Abstract
Neurodegeneration can occur as a result of endogenous oxidative stress. Primary cerebellar granule cells were used in this study to determine if mitochondrial DNA (mtDNA) repair deficiencies correlate with oxidative stress-induced apoptosis in neuronal cells. Granule cells exhibited a significantly higher intracellular oxidative state compared with primary astrocytes as well as increases in reductants, such as glutathione, and redox sensitive signaling molecules, such as AP endonuclease/redox effector factor-1. Cerebellar granule cultures also exhibited an increased susceptibility to exogenous oxidative stress. Menadione (50 μM) produced twice as many lesions in granule cell mtDNA compared with astrocytes, and granule cell mtDNA repair was significantly less efficient. A decreased capacity to repair oxidative mtDNA damage correlates strongly with mitochondrial initiated apoptosis in these neuronal cultures. Interestingly, the mitochondrial activities of initiators for base excision repair (BER), the bifunctional glycosylase/AP lyases as well as AP endonuclease, were significantly higher in cerebellar granule cells compared with astrocytes. The increased mitochondrial AP endonuclease activity in combination with decreased polymerase γ activity may cause an imbalance in oxidative BER leading to an increased production and persistence of mtDNA damage in neurons when treated with menadione. This study provides evidence linking neuronal mtDNA repair capacity with oxidative stress-related neurodegeneration.
INTRODUCTION
Nowhere in the body are the deleterious effects of aging better observed than in the post-mitotic tissues of the brain. Neurodegeneration, among other significant pathologies, can occur as a result of endogenous oxidative stress, and in fact the rate of reactive oxygen species (ROS) production in the vertebrate central nervous system (CNS) has been shown to be inversely proportional to life-span (1,2).
Recently characterized as a ‘crucial nexus’ of ROS production within the cell, mitochondria have been linked to several neurodegenerative disorders as well as normal changes in the CNS that accompany aging (3). This premise relates oxidative stress to aging in long-lived animals through the rate of mitochondrial ROS production and accumulation of mitochondrial DNA (mtDNA) damage. In accordance with this relationship is the observed negative correlation between longevity in mammals and levels of oxidative mtDNA damage in the brain, with no such correlation observed for nuclear DNA (4). In contrast to chromosomal DNA, the increased susceptibility of mtDNA to oxidative damage is due in large part to its close proximity to the cell's main source of free radicals, the mitochondrial respiratory chain (5–9). Indeed, oxidative damage in mtDNA is normally 2–3 times greater than that of nuclear DNA (10). Therefore, the mitochondrial theory of aging suggests that ROS production leads to mtDNA damage and eventually mutations that cause mitochondrial respiratory chain dysfunction that results in increased ROS production, thus a continuation of a ‘vicious cycle’ of mtDNA damage amplifying ROS (11,12).
In addition to the general decline in CNS function associated with normal aging, certain mutations in mitochondrial genes have been implicated in the etiology of various age-related neurodegenerative diseases, such as Parkinson's disease, Alzheimer's disease, Huntington's disease and amyotrophic lateral sclerosis (12–14). The pathogenesis resulting from mtDNA mutations is believed to involve impaired oxidative phosphorylation with a concomitant increase in ROS production (15). Of interest to this study is the neuronal cell's capacity to relieve this ‘vicious cycle’ by removing the causative elements, ROS and mtDNA damage, thereby eliminating the resultant harmful states of oxidative stress and mitochondrial dysfunction. Two factors that might affect mitochondrial dysfunction associated with aging in the CNS are the levels of various free radical scavengers that alter the balance between normal cellular function and oxidative stress as well as the capacity of neuronal cells to repair mtDNA damage, and thus protect against mutations in the mitochondrial genome. Regarding free radical scavengers, there is evidence suggesting that endogenous levels of antioxidants in the brain do not decrease during aging and that experimentally increasing their levels has no effect on longevity (2,16). In fact, tissues that are vulnerable to oxidative stress tend to exhibit much higher levels of antioxidants than other less vulnerable tissues (17,18). However, with respect to mtDNA repair, differential repair of oxidative damage among glial cells has been demonstrated as well as an inverse correlation between mtDNA repair capacity and induction of apoptosis in these cell types. Specifically, astrocytes exhibited greater mtDNA repair capacity with no induction of apoptosis following treatment with DNA damaging agents compared with oligodendrocytes and microglia, both of which exhibited decreased mtDNA repair and increased induction of apoptosis (19,20). These findings suggest the possibility that there exist cell-specific differences in susceptibility to chronic oxidative stress in the aging CNS and warrant further investigation into specific neuronal mtDNA repair processes as well as the relationship of these repair processes with neuronal capacity to endure oxidative stress.
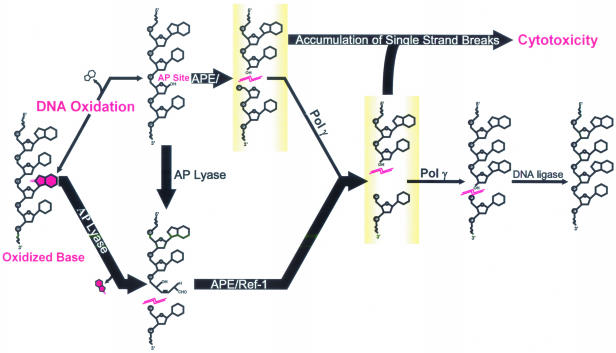
The base excision repair (BER) pathway is the principal mechanism by which mammalian cells repair DNA oxidation damage (21–23) and appears to be the predominant DNA repair system in mitochondria (Figure 1). Little is known about mtDNA repair capacity for oxidative damage among neuronal cell types; however, there is evidence of differential expression of key BER enzymes in the CNS (24–28), suggesting that BER enzyme levels may contribute to neuronal susceptibility to oxidative stress by affecting mtDNA repair efficiency. Therefore, primary cultures of rat cerebellar granule cells were used to test the hypothesis that deficient repair of oxidative mtDNA damage correlates with increased sensitivity to oxidative stress in the CNS, and thus neuronal cells undergo apoptosis in response to oxidative challenge because of a mitochondrial BER pathway imbalance.
Figure 1.
BER of oxidative mtDNA damage in neuronal cells. In addition to oxidized bases, oxidative DNA damage may also include damage to the deoxyribose sugar backbone, which can result in the direct release of bases forming AP sites. The bifunctional glycosylase/AP lyases (AP lyase) excise oxidatively damaged bases, such as 8-oxoguanine, prior to incising the sugar backbone 3′ to the lesion. AP sites are substrates for either AP endonuclease (APE/Ref-1) or the AP lyases and are processed by these two pathway initiators producing single strand breaks: 3′-OH/5′-deoxyribose phosphate (5′-dRP) and 5′-phosphate/3′-fragmented deoxyribose, respectively. The 5′-dRP product of APE/Ref-1 is removed by the dRPase activity of pol γ, and the 3′-fragmented deoxyribose product of the AP lyase is removed by the 3′-diesterase activity of APE/Ref-1. Once the single nucleotide gap has been processed, pol γ can fill the gap. The remaining DNA strand break is sealed by DNA ligase. Pathway arrows are drawn in boldface to indicate the relative high AP lyase and APE activities and low pol γ activity measured in cerebellar granule cell mitochondria. Imbalanced BER can result in accumulation of cytotoxic pathway intermediates.
MATERIALS AND METHODS
Cell cultures
The cerebella of 7-day-old Sprague–Dawley rats were aseptically removed, minced, dissociated into single cell suspensions, and plated into 75 cm2 flasks at a density of 1 × 107 cells/flask (29,30). All experimental procedures were carried out under protocols approved by the Institutional Animal Care and Use Committee of the University of South Alabama and in accordance with the principles outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Cerebellar granule cells were cultured in Eagle's modified essential medium supplemented with 10% FBS, 1% glutamine, 1% potassium chloride, 1.2% glucose and gentamycin sulfate. To prevent growth of non-neuronal cells, 1-day-old cultures were treated with the mitotic inhibitor fluorodeoxyuridine (80 μM). After 1–2 days, granule cell cultures were determined to be 95% pure using immunofluorescence staining with an anti-neurofilament antibody (31). Immunofluorescence staining with biotinylated Griffonia simplicifolia isolectin B4, O4 monoclonal antibody and glial fibrillary acid protein antibody was used to determine the extent of contamination with microglia, oligodendrocytes and astrocytes, respectively. Primary astrocytes were cultured as described previously (19).
Menadione treatment
After 1–2 days in culture, cells were cultured in fresh media then treated with menadione 24 h later. Before exposure to menadione, cells were rinsed with Hanks' balanced salt solution (HBSS). Menadione was dissolved in HBSS to yield the desired concentration(s) while control cells were treated only with HBSS under the same conditions, 1 h in a 5% CO2, 37°C incubated environment. After the 1 h incubation period, cells were either lysed immediately or allowed to repair for 2 or 6 h in culture medium. As described previously (20), cell viability was determined by trypan blue dye exclusion.
Quantification of apoptosis
The percentage of cells in treated and control cultures undergoing apoptosis was measured as described previously (20) using an ApoTag kit (Oncor) and an Annexin-V kit (Trevigen). For these experiments, cells were plated onto poly-lysine pretreated Labtech slides. Experimental cultures were exposed to menadione for 1 h, rinsed with HBSS, and placed in normal culture medium for 6 h. Cultures were then rinsed with ice-cold 1× PBS and the assays conducted according to the manufacturer's instructions. Using fluorescence microscopy, eight random fields of 100 cells were counted, and the percentage of apoptotic cells was expressed (100 × average number of fluorescent cells/total number of cells).
Caspase assays
Cells were induced to undergo apoptosis by 100 μM menadione treatment for 1 h. Following treatment, cells were rinsed with HBSS and culture medium was replenished. At 3 h post-treatment, cells were rinsed with 1× PBS and collected by centrifugation at 200 g for 10 min. Activity was measured using colorimetric assay kits (R&D Systems). Briefly, cells were lysed on ice for 10 min, centrifuged at 10 000 g for 1 min, and the supernatant incubated with caspase-8-specific (4 mM IEDT-pNA) or caspase-9-specific (1 mM LEHD-pNA) substrates. Samples were analyzed spectrophotometrically at 405 nm. Jurkat cells (TIB 152; ATCC) treated with an anti-Fas monoclonal antibody (2 μg/ml for 3 h) served as a positive control for caspase-8 assays while cerebellar granule cells treated with staurosporine (2 μM for 2 h) were used as a caspase-9 positive control.
Intracellular prooxidant levels
Prooxidant production was determined using the oxidation-sensitive 5- (and-6)-carboxy-2′, 7′-dichlorodihydrofluorescein diacetate (C-400, 10 μg/ml) and oxidation-insensitive 5- (and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (C-369, 10 μg/ml) fluorescent dyes (dissolved in Me2SO) obtained from Molecular Probes. The oxidized form of the dye acts as a control for changes in uptake, ester cleavage and efflux, so that any changes in fluorescence seen among cell types with the oxidation-sensitive dye can be directly attributed to changes in dye oxidation (32). Cells were harvested, labeled with the fluorescent dyes for 15 min at 37°C, placed on ice, and analyzed using a FACScan flow cytometer (BD Biosciences, Mountain View, CA) (excitation 488 nm, emission 535 nm). The mean fluorescence intensity of 20 000 cells was analyzed in each sample and corrected for autofluorescence from unlabeled cells.
Measurement of glutathione levels
Following treatment, the cells were scrape-harvested in PBS at 4°C, centrifuged, the PBS was discarded and then the cell pellets were frozen at −80°C. Samples were thawed and whole homogenates were prepared as described previously (33). Total glutathione (GSH + GSSG) and glutathione disulfide (GSSG) were determined using a spectrophotometric recycling assay (33). All biochemical determinations were normalized to protein content using the method of Lowry et al. (34).
Superoxide dismutase activity assays
Total superoxide dismutase (SOD), Mn-SOD and CuZn-SOD activities were measured as described previously (35). Briefly, this assay is a competitive inhibition assay which uses xanthine oxidase-generated superoxide to reduce nitroblue-tetrazolium (NBT) at a constant rate (absorbance of 0.02/min), which is monitored spectrophotometrically at 560 nm. This rate of reduction of NBT is progressively inhibited by increasing amounts of SOD or cellular homogenate containing SOD. The amount of protein that inhibits NBT reduction to 50% of the maximal inhibition is defined as 1 U of SOD activity (7–10 ng = 1 U of activity for purified CuZn-SOD from Diagnostic Data, Mountain View, CA). The assay also utilizes the inhibition of CuZn-SOD by 5 mM cyanide to differentiate between CuZn-SOD and Mn-SOD activities. This concentration of cyanide inhibited 100–150 ng of purified CuZn-SOD without inhibiting the NBT reduction mediated by xanthine oxidase. CuZn-SOD activity was determined by subtracting Mn-SOD activity from total SOD activity. Protein concentration was determined by using the Lowry method (34) and enzymatic activity was expressed in U/mg protein and U/106 cells.
Quantitative Southern blot technique
mtDNA damage and repair was detected using a quantitative Southern blot (QSB) technique in conjunction with a mitochondrial specific PCR-generated probe (36,37). After drug exposure, cells were lysed immediately or allowed to repair for intervals of 2 or 6 h. High-molecular weight DNA was extracted from control and treated cells and digested to completion with BamHI. After heating, alkali treatment, and alkaline gel electrophoresis, QSB was performed using mtDNA-specific probes and the data were analyzed by quantitative densitometry (36,38). Alkali treatment causes strand breaks at any abasic or sugar modified site in DNA resulting in fewer full-length restriction fragments to bind to the mitochondrial-specific probe. The optimal range for studying repair using this technique requires an initial lesion density or break frequency (BF) of ∼0.5–1.5 lesions/104 nt. BF was calculated densitometrically using a Poisson expression, BF = −ln(treated samples/control samples), and expressed as %Repair.
mtDNA probe
The probe used to hybridize to mtDNA was generated by PCR from a mouse mtDNA sequence using the following primers: 5′-GCAGGAACAGGATGAACAGTCT-3′ from the sense strand and 5′-GTATCGTGAAGCACGATGTCAAGGGATGAG-3′ from the antisense strand. The 745 bp PCR product recognizes a 10.8 kb restriction fragment when hybridized to rat mtDNA digested with BamH1 (19).
Enzyme activity assays
These studies employ an in vitro oligonucleotide cleavage assay to assess and compare APE and AP lyase activities in nuclear and mitochondrial lysates. A total of 3–5 150 cm plates of each cell type at confluence were harvested and treated as described previously (39) with 5 ml of ice-cold 0.4% digitonin to selectively lyse the plasma membrane. The homogenate was combined with a mannitol–sucrose buffer and centrifuged to pellet nuclei. The nuclear pellets were washed, and the remaining supernatant was centrifuged at 10 000 g to pellet mitochondria. The resulting mitochondrial and nuclear pellets were then resuspended in 0.2% Triton X-100 lysing solution, and the supernatant protein was used for activity assays and western blots. Protein concentrations were determined using the Bio-Rad protein dye micro-assay according to the manufacturer's recommendations (Bradford method).
The APE assay utilizes a 21mer oligonucleotide containing a tetrahydrofuran apurinic/apyrimidinic (AP) site at the tenth position (THF oligo) (Trevigen, Gaithersburg, MD). The THF oligo was radiolabeled and allowed to anneal with a complementary oligonucleotide to form duplex DNA. Activity assays contained 7.5 pmol of labeled THF oligonucleotide, 5 μl of 10× REC buffer 7 (Trevigen) and equal amounts of protein from either nuclear or mitochondrial extracts from each cell type for a final concentration of 0.025 μg/μl in a total volume of 50 μl. Aliquots (10 μl) were removed at 2.5, 5, 20, 40 and 60 min time points during the 37°C incubation. The zero time points and controls were assayed separately in 10 μl vol containing 1.5 pmol of labeled substrate and 1 μl of 10× REC buffer 7. Purified human APE (1 U; Trevigen) was used as a positive control. The reaction contents were resolved on a 20% TBE-urea gel and visualized by autoradiography. Specific bands were analyzed by using densitometry. Enzyme activity is a measure of percentage of substrate converted over time. The percentage substrate remaining was calculated: 100 × substrate remaining/zero-timepoint substrate. Total enzyme activity is represented as a measure of slope that includes all linear time points.
The AP lyase activity assays were conducted in parallel with the APE assays. The substrate oligonucleotide (Trevigen) used in these experiments contains an AP site at the tenth position (AP oligo) instead of a THF AP site. With the exception of this substrate modification, the AP lyase activity assays were identical to the APE activity assays. Conversion of the THF oligo is specific for APE activity. Both APE and AP lyase activity will convert the AP oligo. Therefore, total AP lyase activity was determined by subtracting the THF oligo conversion from the AP oligo conversion for each time point in the individual extracts analyzed.
Western blot analysis
Western blots were performed using either whole-cell, mitochondrial or cytosolic protein extracts from primary CNS cultures as described previously (20). Protein concentrations were measured using the Bio-Rad protein assay reagent. Protein (15–25 μg) was separated on 4–15% polyacrylamide–SDS gels (Bio-Rad) and then transferred onto Immobilon-P (polyvinylidene diflouride) membranes with a Semiphore transfer cell (Hoefer Scientific) at 100 mA for 2 h. The membranes were blocked with 5% non-fat milk in Tris-buffered saline [10 mM Tris (pH 8) and 150 mM NaCl] containing Tween-20 and incubated with mouse anti-cytochrome c monoclonal antibody (Pharmingen), 8-oxoguanine DNA-glycosylase polyclonal antibody (Novus Biologicals), anti-actin antibody (Sigma), a monoclonal AP endonuclease antibody (a gift from Dr Mark Kelley, Indiana University) and a polymerase gamma β subunit polyclonal antibody (a gift from Dr Daniel Bogenhagen, SUNY at Stony Brook) as well as a polymerase gamma α subunit polyclonal antibody (40). Membranes were incubated for 1 h with anti-mouse or rabbit IgG secondary antibodies (Jackson ImmunoResearch) at room temperature. Protein bands were visualized by using chemiluminescence (SuperSignal, Pierce).
Polymerase γ activity analysis
A total of 5 × 106 cells were lysed in 1 ml [200 mM NaCl, 100 mM Tris–HCl (pH 7.5), 8% glycerol, 1% NP-40, 0.1 mM EDTA and 0.25 mM β-mercaptoethanol]. The lysate was centrifuged at 14 000 r.p.m. (JA-17 rotor) for 10 min and clear supernatant saved as soluble lysate. Protein concentration of the soluble lysates was determined by Bradford analysis against a BSA standard curve. Polymerase γ (pol γ) activity was assayed in the lysates by a standard reverse transcriptase assay and in immunoprecipitates using pol γ mono-specific antibodies (40). This is a filter based assay in which 1 U is defined as the amount of polymerase to incorporate 1 pmol of dNTP into acid insoluble DNA per hour at 37°C. For determining activity in immunoprecipitates, 230 μg of each soluble lysate in a final volume of 1 ml was mixed end over end for 1 h at 4°C with 25 μl of CD7 antisera immobilized on Protein G–Sepharose (41). After 1 h, the antibody beads were washed three times with 1 ml each by centrifugation and resuspended. 5 and 15 μl of beads were assayed directly for pol γ activity. Pol γ activity was derived from three independent determinations.
Data analysis
Statistical analysis was performed using one-way ANOVA with post hoc comparison by Tukey–Kramer multiple comparisons test. Intergroup differences with a P-value of <0.05 were considered significant.
RESULTS
Oxidative challenge induces apoptosis in cerebellar granule cells
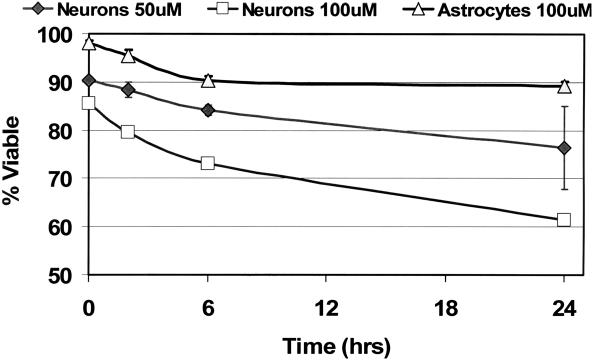
Chronic oxidative stress in neuronal cells has been linked to age-associated neurodegeneration; therefore, experiments were conducted to determine the sensitivity of cerebellar granule cells to the redox cycler, menadione. Granule cell (neuronal) cultures tolerated 1 h treatments with 50 μM menadione relatively well; 84% of the cells were viable after 6 h (Figure 2). However, trypan blue dye exclusion assays revealed that cell viability in these cultures drops to ∼70% after 6 h following treatment with 100 μM menadione. Throughout this investigation, primary astrocytes were used as a standard for a CNS cell type that is resistant to oxidative challenge compared with other glial and neuronal cell types. In contrast to the granule cell cultures, >90% of cells in primary astrocyte cultures were viable at 6 h post-treatment with 100 μM menadione (20).
Figure 2.
Cerebellar granule cells exhibit increased sensitivity to oxidative challenge. Cell cultures were treated as indicated with either 50 or 100 μM menadione for 1 h and viability was assessed after 0, 2, 6 and 24 h. Untreated cultures exhibited viable percents >95. Percent viable = 100 × the number of cells that exclude trypan blue dye/total cell number.
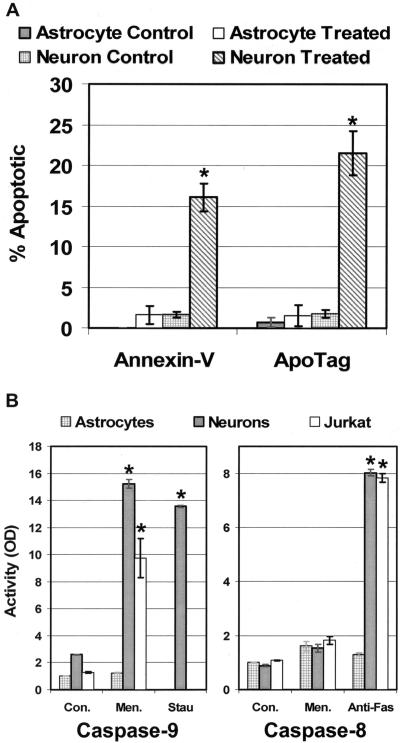
To determine whether the relative increase in sensitivity to oxidative stress observed in cerebellar granule cells is due to programmed cell death, cells were treated with 100 μM menadione and several markers of apoptosis were monitored. TUNEL and Annexin-V assays confirmed the presence of apoptotic granule cells in these cultures (Figure 3A). These studies showed that neuronal apoptosis increased significantly with ∼20% of the cerebellar granule cells undergoing apoptosis 6 h following treatment with menadione. No increase in the percentage of apoptotic cells was detected in treated astrocyte cultures.
Figure 3.
Menadione induced apoptosis was detected in cerebellar granule cells by Annexin-V and TUNEL (ApoTag) assays. (A) Experimental cultures were exposed to 100 μM menadione for 1 h, and assays were performed at 6 h following treatment. The number of positive-staining cells was divided by the total number of cells in the field to yield a percentage of apoptotic cells. (B) Cell lysates taken 3 h following a 100 μM menadione treatment were utilized in colorimetric caspase-9 activity assays. Anti-Fas antibody was used as a positive control for caspase-8 activation. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05) when compared with matched controls.
Menadione redox cycles with complex I of the mitochondrial electron transport chain to produce ROS that damage targets situated primarily in the mitochondrial matrix, such as mtDNA (42,43). Therefore, it seemed likely that menadione would induce apoptosis through a mitochondrial mechanism. Mitochondrial apoptotic pathways are associated with the activation of caspase-9. Additionally, the release of cytochrome c from the mitochondrial intermembrane space into the cytosol is required for caspase-9 activation. Western blot analysis showed the release of cytochrome c into the cytosol of granule cells following treatment with 100 μM menadione (data not shown). Caspase-9 activity was measured in these cells to confirm the release of cytochrome c as well as to determine whether oxidative stress in granule cells induces a mitochondrial pathway of apoptosis (Figure 3B). The broad-spectrum protein kinase inhibitor, staurosporine, was utilized as a positive control as well as Jurkat cells, which are known to undergo a caspase-9 mediated apoptosis when oxidatively challenged. Again, all activities were standardized to caspase-9 activity in primary astrocytes. Three hours following treatment with menadione or staurosporine, caspase-9 activity was significantly enhanced in cerebellar granule cells with no increase in activity detected in astrocytes.
The extracellular signal-mediated Fas-pathway of apoptosis involves activation of caspase-8 (44,45). Some evidence suggests this caspase-8 mechanism may also activate mitochondrial apoptotic machinery (46). Treatment with the anti-Fas antibody induced caspase-8 activity in granule cells; however, treatment with menadione did not cause a similar elevation in caspase-8 activity (Figure 3B). These results indicate that cerebellar granule cells are likely undergoing a caspase-8 independent mitochondrial initiated apoptosis in response to oxidative challenge.
Granule cells exhibit an increased intracellular oxidative state as well as increased levels of antioxidants
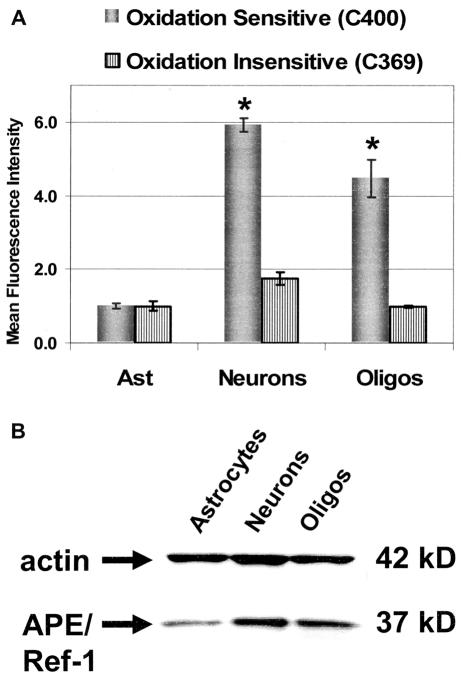
Chronic increased oxidative stress in mitochondria may be an important factor contributing to the increased sensitivity of neuronal cells to oxidative challenge. Therefore, steady-state levels of intracellular prooxidants were determined for various CNS cell types using an oxidation-sensitive probe (Figure 4A). In addition to primary cerebellar granule cell and astrocyte cultures, our laboratory also routinely cultures primary oligodendrocytes. Therefore, primary oligodendrocytes, which are known to be sensitive to oxidative stress compared with astrocytes (20), were included in these experiments. Oxidation of the intracellular oxidation-sensitive probe results in increased mean fluorescent intensity (MFI). These studies revealed that granule cells and oligodendrocytes have significantly elevated steady-state levels of intracellular prooxidants with cerebellar granule cells exhibiting an MFI 6-fold greater than that of astrocytes. Since the measured MFI among these cell cultures stained with an oxidation- insensitive analog was more similar, it seems likely that the significant increase in C-400 oxidation is indicative of an increased oxidative state in both cerebellar granule cells and oligodendrocytes compared with astrocytes.
Figure 4.
Increased intracellular steady-state levels of prooxidants were detected in granule cells and oligodendrocytes. (A) The relative steady-state levels of intracellular prooxidants in primary CNS cell types were measured using the C-400 oxidation-sensitive fluorescent dye. Mean fluorescence intensity of 20 000 cells analyzed by flow cytometry is shown normalized to astrocytes. An oxidation-insensitive analog of C-400 (C-369) was used to control for any differences in dye uptake, ester cleavage and efflux among these cell types. (B) Whole-cell protein extracts from primary CNS cell types were analyzed by western blot analysis using anti-actin and monoclonal APE/Ref-1 antibodies. Analysis using densitometry revealed APE/Ref-1/actin ratios of 0.3, 0.8 and 0.7 for astrocytes, cerebellar granule cells and oligodendrocytes, respectively. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with astrocytes.
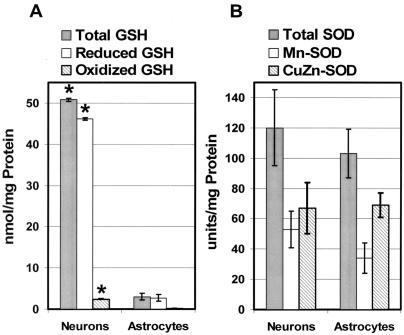
ROS-mediated redox responsive signal cascades in mammalian cells have been shown to contribute to redox homeostasis by increasing expression of antioxidants (47); thus, an increased steady-state level of intracellular prooxidants is usually accompanied by compensatory increases in levels of various free radical scavengers or reductants. To determine whether the increased sensitivity of cerebellar granule cells to oxidative stress is due to antioxidant or reductant deficiencies, redox effector factor-1 (APE/Ref-1) and glutathione (GSH) levels as well as SOD activities were determined for different CNS cell types. APE/Ref-1 is an important reductant for cell signaling (48) that appears to be expressed at higher levels in granule cells and oligodendrocytes compared with astrocytes (Figure 4B). The oxidized GSH (GSSG) to reduced GSH ratio is generally accepted as an indicator of cellular redox state. Though the intracellular GSSG/GSH ratios in granule cells and astrocytes are similar, total GSH levels in cerebellar granule cells are significantly higher indicating an increased metabolic demand for this cellular antioxidant (Figure 5A) (47). Mn-SOD is an antioxidant enzyme that catalyzes the dismutation of superoxide () to hydrogen peroxide and oxygen in mitochondria. SOD activities were measured in astrocyte and granule cell lysates and found to be similar (Figure 5B). Therefore, the sensitivity of cerebellar granule cells to oxidative insult does not appear to stem from decreased levels of antioxidants; in fact, these neuronal cells exhibit increased levels of two important reductants involved in cell signaling and hydroperoxide metabolism (APE/Ref-1 and GSH).
Figure 5.
GSH levels were significantly increased in granule cells compared with astrocytes. Control cultures of astrocytes and cerebellar granule cells (Neurons) were harvested and homogenates were prepared and analyzed as described in Materials and Methods. (A) Both oxidized GSH as well as reduced GSH levels were significantly higher in granule cells compared with astrocytes. (B) SOD activities did not vary significantly between astrocytes and granule cells. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with astrocytes.
Increased susceptibility to mtDNA damage as well as decreased mtDNA repair capacity correlates with sensitivity to oxidative stress in cerebellar granule cell cultures
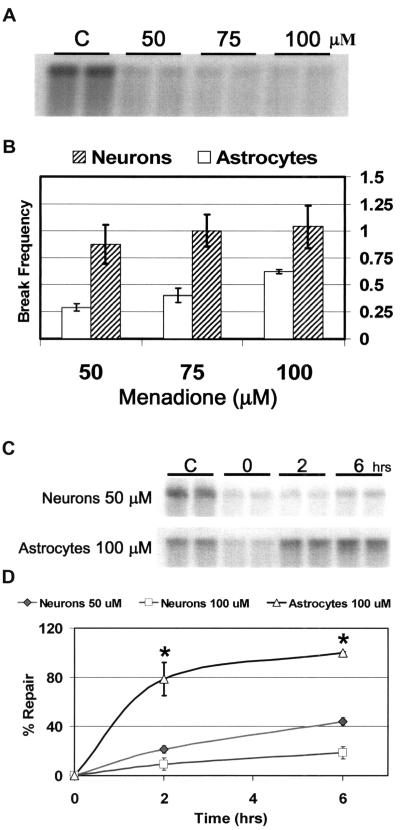
Menadione as well as other sources of oxidative stress can damage the mitochondrial genome and lead to errors in transcription, mutations, recombinations and rearrangements. The capacity of a cell to resist these transformations depends largely upon its capacity to repair these oxidative mtDNA lesions. Initial studies quantified mtDNA damage following exposure of granule cells to increasing concentrations of menadione (Figure 6A). Cells were treated with 50, 75 and 100 μM menadione for 1 h, and the damage was visualized using quantitative Southern blot analysis (QSB). QSB is a DNA repair assay that can detect the production and persistence of damaged sugars as well as BER pathway intermediates, which include AP sites and single-strand breaks (49). Because alkali treatment causes strand breaks at abasic sites or modified sugars, fewer full-length restriction fragments are present to bind the mitochondrial probe. Therefore, the decreased intensity of the 10.8 kb band indicates increased levels of mtDNA damage. Densitometric analysis of these data revealed that treatment with 50 μM menadione generated 0.9 breaks per restriction fragment in cerebellar granule cells (Figure 6B), which is sufficient damage to conduct repair studies using QSB. All of the three concentrations of menadione used in these experiments resulted in higher levels of mtDNA damage in granule cells compared with astrocytes. No nuclear DNA damage was detected at these concentrations using either QSB or quantitative extended length PCR (data not shown).
Figure 6.
Cerebellar granule cells exhibit increased susceptibility to mtDNA damage. (A) High molecular weight DNA from primary cultures of cerebellar granule cells (Neurons) was isolated and restricted to completion with BamH1 immediately following treatment with varying concentrations of menadione (50, 75 and 100 μM). Control cultures (C) were incubated in drug diluent only (HBSS). Duplicate samples were exposed to 0.1 N NaOH before QSB analysis and hybridized with a PCR-generated mitochondrial probe. (B) Break frequency (BF) for menadione treated cerebellar granule cells and astrocytes was calculated densitometrically using a Poisson expression: BF = −ln(OD treated samples/OD control samples). (C) High molecular weight DNA was isolated from cultures immediately (0) or at 2 and 6 h following menadione treatment. Cerebellar granule cells (Neurons) were treated with 50 μM menadione and astrocytes with 100 μM menadione to generate comparable mtDNA damage. (D) Autoradiographic bands from QSB were densitometrically analyzed to calculate BF, and repair was expressed as percent over time. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with matched neuron samples.
The mtDNA repair capacity of cerebellar granule cells was determined by either harvesting cells immediately after a 1 h menadione treatment or allowing time for repair. Compared with control samples (C), 0 h samples show substantially fewer 10.8 kb fragments as indicated by a decrease in the intensity of the corresponding restriction band (Figure 6C). Only 46.5% of this initial mtDNA damage was repaired by 6 h after treatment with 50 μM menadione (Figure 6D). Previously, we have shown that repair proficient cells such as RINr38 cells and primary astrocytes remove all menadione generated oxidative lesions by 6 h (20,37). Astrocytes treated with 100 μM menadione exhibited significantly higher repair capacity compared with cerebellar granule cells treated with either 50 or 100 μM menadione. This mtDNA repair deficiency in cerebellar granule cells was also confirmed at the single nucleotide level using ligation-mediated PCR (LMPCR) (data not shown). The band intensity from astrocyte DNA samples taken at 6 h following menadione treatment appears to be greater than that of control samples (Figure 6C). This apparent overshoot of DNA repair has been reported previously in oxidation repair studies of primary rat astrocytes as well as a rat insulinoma cell line (20,37) and attributed to a treatment induced mtDNA repair adaptation that repairs not only 100% of the experimentally induced mtDNA damage, but also preexisting endogenous damage.
An imbalance in the mitochondrial BER pathway may contribute to decreased mtDNA repair efficiency in neuronal cells
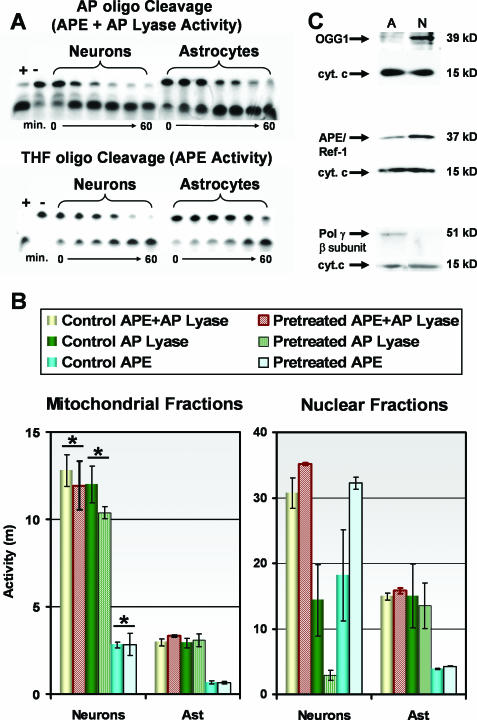
A decreased capacity to repair oxidatively damaged mtDNA appears to correlate with high sensitivity in cerebellar granule cells to oxidative stress. Mitochondrial BER of oxidative damage is initiated by the bifunctional glycosylase/AP lyases (AP lyase), such as 8-oxoguanine DNA-glycosylase (OGG1) (Figure 1). AP lyases excise oxidatively damaged bases prior to incising the sugar backbone 3′ to the lesion. In addition to oxidized bases, oxidative DNA damage also may include damage to the deoxyribose sugar backbone, which can result in the direct release of bases forming AP sites that are substrates for AP endonuclease (APE/Ref-1) (50,51). Because of this free radical induced AP site formation, APE/Ref-1 as well as the AP lyases are considered here to be BER pathway initiators for oxidative DNA damage. To determine the role of these initiators in neuronal mtDNA repair, the relative AP lyase and APE activities were assessed for a proficient BER pathway (astrocyte mitochondrial extracts) as well as a deficient BER pathway (cerebellar granule cell mitochondrial extracts). The rate at which mitochondrial lysates cleaved an APE-specific [tetrahydrofuran AP site containing (THF) oligonucleotide] substrate determined APE activity. Because pure AP sites in DNA can be processed by either APE/Ref-1 or AP lyases, mitochondrial AP lyase activity was differentiated from APE activity by subtracting the THF oligo rate of cleavage from the AP oligo rate of cleavage (Figure 7A). Interestingly, the mitochondrial activity of both pathway initiators was significantly higher in cerebellar granule cell (neuronal) cultures compared with astrocytes (Figure 7B). To determine whether AP lyase or APE activities in astrocytes are inducible under oxidative stress, cells were pretreated with 100 μM menadione for 1 h, washed with fresh media, and then harvested 2 h later for nuclear and mitochondrial protein fractionation. Mitochondrial APE and AP lyase activities were not induced under these conditions in either cell type (Figure 7B). However, APE activity was elevated in neuronal nuclear extracts taken from pretreated cerebellar granule cell cultures. Western blot analysis confirmed that levels of OGG1 and APE/Ref-1 are higher in cerebellar granule cell mitochondrial extracts compared with astrocytes (Figure 7C).
Figure 7.
Mitochondrial and nuclear APE and AP lyase activities were determined for astrocytes and cerebellar granule cells (Neurons) using oligonucleotide cleavage assays. (A) APE activity was determined by incubating mitochondrial protein extracts with a labeled oligonucleotide containing a tetrahydrofuran abasic site (THF oligo). The reaction contents (21mer substrate and 9mer product) were resolved on a 20% TBE-urea gel. A substrate oligonucleotide containing a pure abasic site (AP oligo), which is cleaved by both APE and AP lyase activity, was used in parallel with the THF oligo. Therefore, total AP lyase activity was determined by subtracting the THF oligo cleavage from the AP oligo cleavage for each time point. Purified human APE (1 U) was used as a positive control (+). (B) Enzyme activity was calculated by densitometry as a measure of percent substrate cleaved over time and represented as a measure of slope (m). Treated samples were extracted from cell cultures 2 h following exposure to 100 μM menadione. Both AP lyase and APE activities were significantly higher in granule cell cultures compared with astrocytes. Treatment of cultures with menadione had no effect on mitochondrial activities in either cell type; however, APE activity was induced by menadione treatment in neuron nuclear extracts. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05) compared with matched astrocyte mitochondrial lysates. (C) Western blot analysis was performed to determine the relative levels of key mitochondrial BER enzymes in astrocytes (A) and cerebellar granule cells (N). Anti-cytochrome c antibody was used to confirm equal loading of mitochondrial extracts.
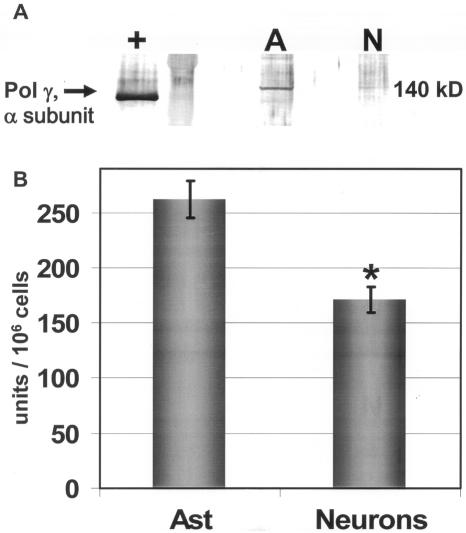
If OGG1 or APE/Ref-1 were rate-limiting in neuronal mtDNA repair, then the mitochondrial activities of these initiators would be contradictory with respect to the decreased mtDNA repair capacity observed in cerebellar granule cells. Therefore, it seemed reasonable that the rate-limiting factor in neuronal BER is downstream, thus pol γ levels were assessed. Western blot analysis revealed that granule cells exhibit substantially lower levels of pol γ (Figures 7C and 8A), and activity assays with both lysates and immunoprecipitates using pol γ-specific antibodies confirmed decreased pol γ activity in cerebellar granule cells compared with astrocytes (Figure 8B) suggesting that decreased levels of pol γ may contribute to deficient mtDNA repair in neuronal cells.
Figure 8.
Pol γ activity was assayed in cerebellar granule cell (Neuron) and astrocyte lysates by a reverse transcriptase assay. (A) Western blot analysis with pol γ antibodies revealed increased levels of the catalytic α subunit of pol γ in astrocyte whole-cell lysates (A) compared with granule cell (N) lysates. Recombinant human pol γ (+) (100 ng) was used as a positive control. Recombinant pol γ runs faster in the gel because of the histidine tag and the absence of a mitochondrial targeting sequence. (B) Pol γ activity in astrocytes was 1.6-fold higher compared with granule cells as determined in anti-pol γ-specific antibody immunoprecipitates as well as lysates. One unit is defined as the amount of polymerase required to incorporate 1 pmol of dNTP into acid insoluble DNA/h at 37°C. Activity is presented as U/106 cells. The average results ± SE from three or more separate experiments are shown. An asterisk (*) indicates a significant difference (P < 0.05).
DISCUSSION
The normal physiological function of the brain requires that neuronal tissues carry a heavy oxidative load. The increased rates of transcription and translation required for the specific duties of the CNS translate into high metabolic rates, and thus increased ROS production. Generation of ROS at complex I in the mitochondrial respiratory chain has been linked to age-associated changes in the CNS mediated through mtDNA damage and the accumulation of mtDNA mutations (3,42,43). In order to maintain ‘redox homeostasis’, neuronal cells must balance the effects of increased oxidative stress with various antioxidants or reductants such as SOD and GSH as well as maintain the lowest advantageous rate of mutation in the mitochondrial genome by efficiently repairing oxidative mtDNA damage.
Primary cerebellar granule cells were used as a neuronal model in these studies to assess whether mtDNA repair deficiencies correlate with oxidative stress-induced apoptosis in neuronal cells. Neuronal characteristics were observed in these primary cultures, which exhibited a very high oxidative state relative to astrocytes (Figure 4A) along with compensatory increases in reductants, such as APE/Ref-1 and GSH (Figures 4B and 5A). Also apparent in the cerebellar granule cultures was a relative increase in susceptibility to menadione-mediated oxidative stress. The same dose of menadione produced twice as many lesions in granule cell mtDNA compared with astrocytes (Figure 6B), and granule cells repaired the damage at a significantly reduced rate (Figure 6D). The increased production and persistence of oxidative mtDNA damage correlates well with mitochondrial initiated apoptosis in these primary cultures (Figure 3) and provides evidence linking neuronal mtDNA repair capacity to age-related neurodegeneration.
The mitochondrial genome in humans consists of a 16.6 kb molecule that encodes two rRNAs, 22 tRNAs and 13 peptides. These peptides are important constituents of the electron transport chain, and errors in mtDNA leading to erroneous translation of these subunits result in decreased ATP production, increased production of damaging ROS, and may ultimately lead to the initiation of apoptosis (52–54). The BER pathway is the principal mechanism by which mammalian cells repair mtDNA oxidation damage (21–23) (Figure 1). Interestingly, the intermediate single strand break products generated in this pathway are more cytotoxic than the oxidative lesions themselves. Under optimal conditions where the pathway initiators are rate-limiting, pathway intermediates exist for very short periods of time, always in the process of conversion, and never left unattended. Once a damaged base enters into the pathway, repair proceeds rapidly to completion through a series of hand-offs from one BER enzyme to the next, with subsequent enzyme activities in relative abundance. The concept of cytotoxic intermediates becomes important when there is a pathway imbalance. Previous results from several laboratories have revealed that experimentally increasing the pathway initiator to polymerase ratio in the nucleus leads to decreased nuclear BER efficiency and increased sensitivity to DNA damaging agents (55).
This study of primary neuronal cells has uncovered a natural imbalance in mitochondrial BER that correlates with decreased mtDNA repair efficiency as well as increased sensitivity to oxidative stress. Relative to the repair proficient pathway of astrocytes, cerebellar granule cells exhibited decreased pol γ activity (Figure 8B) along with increased mitochondrial BER initiator activities: APE and the AP lyases (Figure 7B). Pol γ, which is composed of two subunits, a 125–140 kDa catalytic α-subunit and a 35–55 kDa β-subunit, accounts for ∼1% of cellular polymerase activity and is the only DNA polymerase located in the mitochondria (56). With respect to the high oxidative state of cerebellar granule cells, it is interesting that pol γ is detected as a major oxidized protein in the mitochondrial matrix and that its level of oxidation inversely correlates with activity (57). The findings from this study suggest that pol γ is rate-limiting in the neuronal mitochondrial BER pathway. Increased initiator activities do not cause an overall increase in the rate of mtDNA repair in cerebellar granule cells, but do change the nature of the damage from a less cytotoxic oxidatively damaged base, waiting to enter into the pathway, to a more cytotoxic strand break waiting for pol γ and then DNA ligase to carry the process through to completion. Therefore, it is likely the persistence and buildup of cytotoxic BER pathway intermediates in mtDNA that induce apoptosis in these neuronal cells.
8-Oxo-7,8-dihydroguanine (8oxoG) is one of the more common DNA lesions generated by oxidative attack. 8-Oxoguanine DNA-glycosylase (OGG1) is a bifunctional DNA-glycosylase possessing an AP lyase activity, which specifically removes 8oxoG opposite C (58). In vitro reconstitutions of 8oxoG repair suggest that OGG1 activity may be product inhibited and thus APE/Ref-1 may preclude the AP lyase activity of OGG1 (59). Indicative of this may be the decrease in AP lyase activity opposite the induction of APE/Ref-1 observed in treated neuronal nuclear extracts (Figure 7B). Based on these findings, it is more likely that the accumulating cytotoxic BER pathway intermediate is the 3′-OH/5′-deoxyribose phosphate product of APE/Ref-1 (Figure 1).
Apurinic/apyrimidinic endonuclease/redox effector factor-1 (APE/Ref-1) is a multifunctional enzyme that influences a variety of cellular processes (48). In addition to its DNA repair function, which includes 5′-AP site incision as well as 3′-diesterase activity, APE/Ref-1 also functions as a major cellular reduction/oxidation (redox) factor that maintains many important transcription factors, such as AP-1, NF-κB and p53 in a reduced (active) state. This study reveals that a high cellular oxidative state in primary CNS cell types correlates with increased expression of APE/Ref-1 (Figure 4). Recent attempts to link increased levels of APE/Ref-1 with protection in neuronal cells have fallen short of showing definitely which of the APE/Ref-1 roles is most at play in the management of oxidative stress (26,60). Compared with astrocytes, primary cerebellar granule cells provide a neuronal model that exhibits increased sensitivity to oxidative challenge, decreased mtDNA repair and yet, increased mitochondrial and nuclear APE activity. Therefore, it is very unlikely that APE is rate-limiting in granule cell BER and just as unlikely that increases in APE activity will increase DNA repair capacity, particularly in the mitochondria. Instead, these studies suggest that increased APE/Ref-1 levels, as a result of redox signaling, are necessary in a highly oxidizing intracellular environment to maintain important transcription factors in a reduced, active state and are thus compensatory for maintaining redox homeostasis in neuronal cell types. Therefore, consequential to the neuronal cell's requirement for increased Ref-1 activity, increased APE activity may cause a mitochondrial BER imbalance.
In conclusion, the results from this study suggest a possible link between oxidative stress-related neurodegeneration and decreased mtDNA repair capacity in neuronal cells. Although decreased antioxidant levels were not correlated with increased sensitivity of CNS cell types to oxidative challenge, a strong correlation between decreased mtDNA repair efficiency and induction of apoptosis in cerebellar granule cells was observed. Increased APE/Ref-1 activity in combination with decreased pol γ activity in mitochondria may play a role in decreased mtDNA repair efficiency in neuronal cells by causing a BER imbalance that results in a buildup and persistence of cytotoxic pathway intermediates. Therefore, cell type-specific BER pathway kinetics are important considerations when designing potential mtDNA repair altering therapies for age-related neurodegeneration. Increasing the expression of pol γ in the CNS may be a useful strategy for minimizing the harmful effects of oxidative stress by increasing mtDNA repair capacity in neuronal cells without altering the mtDNA repair kinetics of repair proficient cell types such as astrocytes.
Acknowledgments
The authors thank Dr Ryuji Higashikubo for assistance with experiments involving flow cytometry. The authors also thank J. E. Sim for technical assistance with the measurements of cellular antioxidants. This research was supported by National Institutes of Health Grants ES03456, ES05865, NS047208, CA100045 and AG19602. The costs of publication of this article were defrayed in part by the payment of page charges. Therefore, this article must be hereby marked ‘advertisement’ in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. Funding to pay the Open Access publication charges for this article was provided by NIH; ES 05865 and NIH; NS047208.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hinerfeld D., Traini M.D., Weinberger R.P., Cochran B., Doctrow S.R., Harry J., Melov S. Endogenous mitochondrial oxidative stress: neurodegeneration, proteomic analysis, specific respiratory chain defects and efficacious antioxidant therapy in superoxide dismutase 2 null mice. J. Neurochem. 2004;88:657–667. doi: 10.1046/j.1471-4159.2003.02195.x. [DOI] [PubMed] [Google Scholar]
- 2.Barja G. Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biol. Rev. Camb. Philos. Soc. 2004;79:235–251. doi: 10.1017/s1464793103006213. [DOI] [PubMed] [Google Scholar]
- 3.Melov S. Modeling mitochondrial function in aging neurons. Trends Neurosci. 2004;27:601–606. doi: 10.1016/j.tins.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Barja G., Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- 5.Wunderlich V., Schutt M., Bottger M., Graffi A. Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea. Biochem. J. 1970;118:99–109. doi: 10.1042/bj1180099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niranjan B.G., Bhat N.K., Avadhani N.G. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- 7.Rushmore T., Snyder R., Kalf G. Covalent binding of benzene and its metabolites to DNA in rabbit bone marrow mitochondria in vitro. Chem. Biol. Interact. 1984;49:133–154. doi: 10.1016/0009-2797(84)90057-7. [DOI] [PubMed] [Google Scholar]
- 8.Levy G.N., Brabec M.J. Binding of carbon tetrachloride metabolites to rat hepatic mitochondrial DNA. Toxicol. Lett. 1984;22:229–234. doi: 10.1016/0378-4274(84)90071-7. [DOI] [PubMed] [Google Scholar]
- 9.Backer J.M., Weinstein I.B. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209:297–299. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 10.Hudson E.K., Hogue B.A., Souza-Pinto N.C., Croteau D.L., Anson R.M., Bohr V.A., Hansford R.G. Age-associated change in mitochondrial DNA damage. Free Radic. Res. 1998;29:573–579. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- 11.Spitz D.R., Azzam E.I., Li J.J., Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: a unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 12.Wallace D.C. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 13.Taylor R. Mitochondrial DNA may hold a key to human degenerative diseases. J. NIH Res. 1992;362:237–248. [Google Scholar]
- 14.Ikebe S., Tanaka M., Ohno K., Sato W., Hattori K., Kondo T., Mizuno Y., Ozawa T. Increase of deleted mitochondrial DNA in the striatum in Parkinson's disease and senescence. Biochem. Biophys. Res. Commun. 1990;170:1044–1048. doi: 10.1016/0006-291x(90)90497-b. [DOI] [PubMed] [Google Scholar]
- 15.Schapira A.H. Inborn and induced defects of mitochondria. Arch. Neurol. 1998;55:1293–1296. doi: 10.1001/archneur.55.10.1293. [DOI] [PubMed] [Google Scholar]
- 16.Benzi G., Moretti A. Age- and peroxidative stress-related modifications of the cerebral enzymatic activities linked to mitochondria and the glutathione system. Free Radic Biol. Med. 1995;19:77–101. doi: 10.1016/0891-5849(94)00244-e. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Campo R., Lopez-Torres M., Cadenas S., Rojas C., Barja G. The rate of free radical production as a determinant of the rate of aging: evidence from the comparative approach. J. Comp. Physiol. B. 1998;168:149–158. doi: 10.1007/s003600050131. [DOI] [PubMed] [Google Scholar]
- 18.Barja G. Free radicals and aging. Trends Neurosci. 2004;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Ledoux S.P., Shen C.C., Grishko V.I., Fields P.A., Gard A.L., Wilson G.L. Glial cell-specific differences in response to alkylation damage. Glia. 1998;24:304–312. [PubMed] [Google Scholar]
- 20.Hollensworth S.B., Shen C., Sim J.E., Spitz D.R., Wilson G.L., LeDoux S.P. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic. Biol. Med. 2000;28:1161–1174. doi: 10.1016/s0891-5849(00)00214-8. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl T., Wood R.D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 22.Seeberg E., Eide L., Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 23.Wilson S.H. Mammalian base excision repair and DNA polymerase beta. Mutat. Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 24.Karahalil B., Hogue B.A., de Souza-Pinto N.C., Bohr V.A. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 25.Wilson T.M., Rivkees S.A., Deutsch W.A., Kelley M.R. Differential expression of the apurinic/apyrimidinic endonuclease (APE/ref-1) multifunctional DNA base excision repair gene during fetal development and in adult rat brain and testis. Mutat. Res. 1996;362:237–248. doi: 10.1016/0921-8777(95)00053-4. [DOI] [PubMed] [Google Scholar]
- 26.Kimura H., Dong X., Yagita K., Okamura H. Brain expression of apurinic/apyrimidinic endonuclease (APE/Ref-1) multifunctional DNA repair enzyme gene in the mouse with special reference to the suprachiasmatic nucleus. Neurosci. Res. 2003;46:443–452. doi: 10.1016/s0168-0102(03)00124-x. [DOI] [PubMed] [Google Scholar]
- 27.Verjat T., Dhenaut A., Radicella J.P., Araneda S. Detection of 8-oxoG DNA glycosylase activity and OGG1 transcripts in the rat CNS. Mutat. Res. 2000;460:127–138. doi: 10.1016/s0921-8777(00)00022-7. [DOI] [PubMed] [Google Scholar]
- 28.Araneda S., Mermet N., Verjat T., Angulo J.F., Radicella J.P. Expression of Kin17 and 8-OxoG DNA glycosylase in cells of rodent and quail central nervous system. Brain Res. Bull. 2001;56:139–146. doi: 10.1016/s0361-9230(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 29.Cohen J., Wilkin P. Neural Cell Culture: A Practical Approach. Oxford: IRL Press; 1995. [Google Scholar]
- 30.Gard A.L., Warrington A.E., Pfeiffer S.E. Direct microculture enzyme-linked immunosorbent assay for studying neural cells: oligodendrocytes. J. Neurosci. Res. 1988;20:46–53. doi: 10.1002/jnr.490200108. [DOI] [PubMed] [Google Scholar]
- 31.Shaw G., Debus E., Weber K. The immunological relatedness of neurofilament proteins of higher vertebrates. Eur. J. Cell Biol. 1984;34:130–136. [PubMed] [Google Scholar]
- 32.Ahmad I.M., Aykin-Burns N., Sim J.E., Walsh S.A., Higashikubo R., Buettner G.R., Venkataraman S., Mackey M.A., Flanagan S.W., Oberley L.W., et al. Mitochondrial O2*− and H2O2 mediate glucose deprivation-induced stress in human cancer cells. J. Biol. Chem. 2005;280:4254–4263. doi: 10.1074/jbc.M411662200. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y.J., Galoforo S.S., Berns C.M., Chen J.C., Davis B.H., Sim J.E., Corry P.M., Spitz D.R. Glucose deprivation-induced cytotoxicity and alterations in mitogen-activated protein kinase activation are mediated by oxidative stress in multidrug-resistant human breast carcinoma cells. J. Biol. Chem. 1998;273:294–299. doi: 10.1074/jbc.273.9.5294. [DOI] [PubMed] [Google Scholar]
- 34.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 35.Spitz D.R., Oberley L.W. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal. Biochem. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- 36.LeDoux S.P., Wilson G.L., Beecham E.J., Stevnsner T., Wassermann K., Bohr V.A. Repair of mitochondrial DNA after various types of DNA damage in chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 37.Driggers W.J., LeDoux S.P., Wilson G.L. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J. Biol. Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- 38.LeDoux S.P., Patton N.J., Nelson J.W., Bohr W.A., Wilson G.L. Preferential DNA repair of alkali-labile sites within the active insulin gene. J. Biol. Chem. 1990;265:14875–14880. [PubMed] [Google Scholar]
- 39.Dobson A.W., Xu Y., Kelley M.R., LeDoux S.P., Wilson G.L. Enhanced mitochondrial DNA repair and cellular survival after oxidative stress by targeting the human 8-oxoguanine glycosylase repair enzyme to mitochondria. J. Biol. Chem. 2000;275:37518–37523. doi: 10.1074/jbc.M000831200. [DOI] [PubMed] [Google Scholar]
- 40.Longley M.J., Ropp P.A., Lim S.E., Copeland W.C. Characterization of the native and recombinant catalytic subunit of human DNA polymerase gamma: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 41.Ropp P.A., Copeland W.C. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 42.Genova M.L., Ventura B., Giuliano G., Bovina C., Formiggini G., Parenti Castelli G., Lenaz G. The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron–sulfur cluster N2. FEBS Lett. 2001;505:364–368. doi: 10.1016/s0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 43.Kudin A.P., Bimpong-Buta N.Y., Vielhaber S., Elger C.E., Kunz W.S. Characterization of superoxide-producing sites in isolated brain mitochondria. J. Biol. Chem. 2004;279:4127–4135. doi: 10.1074/jbc.M310341200. [DOI] [PubMed] [Google Scholar]
- 44.Chinnaiyan A.M., O'Rourke K., Tewari M., Dixit V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 45.Kamada S., Washida M., Hasegawa J., Kusano H., Funahashi Y., Tsujimoto Y. Involvement of caspase-4(-like) protease in Fas-mediated apoptotic pathway. Oncogene. 1997;15:285–290. doi: 10.1038/sj.onc.1201192. [DOI] [PubMed] [Google Scholar]
- 46.Le D.A., Wu Y., Huang Z., Matsushita K., Plesnila N., Augustinack J.C., Hyman B.T., Yuan J., Kuida K., Flavell R.A., et al. Caspase activation and neuroprotection in caspase-3-deficient mice after in vivo cerebral ischemia and in vitro oxygen glucose deprivation. Proc. Natl Acad. Sci. USA. 2002;99:15188–15193. doi: 10.1073/pnas.232473399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 48.Evans A.R., Limp-Foster M., Kelley M.R. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 49.LeDoux S.P., Wilson G.L., Bohr V.A. Mitochondrial DNA Repair and Cell Injury. San Diego, CA: Academic Press; 1993. [Google Scholar]
- 50.Nakamura J., Swenberg J.A. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]
- 51.Haring M., Rudiger H., Demple B., Boiteux S., Epe B. Recognition of oxidized abasic sites by repair endonucleases. Nucleic Acids Res. 1994;22:2010–2015. doi: 10.1093/nar/22.11.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papa S., Skulachev V.P. Reactive oxygen species, mitochondria, apoptosis and aging. Mol. Cell. Biochem. 1997;174:305–319. [PubMed] [Google Scholar]
- 53.Singh K.K., Russell J., Sigala B., Zhang Y., Williams J., Keshav K.F. Mitochondrial DNA determines the cellular response to cancer therapeutic agents. Oncogene. 1999;18:6641–6646. doi: 10.1038/sj.onc.1203056. [DOI] [PubMed] [Google Scholar]
- 54.Green D., Kroemer G. The central executioners of apoptosis: caspases or mitochondria? Trends Cell Biol. 1998;8:267–271. doi: 10.1016/s0962-8924(98)01273-2. [DOI] [PubMed] [Google Scholar]
- 55.Sobol R.W., Kartalou M., Almeida K.H., Joyce D.F., Engelward B.P., Horton J.K., Prasad R., Samson L.D., Wilson S.H. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J. Biol. Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 56.Copeland W.C., Longley M.J. DNA polymerase gamma in mitochondrial DNA replication and repair. Scientific World J. 2003;3:34–44. doi: 10.1100/tsw.2003.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graziewicz M.A., Day B.J., Copeland W.C. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res. 2002;30:2817–2824. doi: 10.1093/nar/gkf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fortini P., Pascucci B., Parlanti E., D'Errico M., Simonelli V., Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat. Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Vidal A.E., Hickson I.D., Boiteux S., Radicella J.P. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasko M.R., Guo C., Kelley M.R. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 2005;4:367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]