associated with neuro- and morphogenesis; and neuronal and cell development. Oligodendrocytes exhibited reduced expression of quasi-prime gene sets involved in ion transmembrane transport regulation pathways. These results suggest that the human quasi-prime gene sets in the primary motor cortex are predominantly expressed in nonneuronal cell types that regulate the cell and tissue environment to support and protect neuronal cell development.
Human quasi-prime genes in nonneuronal cells of the primary motor cortex are associated with neurological, behavioral diseases, and cancer
Furthermore, we performed GSEA of differentially expressed quasi-prime genes for cell type–specific disease association analysis using two additional databases: Disease Ontology (DO) (Supplemental Fig. 11A) and DisGeNET (Supplemental Fig. 11B). The GSEA results from both databases were consistent and confirmed our previous findings of cognitive, developmental, behavioral, and cancer associations. We found that the activated biological pathways associated with quasi-prime genes were mainly associated with cancer, immune system diseases, and viral infectious diseases in the five nonneuronal cell types. These cell types also had underexpressed biological pathways associated with substance abuse and addiction and cognitive, mental, and developmental disability and disorder. Astrocytes had overexpressed biological pathways involved in sensory system and demyelinating diseases, as well as underexpressed biological pathways associated with substance addiction and abuse. Microglia perivascular macrophages had activated biological pathways associated with immune-related cancers and viral infectious disease. These cell types had underexpressed biological pathways linked to attention deficit disorder, autism, and schizophrenia, whereas oligodendrocytes also had suppressed biological pathways linked to schizophrenia and mathematical ability. We also found associations of quasi-prime genes in two neuronal cell types: in somatostatin chondrolectin (Sst Chondl), a GABAergic interneuron, and in layer five extra telencephalic-projecting neurons (L5 ET), a type of glutamatergic neuron found in the neocortex.
Two clusters of cognitive/mental/developmental diseases and cancer/carcinomas among the network graphs of each disease database are visualized (Supplemental Fig. 12). These diseases are known to be associated with the nonneuronal biological pathways of neuronal development, support, and protection derived from GO GSEA. Therefore, human quasi-prime gene sets are most associated with two disease classes and are useful to determine cell-specific mechanistic relationships of biological significance for cognitive/mental/developmental disease and cancer.
Human nucleic quasi-primes are crucial determinants of disease variant distribution and quantitative trait loci
An analysis of gnomAD v2 variants revealed that human quasi-primes exhibit a greater likelihood to be a constrained gene than would be expected by random chance. This is substantiated by thresholds of elevated constraint (pLI ≥ 0.9—P-value: 2.65 × 10−45, odds ratio: 1.66, effect size [Cohen's h]: 0.22; and LOEUF 90% upper < 0.35—P-value: 1.23 × 10−57, odds ratio: 1.77, effect size [Cohen's h]: 0.26) (Supplemental Fig. 7B). Furthermore, we observed an enrichment of pLOF variant types such as frameshift (decile bin: 0–4), splice acceptor (decile bin: 0–5), splice donor (decile bin: 0–5), and stop gained (decile bin: 0–4) variants for human quasi-primes across a diverse range of constraint as indicated by the LOEUF decile (Supplemental Fig. 7C). Our data strongly suggest that human quasi-primes are overrepresented in highly constrained regions with an increased number of pathogenic and pLOF variants (Spearman's correlation—rho: −0.99, P-value: 9.31 × 10−8), further providing evidence towards their potential role in genetic diseases.
We investigated the likelihood of variants identified through genome-wide association studies (GWAS). The risk of disease or specific traits exhibits a greater likelihood to coincide with or to be located in regions near human quasi-prime loci. We generated a simulated control human quasi-prime set, containing sequences within 10 kb from the original locus and with the same GC content for comparison. Our findings indicate that variants derived from GWAS exhibit a 3.78-fold increased likelihood of directly overlapping human quasi-prime loci compared with matched controls (binomial test, P-value < 2 × 10−13) and show a 4.43-fold enrichment relative to the background vicinity (Fig. 7A). These results suggest that human quasi-prime sequences possess an increased presence of variants that exhibit substantial associations with human diseases and traits.
Figure 7.
Analysis of disruptive and regulatory variants. (A) Enrichment of human quasi-primes at and around GWAS variant loci. (B) Enrichment of human quasi-primes at and around eQTL loci. Each line represents a tissue. (C) Enrichment of human quasi-primes at and around multitissue eQTL loci. Enrichment of human quasi-primes at eQTL loci. (D) Enrichment of human quasi-primes at and around sQTL loci. (E) Enrichment of human quasi-primes at and around mQTL loci. (F) Enrichment of human quasi-primes at and around DisGeNET-derived loci. Gray lines represent the enrichment for simulated controls of human quasi-prime.
Additionally, we examined expression quantitative trait loci (eQTL) from 49 tissues using the GTEx (The GTEx Consortium 2020) to provide additional evidence for the functional roles of human quasi-prime loci. We observe that across the tissues examined, single-tissue eQTLs are enriched at human quasi-prime loci relative to controls (t-test, P-value = 3.5 × 10−85). Across the tissues examined, there is on average a 4.34-fold enrichment of eQTLs relative to the surrounding regions (Fig. 7B), and they are 3.92 times more likely to overlap the quasi-prime loci relative to the matched controls (binomial test, P-value < 5 × 10−8). Similar results were observed when examining eQTLs found across multiple tissues, reporting a 4.35-fold enrichment at human quasi-prime sequences over the surrounding regions (Fig. 7C).
We next examined if human quasi-primes are also enriched for genetic variants that affect alternative splicing or methylation. To that end, we analyzed splicing QTLs (sQTLs) across the 49 tissues available in the GTEx. We find that across the examined tissues, sQTLs are 4.75-fold more likely to overlap human quasi-prime loci than their matched controls (t-test, P-value = 4.67 × 10−13) and are 4.29-fold enriched over their surrounding sequences (Fig. 7D). Methylation QTLs were also examined across nine tissues, and we observed 5.37-fold more mQTLs at human quasi-primes relative to the matched controls (t-test, P-value < 0.001) (Fig. 7E). We therefore conclude that expression, splicing, and methylation QTLs are significantly enriched at human quasi-prime loci.
DisGeNet provides curated variant-disease associations derived from multiple sources (Piñero et al. 2017, 2020). We examined whether variants associated with human diseases are enriched at human quasi-prime sequences. Human quasi-prime loci are observed to be 2.5-fold more likely to overlap with disease variants than expected by chance using the simulated control loci (Fisher's two-sided test, P-value < 0.001), and they are 3.86-fold enriched over surrounding regions (Fig. 7F). The identified variants include multiples that disrupt transcription factor binding. For instance, we find that the variant rs11099601, which overlaps a human quasi-prime, is found in the vicinity of the promoter regions of HELQ, MRPS18C, and ABRAXAS1 genes and affects the expression of all three genes in breast carcinoma, as shown previously, likely by disrupting transcription factor binding (Hamdi et al. 2016). Future massively parallel reporter assays or CRISPR-Cas9 disruptions of quasi-prime sequences with CRISPR-Cas9 will be interesting to systematically examine the cis-regulatory effects and mechanisms at human quasi-primes loci. These results further validate our previous associations and provide support for the importance of studying human quasi-primes to gain insights into human traits and diseases.
Discussion
In this work, we have analyzed 45,076 reference genomes and identified the shortest nucleic k-mers that are unique to each species’ genome, which we refer to as nucleic quasi-primes. This extends our recent work on the detection of peptide quasi-primes (Mouratidis et al. 2023) and also provides multiple biological insights and potential future applications. We also identify primes, the set of shortest k-mers that are absent from all studied genomes, which, similarly to quasi-primes, exhibit an unusually high GC content and are enriched for CpGs. Because CpGs are hypermutable (Sved and Bird 1990; Fryxell and Moon 2005), we postulate that their absence is at least in part driven by the higher mutation rate. Further examination of genome primes in future studies and investigation of potential biological functions would be of high interest, especially to understand if their presence is detrimental to organismal fitness.
We perform a case study on the set of human nucleic quasi-primes and find that they are enriched in genes that are highly expressed in regions associated with brain development and function. We also discover associations between quasi-prime-containing genes and neurological disease–associated genes, including schizophrenia, bipolar disorder, intellectual disability, autism, addiction, and drug abuse. These findings are supported by the expansion of brain size and cognitive tasks following the divergence of humans from other primates (Florio et al. 2017). The faster turnover rate of CpG sequences within human quasi-primes can result in evolutionary adaptations and can account for human-specific traits and molecular phenotypes. Characteristics of schizophrenia, which is the disease with the strongest association with human quasi-prime-containing genes, have not been observed in other species (Burns 2004). Similarly, a strong association of scoliosis, a bipedal-specific disorder (de Reuver et al. 2021), and of hepatitis/hepatitis B, viral disease derived from a humanoid host pathogen (Devaux et al. 2019), were observed (Fig. 5), indicating that human quasi-primes can identify genomic regions associated with species-specific traits. This positions quasi-prime as a potential tool for the study of species-specific traits. Identification of quasi-primes can therefore provide insights into the acquisition of new traits and identify regions of recent evolution.
Similarly to nucleic primes, human quasi-primes have high GC content and CpG sites. Additionally, human quasi-primes are enriched for mQTLs. We therefore speculate that the presence of epigenetic changes found within human nucleic quasi-primes could partially account for their human-specific roles. Genes within the human genome exhibiting a high degree of constraint are predisposed to being classified as quasi-prime genes. Furthermore, highly constrained quasi-prime genes demonstrate a significant enrichment for variants that are pathogenic and potentially lead to loss of function. In future studies, it will be of interest to use nucleic quasi-primes to further explore and understand the functional genomic elements that are linked to phenotypic changes and the evolution of species-specific traits.
Quasi-prime loci can also be utilized to gain insights in cell type–specific associations. In a human case study of the primary motor cortex at single-cell resolution, we highlight biological and disease associations among brain cell types to functionally profile individual cell types via human quasi-prime genes. Genes containing quasi-primes account for notable gene expression diversity and heterogeneity across cells in the primary motor cortex. This heterogeneity may be crucial for the tissue's overall function, enabling it to respond to diverse stimuli or perform specialized tasks. The most variable quasi-prime genes are related to neuronal signaling, tissue reorganization, transferase activity, proteoglycan binding, and RNA-editing activity. It is worth noting the activation of biological pathways related to cellular senescence, cell proliferation, and immune system pathways and the suppression of pathways related to cell/tissue development, organization, signaling, and transport among the most variable expressed genes in nonneuronal cells. We therefore indicate that unique species-specific gene sets can deliver cell-specific information, highlighting a connection to a heritable disease mechanism. We also observe that human quasi-primes are hotspots for pathogenic variants and for variants that significantly alter gene expression or result in alternative splicing or variable DNA methylation. This can lead to further research regarding evolutionary adaptations in humans and can enable an improved understanding of human diseases.
The identification of taxonomy-specific k-mers, which we termed taxonomic quasi-primes, could enable the examination of genome evolution and trait development in individual taxa. We exemplify this by analyzing the mammalian genomes available from the Zoonomia project (Zoonomia Consortium 2020). We find that mammalian taxonomy-specific k-mers can be identified, and they enable the clustering of closely related mammalian species. Therefore, species-specific quasi-primes could have applications for examining more recent trait evolution in a recent lineage or organism, whereas the usage of taxonomic quasi-primes could be used in future work to investigate the emergence of traits in a common ancestor of organisms belonging to the same taxonomy.
Additionally, several applications using nucleic quasi-primes are possible and can be the target of future studies. One of these is the usage of nucleic quasi-primes for the high-throughput detection of multiple organisms in diverse settings, including pathogen detection in clinical settings or in food safety applications such as foodborne outbreaks from microbial contamination (Tringe and Rubin 2005). Nucleic quasi-primes could also enable the detection of invasive or rare species with consequences in conservation. Detection based on nucleic quasi-primes could be coupled with other existing technologies, including CRISPR nucleases that cut in species-specific genomic loci, (Kellner et al. 2019) or adaptive sequencing, which enriches for a set of short sequences (Loose et al. 2016). We note that future work is required to examine the impact of DNA polymorphisms at quasi-prime loci (Teama 2018). Additionally, population variants that disrupt or introduce human quasi-primes could be used as a unique identification signature of individuals and could result in a new type of DNA fingerprinting method. Finally, genome primes are also of high practical interest. Applications can include their usage as genetic barcodes to track cells or organisms. Another potential application may involve their usage as highly specific CRISPR-Cas9 landing pads in genetic engineering and therapeutic applications.
Early genome assemblies often contain gaps in which sequences are unknown owing to technical limitations. Initial genome assemblies might have ambiguous or poorly resolved regions because of complex genetic structures or repetitive DNA sequences such as tandem repeats that require longer read sequencing. As sequencing technologies improve and sequencing error rates decline, errors in the initial assemblies can be identified and corrected. Some regions, especially those that are highly repetitive (like centromeres and telomeres), might be missing or incomplete in earlier versions. Therefore, regular updates of the quasi-prime lists will result in improved quality of the described approach in the future. As the representation of genomic diversity in nature increases with the sequencing of more organismal genomes, the identification of nucleic quasi-primes will provide succinct genomic fingerprints for every available organism.
Methods
Reference genomes used
Collection of reference genomes was performed for the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and RefSeq databases as well as 104 reference genomes from the UCSC Genome Browser website (Supplemental Table 1). In a selected case study on quasi-primes across mammalian genomes, we also used the complete genomes available from the Zoonomia project (Zoonomia Consortium 2020). In the case of duplicate genomes across the different databases, we retained the reference genome from a single source, prioritizing Zoonomia, followed by UCSC, by RefSeq, and finally by GenBank. All genomes used are found in Supplemental Table 1. NCBI RefSeq gene annotation was used for all non-human primates.
k-mer extraction
k-mer extraction was performed for k-mer lengths up to 17 bp, utilizing Jellyfish version 2.2.10 with the -C flag to count both canonical k-mers and their reverse complements as the same k-mer. We then calculated for each k-mer its reverse complement and included it in the final k-mer extraction results for each reference genome. This approach ensures that each k-mer, its reverse complement, is present only once. Only DNA letters “A,” “C,” “G,” and “T” were considered in the analyses, and other IUPAC letters, including “N,” were ignored.
Genome simulations
Simulated genomes were generated using uShuffle (Jiang et al. 2008). For every reference genome, we generated a simulated control genome controlling for dinucleotide content, in which each chromosome in the simulated genome had the same dinucleotide content as the original genome. Therefore, the controls in this study accounted for mono- and dinucleotide biases (including GC content) in any given species. For each simulated genome, oligonucleotide k-mer extraction was performed for k-mer lengths up to and including 17 bp and compared against the number of k-mers identified in the reference genomes (Fig. 2B). For each reference genome and each simulated genome, the number of k-mers identified was compared, and significance was estimated using binomial tests with Bonferroni corrections.
To examine the impact of increasing the number of available organismal genomes on our findings, we first sorted the available genomes by size and distributed them into five equal-sized bins. We then selected genomes incrementally from these bins to create subsets representing 5%, 10%, 25%, 50%, and 75% of the total data set, ensuring that each larger subset included all genomes from the previous one. This selection process was repeated across three independent simulations. For each subset in every simulation, we applied our quasi-prime detection algorithm, focusing on sequences of length 16. To model the behavior of quasi-primes as the data set expanded, we fitted an inverse function to the simulation results. Extrapolating from this model, we predicted the number of quasi-primes in a data set of 500,000 genomes.
Nucleic quasi-prime definition
Let us define the DNA alphabet L = {A, T, C, G}. We define a DNA sequence of length n as A = a1a2a3 … an, where ai ∈ L. We denote its reverse complement sequence , where
A genome GN is a set of DNA sequences representing both strands of chromosomes. We define C = {GN1, GN2, GN3, …, GNk} the set of the k genomes analyzed, where GNi is the genome of the ith species analyzed.
For the purpose of this paper, a DNA k-mer is defined as a short DNA sequence K = k1k2 … kl with length l ∈ ℕ and 1 | 17. We define K ∈ GN if and only if there is DNA sequence A = a1a2a3 … an ∈ GN and 1 ≤ i ≤ n − l, such as aiai+1 … ai+l−1 = k1k2 … kl.
We define the set of quasi-primes of a genome GNi as Q = {K |K ∈ GNi ∧ K ∉ GNj for j ≠ i}.
Taxonomic quasi-prime definition
Let us define a taxonomy Ti as a specific set of m genomes, where m ∈ ℕ, denoted as Ti = {GN1i, GN2i, …, GNmi}. We define S = {T1, T2, …,Tn}, where n ≥ 2 is a set of taxonomies.
A DNA k-mer K of length l where 1 ≤ l ≤ 16 is defined as a taxonomic quasi-prime for taxonomy Ti of taxonomic set S if and only if
K is found in at least one genome GNxiTi.
K is absent in all genomes belonging to any taxonomy Tj where Tj ∈ S and j ≠ i.
The quasi-prime sets derived are available at kmerDB (https://kmer.pennstatehealth.net/kMerDB/), where we have developed a browser for the data set. We provide a downloadable analysis of detailed frequency and annotation information for each species.
Quasi-prime genomic analyses
Identification of the genomic locations of quasi-primes was performed using a custom Python script “quasi_primes_counts.py” (see “Software availability” section; Supplemental Code). The computational size for extracting 16mer quasi-primes was 5.1 TB storage space, with Max RSS (RAM for job) of 38 GB, and total run time of 4 days (not parallelized jobs) for a total number of 45,076 complete genomes. To minimize memory usage, we encoded the k-mers by using two bits for each character (A, T, C, G), fitting each k-mer into a uint32 structure. Each k-mer was stored as a key in a hashmap with a uint32 structure. The value associated with each key is the species’ ID, represented as an integer (mapped to an integer for each species).
Even though the number of reference genomes after a threshold does not alter the total memory, the run time is increased. The time complexity of the algorithm is O(n), where n is the number of reference genomes processed, so we observe a linear increment in run time as n increases. The space complexity is O(4k), where k refers to the k-mer length, so it is independent of the number of genomes. The linear time complexity and constant space complexity (with respect to the number of genomes) ensure that our algorithm can scale efficiently in the future, as the volume of available genomic data increases significantly. If, nevertheless, for some applications it is desirable to ensure an even faster processing time, we have the option of parallelizing the process. We can split our data set into parts, perform the process of detecting quasi-primes for each individual part, and then merge the results, which would allow the rapid identification of quasi-primes for an arbitrarily large data set as long as a sufficient number of computational nodes is available.
GENCODE annotation (v43) was used to derive gene information (Frankish et al. 2023). Genomic subcompartments, namely, genic regions, intronic regions, coding regions, and 5′ and 3′ UTRs, as well as 2500 bp regions upstream of the TSS were derived with the UCSC Table Browser (Nassar et al. 2023). Cis-regulatory elements as defined by ENCODE were used for the analyses (The ENCODE Project Consortium et al. 2020). Cis-regulatory elements included CTCF-only, CTCF-bound, PLS, DNase-H3K4me3, dELS, and pELS terms. BEDTools utilities v2.21.0 (Quinlan 2014) were used to perform the analyses and estimate the density of human nucleic quasi-primes across each genomic element (Fig. 4B,C). Human accelerated regions were derived from Doan et al. (2016), and liftOver was used to transfer them to GRCh38 (hg38) reference genome coordinates. The intersection between human quasi-prime genomic loci and human accelerated regions was performed with the function intersect from BEDTools (Quinlan 2014).
Bulk RNA-seq analysis
Consensus normalized expressions were downloaded from The Human Protein Atlas (RNA consensus tissue gene data) (Pontén et al. 2008) to plot expression levels of quasi-prime genes across 50 tissues. The downloaded data were based on The Human Protein Atlas version 23.0 and Ensembl version 109 (Fig. 4D; Martin et al. 2023).
GO term analysis
A GO term analysis was performed for genes that contained at least one human quasi-prime sequence in the reference human genome with ShinyGO for GO biological process, GO molecular process, and GO molecular function (Fig. 4E,G; Ge et al. 2020).
Ingenuity pathway analysis
The list of quasi-prime genes was uploaded to the IPA platform (Qiagen), and a pathway analysis for human species only was performed (Fig. 5A). The significance values (P-value of overlap) for the pathways was calculated by the right-tailed Fisher's exact test.
Zoonomia analysis
246 mammalian genomes were downloaded from the Zoonomia project to identify nucleic quasi-primes and taxonomic quasi-primes. A Jaccard index was calculated using seaborn for clustering of mammalian taxonomic quasi-primes.
DisGeNET enrichment analysis
The 2492 quasi-prime genes were used in a DisGeNET enrichment analysis (Piñero et al. 2020) to determine which diseases the list of genes is most associated with the “curated” (Fig. 5B) and the “all” (Fig. 5C) databases using the disease_enrichment(vocabulary=“HGNC”) method. The Unified Medical Language System (UMLS) (Bodenreider 2004) was used to identify relevant diseases to query in the disease2gene() method from DisGeNET. The “MRCONSO.RRF” file was downloaded from the UMLS database. The file was filtered to retain values from the English language and MSH database while removing duplicated CUI values and any disease names with numbers. This database was subset to reflect disease names considering relevant keywords (Supplemental Table 2). Additional CUIs were added to reflect diseases, disease classes, and potential human-specific diseases of interest (Supplemental Table 2). The resulting list of CUIs were split into multiple disease2gene() DisGeNET queries as the maximum allowed per query at the present time is 447 diseases. A cutoff of 0.3 was used when querying the DisGeNET “all” database. The resulting DisGeNET S4 classes were combined and were subsequently stratified to reflect disease enrichment results for genes associated with quasi-prime regions. The count of individual quasi-prime associated genes per disease was calculated and displayed as a bar plot (Fig. 5D). A ratio of the amount of quasi-prime associated genes per disease and the total genes associated with a disease was calculated. Diseases with six or fewer associated quasi-prime genes were filtered from this analysis. A heatmap representing the class of proteins that the genes located in quasi-prime regions are most associated with per disease was subsequently generated.
Single-cell RNA-seq analyses
The human M1 primary motor cortex data set, available from Allen Institute for Brain Science (Bakken et al. 2021), was used to analyze the relationship of quasi-prime defined genes from GO term analysis among cell types found in the brain. The human M1 primary motor cortex data set was analyzed using the Seurat (version 4.3.0.1) package in R (Supplemental Fig. 9; Satija et al. 2015; Butler et al. 2018; Stuart et al. 2019; Hao et al. 2021). The data were normalized using the shifted logarithm (for comparison of normalization strategies, see Supplemental Fig. 10A); FindVariableFeatures() was used to generate the 2000 most variable genes (Fig. 6A); and the data were scaled using scaleData(). FindAllMarkers(features=“quasi-prime-genes,” thresh.use=1, min.pct=0.25) was used to calculate the differentially expressed quasi-prime genes (DEGs) between each cell type. DEGs were thresholded by P-adjusted value < 0.05 and used for analysis (Fig. 6B; Supplemental Fig. 10). The resulting DEGs were converted to Entrez format and used in GSEA of the GO (Fig. 6C,D), DisGeNET, and DO (Supplemental Fig. 11A,B) databases using the comparecluster() method in the ClusterProfiler package (version 3.17) (Yu et al. 2012; Wu et al. 2021). Gene sets with a minimum of three genes were included in each analysis.
Variant analysis
The GWAS catalog was derived from https://www.ebi.ac.uk/gwas/api/search/downloads/full (MacArthur et al. 2017). Multitissue and single-tissue eQTLs were derived from the GTEx Consortium (v8) (The GTEx Consortium 2020). sQTLs were derived from the GTEx Consortium (The GTEx Consortium 2020; Garrido-Martín et al. 2021). mQTLs were derived for nine tissues from the GTEx Consortium (The GTEx Consortium 2020; Oliva et al. 2023). For the mQTL analysis, only events with Q-value < 0.05 were analyzed. DisGeNET variants were derived from https://www.disgenet.org/static/disgenet_ap1/files/downloads/variant_associations.tsv.gz (Piñero et al. 2020). A set of simulated control human quasi-primes were generated; in each human quasi-prime, a locus that was within 10 kb from the original locus and had the length and GC content was randomly selected. Enrichment for genomic variants was calculated as described in Figure 7 (Georgakopoulos-Soares et al. 2018).
We conducted a variant analysis to investigate the enrichment of quasi-prime regions within variant regions in RegulomeDB (Dong et al. 2023). We simulated control regions to compare the potential enrichment. The Bioframe Python package (v0.4.1) (Open2C et al. 2024) was used to determine the overlap between quasi-prime regions and RegulomeDB regions and between control regions and RegulomeDB regions, respectively. A binomial test was then used to test for enrichment between the groups (SciPy v1.11.2). The enrichment (represented as odds ratio) of each variant characteristic for these overlapped variant groups from RegulomeDB was subsequently plotted (Supplemental Fig. 13A,B).
The gnomAD constraint database (Chg38) was downloaded from https://storage.googleapis.com/gcp-public-data‐‐gnomad/release/2.1.1/constraint/gnomad.v2.1.1.lof_metrics.by_gene.txt.bgz (Karczewski et al. 2020) and subsequently queried to evaluate the enrichment (expressed as odds ratio) of human quasi-prime genes within regions of high constraint. A hypergeometric enrichment test was conducted to compute a P-value and effect size for genes deemed highly constrained (pLI ≥ 0.9 or an oe_lof_upper < 0.35) (Supplemental Fig. 7B).
All pLOF variants were obtained from gnomAD (Karczewski et al. 2020; https://storage.googleapis.com/gcp-public-data‐‐gnomad/papers/2019-flagship-lof/v1.0/gnomad.v2.1.1.all_lofs.txt.bgz). The data frames were merged based on the position of the variant within the constraint/genic region. The enrichment of quasi-prime genes, based on LOEUF decile (Karczewski et al. 2020) bins for each variant type, was also graphically depicted (Fig. 7C). A Spearman's correlation analysis was conducted to examine any potential correlational relationship between the constraint metric LOEUF decile and variant types as a sum for quasi-prime enrichment.
Software availability
All code used in this manuscript is available at GitHub (https://github.com/Georgakopoulos-Soares-lab/dna-quasi-primes) and as Supplemental Code.
Supplemental Material
Acknowledgments
This study was funded by the startup funds of I.G.-S. from the Penn State College of Medicine and by the Huck Innovative and Transformational Seed Fund (HITS) award from the Huck Institutes of the Life Sciences at Penn State University. Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM155468. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. G.A.P. was supported by funds from Fondation Santé; Onassis Foundation; Hellenic Foundation for Research and Innovation (16718-PRPFOR and TAEDR-0539180).
Author contributions: I.M. and I.G.-S. conceived the concept of DNA quasi-primes. I.G.-S. supervised the work. I.M., M.A.K., N.C., C.S.Y.C., M.P., D.V.C., and I.G.-S. wrote the code. I.M., M.A.K., N.C., C.S.Y.C., M.P., D.V.C., and I.G.-S. performed the analyses. M.A.K., C.S.Y.C., M.P., D.V.C., and I.G.-S. generated the visualizations. I.M., M.A.K., N.C., C.S.Y.C., and I.G.-S. wrote the manuscript with help from all other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.280070.124.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Alileche A, Goswami J, Bourland W, Davis M, Hampikian G. 2012. Nullomer derived anticancer peptides (NulloPs): differential lethal effects on normal and cancer cells in vitro. Peptides 38: 302–311. 10.1016/j.peptides.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Al-Salam A, Irwin DM. 2017. Evolution of the vertebrate insulin receptor substrate (Irs) gene family. BMC Evol Biol 17: 148. 10.1186/s12862-017-0994-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, et al. 2021. Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598: 111–119. 10.1038/s41586-021-03465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bize A, Midoux C, Mariadassou M, Schbath S, Forterre P, Da Cunha V. 2021. Exploring short k-mer profiles in cells and mobile elements from Archaea highlights the major influence of both the ecological niche and evolutionary history. BMC Genomics 22: 186. 10.1186/s12864-021-07471-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. 2004. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 32: D267–D270. 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandies P, Peel E, Hogg CJ, Belov K. 2019. The value of reference genomes in the conservation of threatened species. Genes (Basel) 10: 846. 10.3390/genes10110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JK. 2004. An evolutionary theory of schizophrenia: cortical connectivity, metarepresentation, and the social brain. Behav Brain Sci 27: 831–855; discussion 855–885. 10.1017/S0140525X04000196 [DOI] [PubMed] [Google Scholar]
- Bussi Y, Kapon R, Reich Z. 2021. Large-scale k-mer-based analysis of the informational properties of genomes, comparative genomics and taxonomy. PLoS One 16: e0258693. 10.1371/journal.pone.0258693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chor B, Horn D, Goldman N, Levy Y, Massingham T. 2009. Genomic DNA k-mer spectra: models and modalities. Genome Biol 10: R108. 10.1186/gb-2009-10-10-r108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin Tree of Life Project Consortium. 2022. Sequence locally, think globally: the Darwin Tree of Life Project. Proc Natl Acad Sci 119: e2115642118. 10.1073/pnas.2115642118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reuver S, IJsseldijk LL, Homans JF, Willems DS, Veraa S, van Stralen M, Kik MJL, Kruyt MC, Gröne A, Castelein RM. 2021. What a stranded whale with scoliosis can teach us about human idiopathic scoliosis. Sci Rep 11: 7218. 10.1038/s41598-021-86709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, García-Cobos S, Kooistra-Smid AMD, Raangs EC, Rosema S, Veloo ACM, et al. 2017. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol 243: 16–24. 10.1016/j.jbiotec.2016.12.022 [DOI] [PubMed] [Google Scholar]
- Devaux CA, Mediannikov O, Medkour H, Raoult D. 2019. Infectious disease risk across the growing human-non human primate interface: a review of the evidence. Front Public Health 7: 305. 10.3389/fpubh.2019.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan RN, Bae B-I, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, et al. 2016. Mutations in human accelerated regions disrupt cognition and social behavior. Cell 167: 341–354.e12. 10.1016/j.cell.2016.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Zhao N, Spragins E, Kagda MS, Li M, Assis P, Jolanki O, Luo Y, Cherry JM, Boyle AP, et al. 2023. Annotating and prioritizing human non-coding variants with RegulomeDB v.2. Nat Genet 55: 724–726. 10.1038/s41588-023-01365-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris E, Abegglen LM, Schiffman JD, Gregg C. 2018. Accelerated evolution in distinctive species reveals candidate elements for clinically relevant traits, including mutation and cancer resistance. Cell Rep 22: 2742–2755. 10.1016/j.celrep.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M, Borrell V, Huttner WB. 2017. Human-specific genomic signatures of neocortical expansion. Curr Opin Neurobiol 42: 33–44. 10.1016/j.conb.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Foley NM, Mason VC, Harris AJ, Bredemeyer KR, Damas J, Lewin HA, Eizirik E, Gatesy J, Karlsson EK, Lindblad-Toh K, et al. 2023. A genomic timescale for placental mammal evolution. Science 380: eabl8189. 10.1126/science.abl8189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Carbonell-Sala S, Diekhans M, Jungreis I, Loveland JE, Mudge JM, Sisu C, Wright JC, Arnan C, Barnes I, et al. 2023. GENCODE: reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res 51: D942–D949. 10.1093/nar/gkac1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryxell KJ, Moon W-J. 2005. CpG mutation rates in the human genome are highly dependent on local GC content. Mol Biol Evol 22: 650–658. 10.1093/molbev/msi043 [DOI] [PubMed] [Google Scholar]
- Garrido-Martín D, Borsari B, Calvo M, Reverter F, Guigó R. 2021. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat Commun 12: 727. 10.1038/s41467-020-20578-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Jung D, Yao R. 2020. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36: 2628–2629. 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos-Soares I, Morganella S, Jain N, Hemberg M, Nik-Zainal S. 2018. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res 28: 1264–1271. 10.1101/gr.231688.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos-Soares I, Barnea OY, Mouratidis I, Chan CSY, Bradley R, Mahajan M, Sims J, Cintron DL, Easterlin R, Kim JS, et al. 2021a. Leveraging sequences missing from the human genome to diagnose cancer. medRxiv 10.1101/2021.08.15.21261805 [DOI]
- Georgakopoulos-Soares I, Yizhar-Barnea O, Mouratidis I, Hemberg M, Ahituv N. 2021b. Absent from DNA and protein: genomic characterization of nullomers and nullpeptides across functional categories and evolution. Genome Biol 22: 245. 10.1186/s13059-021-02459-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami J, Davis MC, Andersen T, Alileche A, Hampikian G. 2013. Safeguarding forensic DNA reference samples with nullomer barcodes. J Forensic Leg Med 20: 513–519. 10.1016/j.jflm.2013.02.003 [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2020. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369: 1318–1330. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdi Y, Soucy P, Adoue V, Michailidou K, Canisius S, Lemaçon A, Droit A, Andrulis IL, Anton-Culver H, Arndt V, et al. 2016. Association of breast cancer risk with genetic variants showing differential allelic expression: identification of a novel breast cancer susceptibility locus at 4q21. Oncotarget 7: 80140–80163. 10.18632/oncotarget.12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampikian G, Andersen T. 2007. Absent sequences: nullomers and primes. Pac Symp Biocomput 2007: 355–366. 10.1142/9789812772435_0034 [DOI] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell 184: 3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS. 2014. Exploring the genesis and functions of human accelerated regions sheds light on their role in human evolution. Curr Opin Genet Dev 29: 15–21. 10.1016/j.gde.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Jagadeesan B, Gerner-Smidt P, Allard MW, Leuillet S, Winkler A, Xiao Y, Chaffron S, Van Der Vossen J, Tang S, Katase M, et al. 2019. The use of next generation sequencing for improving food safety: translation into practice. Food Microbiol 79: 96–115. 10.1016/j.fm.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Anderson J, Gillespie J, Mayne M. 2008. uShuffle: a useful tool for shuffling biological sequences while preserving the k-let counts. BMC Bioinformatics 9: 192. 10.1186/1471-2105-9-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. 2019. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 14: 2986–3012. 10.1038/s41596-019-0210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulouras G, Frith MC. 2021. Significant non-existence of sequences in genomes and proteomes. Nucleic Acids Res 49: 3139–3155. 10.1093/nar/gkab139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, John Kress W, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP, et al. 2018. Earth BioGenome Project: sequencing life for the future of life. Proc Natl Acad Sci 115: 4325–4333. 10.1073/pnas.1720115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Richards S, Aiden EL, Allende ML, Archibald JM, Bálint M, Barker KB, Baumgartner B, Belov K, Bertorelle G, et al. 2022. The Earth BioGenome Project 2020: starting the clock. Proc Natl Acad Sci 119: 4. 10.1073/pnas.2115635118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Malla S, Stout M. 2016. Real-time selective sequencing using nanopore technology. Nat Methods 13: 751–754. 10.1038/nmeth.3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyčka M, Bubeník M, Závodník M, Peska V, Fajkus P, Demko M, Fajkus J, Fojtová M. 2024. TeloBase: a community-curated database of telomere sequences across the tree of life. Nucleic Acids Res 52: D311–D321. 10.1093/nar/gkad672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. 2017. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog). Nucleic Acids Res 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maljkovic Berry I, Melendrez MC, Bishop-Lilly KA, Rutvisuttinunt W, Pollett S, Talundzic E, Morton L, Jarman RG. 2020. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis 221: S292–S307. 10.1093/infdis/jiz286 [DOI] [PubMed] [Google Scholar]
- Martin FJ, Ridwan Amode M, Aneja A, Austine-Orimoloye O, Azov AG, Barnes I, Becker A, Bennett R, Berry A, Bhai J, et al. 2023. Ensembl 2023. Nucleic Acids Res 51: D933–D941. 10.1093/nar/gkac958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471: 216–219. 10.1038/nature09774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery A, Tsiatsianis GC, Mouratidis I, Chan CSY, Athanasiou M, Papanastasiou AD, Kantere V, Syrigos N, Vathiotis I, Syrigos K, et al. 2024. Utilizing nullomers in cell-free RNA for early cancer detection. Cancer Gene Ther 31: 861–870. 10.1038/s41417-024-00741-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratidis I, Chan CSY, Chantzi N, Tsiatsianis GC, Hemberg M, Ahituv N, Georgakopoulos-Soares I. 2023. Quasi-prime peptides: identification of the shortest peptide sequences unique to a species. NAR Genom Bioinform 5: lqad039. 10.1093/nargab/lqad039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar LR, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee BT, et al. 2023. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res 51: D1188–D1195. 10.1093/nar/gkac1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Rodney Brister J, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M, Demanelis K, Lu Y, Chernoff M, Jasmine F, Ahsan H, Kibriya MG, Chen LS, Pierce BL. 2023. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat Genet 55: 112–122. 10.1038/s41588-022-01248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Open2C, Abdennur N, Fudenberg G, Flyamer IM, Galitsyna AA, Goloborodko A, Imakaev M, Venev S. 2024. Bioframe: operations on genomic intervals in Pandas dataframes. Bioinformatics 40: btae088. 10.1093/bioinformatics/btae088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Dong JC, Trost B, Richardson JS, Tohme S, Babiuk S, Kusalik A, Kung SKP, Kobinger GP. 2012. Pentamers not found in the universal proteome can enhance antigen specific immune responses and adjuvant vaccines. PLoS One 7: e43802. 10.1371/journal.pone.0043802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz F, Furlong LI. 2017. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 45: D833–D839. 10.1093/nar/gkw943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. 2020. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 48: D845–D855. 10.1093/nar/gkz1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168. 10.1371/journal.pgen.0020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontén F, Jirström K, Uhlen M. 2008. The Human Protein Atlas: a tool for pathology. J Pathol 216: 387–393. 10.1002/path.2440 [DOI] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O'Neill K, Robbertse B, et al. 2020. NCBI taxonomy: a comprehensive update on curation, resources and tools. Database 2020: baaa062. 10.1093/database/baaa062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre G-P, Bank C, Brännström Å, et al. 2014. Genomics and the origin of species. Nat Rev Genet 15: 176–192. 10.1038/nrg3644 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lu F, Luo Y, Bie L, Xu L, Wang Y. 2023. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res 51: W397–W403. 10.1093/nar/gkad313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A. 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci 87: 4692–4696. 10.1073/pnas.87.12.4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teama S. 2018. DNA polymorphisms: DNA-based molecular markers and their application in medicine. In Genetic diversity and disease susceptibility. IntechOpen, London. [Google Scholar]
- Tringe SG, Rubin EM. 2005. Metagenomics: DNA sequencing of environmental samples. Nat Rev Genet 6: 805–814. 10.1038/nrg1709 [DOI] [PubMed] [Google Scholar]
- Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, et al. 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb.) 2: 100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoonomia Consortium. 2020. A comparative genomics multitool for scientific discovery and conservation. Nature 587: 240–245. 10.1038/s41586-020-2876-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.