Abstract
Gene duplication is a crucial mechanism of evolutionary innovation. A substantial fraction of eukaryotic genomes consists of paralogous gene families. We assess the extent of ancestral paralogy, which dates back to the last common ancestor of all eukaryotes, and examine the origins of the ancestral paralogs and their potential roles in the emergence of the eukaryotic cell complexity. A parsimonious reconstruction of ancestral gene repertoires shows that 4137 orthologous gene sets in the last eukaryotic common ancestor (LECA) map back to 2150 orthologous sets in the hypothetical first eukaryotic common ancestor (FECA) [paralogy quotient (PQ) of 1.92]. Analogous reconstructions show significantly lower levels of paralogy in prokaryotes, 1.19 for archaea and 1.25 for bacteria. The only functional class of eukaryotic proteins with a significant excess of paralogous clusters over the mean includes molecular chaperones and proteins with related functions. Almost all genes in this category underwent multiple duplications during early eukaryotic evolution. In structural terms, the most prominent sets of paralogs are superstructure-forming proteins with repetitive domains, such as WD-40 and TPR. In addition to the true ancestral paralogs which evolved via duplication at the onset of eukaryotic evolution, numerous pseudoparalogs were detected, i.e. homologous genes that apparently were acquired by early eukaryotes via different routes, including horizontal gene transfer (HGT) from diverse bacteria. The results of this study demonstrate a major increase in the level of gene paralogy as a hallmark of the early evolution of eukaryotes.
INTRODUCTION
Gene duplication is one of the central avenues of biological innovation. The evolutionary potential of duplication was presciently recognized by the founders of Evolutionary genetics, Fisher (1), Haldane (2), Muller (3) and Bridges (4), and was put into a coherent framework by Ohno in his tellingly entitled 1970 book ‘Evolution by Gene Duplication’ (5). Ohno posited that, after a duplication, one of the two identical copies of a gene becomes free of selective constraints and prone to accumulating mutations that would have been wiped out by purifying selection before the duplication. Although, the most common fate of this copy will be mutational inactivation, pseudogenization, and eventual elimination, some of the duplicates would be fixed by virtue of a beneficial mutation(s) leading to a new function (neofunctionalization). In the genomic era, analyses of the selection mode during gene evolution after duplication indicated that paralogs are subjected to purifying selection from the moment of duplication (6–10), suggesting that Ohno's neofunctionalization model was likely to be an over-simplification. Accordingly, the more realistic model of subfunctionalization have been proposed whereby each of the paralogs retains and, possibly, enhances a subset of the original, multiple functions of the ancestral gene (7,8). Conceivably, paralogs take both the path of neofunctionalization and, more often, that of subfunctionalization (11) or, according to the latest analyses, the two models may apply to different phases in the evolution of the same paralogous family (12).
Undoubtedly, gene duplication has been a major aspect of genome evolution throughout the entire history of life. Comparative-genomic analysis shows that a considerable number of duplications are (nearly) universal in modern life forms, hence predating the last universal common ancestor (LUCA). Examples include several translation factors, aminoacyl-tRNA synthetases, helicases and other widespread protein families (13–17). On the other end of the evolutionary spectrum, most of the sequenced genomes, particularly those of complex eukaryotes, contain numerous paralogs with highly similar sequences which must have evolved as a result of recent gene duplications (9,10,18,19). Gene amplification is a common response to various stress factors in bacteria and yeast (20,21), and to drug treatment in cancer cells (22,23). It is widely believed that these adaptive responses mimic the general course of evolution whereby lineage-specific expansion of paralogous gene families is one of the major mechanisms of adaptive evolution (24–26).
Given the apparent crucial role of gene duplication in biological innovation, it appears likely that increase in duplication rate is specifically associated with major evolutionary transitions (27). The origin of the eukaryotic cell, with its complexity dramatically surpassing that of prokaryotes, is one of the most dramatic in the series of such transitions, second, perhaps, only to the origin of cellular organization itself (28). Several well-characterized cases suggest that diversification through gene duplication, indeed, was a crucial factor during early evolution of eukaryotes. In particular, certain central components of cellular information-processing systems, e.g. the core RNA polymerase subunits (29), replicative DNA polymerases (30) and the MCM licensing factors of DNA replication (31), which are encoded by a single gene in most prokaryotes, are represented by several paralogs in (apparently) all eukaryotes. The same pattern is seen among certain molecular chaperones, such as the TCP complex subunits (32), and core components of the protein degradation machinery, the proteasome subunits (33,34), and the RNA degradation machinery, the exosome subunits (35).
Homologous genes may appear in a genome not only via gene duplication but also as a result of horizontal gene transfer (HGT) (36). Formally, such genes do not fit the definition of paralogy and, accordingly, have been dubbed to pseudoparalogs (37). However, functional implications of pseudoparalogy may not be too different from those of genuine paralogy, both phenomena providing for functional diversification and increase in the level of organizational complexity during evolution (accordingly, in what follows, we use the term paralogy broadly, to include pseudoparalogs, unless otherwise specified). Perhaps, the early phase of eukaryotic evolution was particularly conducive to pseudoparalogy through gene transfer to the nuclear genome from the proto-mitochondrial endosymbiont, to other, transient endosymbionts and, possibly, via other routes as well (38–40).
Taken together, these observations suggest that a dramatic increase in the level of paralogy and pseudoparalogy (to which we will refer as paralogization for the sake of brevity) through extensive gene duplication and HGT might have been a crucial aspect of the emergence of the eukaryotic cell. With the accumulation of multiple complete genome sequences of unicellular and multicellular eukaryotes, it has become possible to put this hypothesis to test through comprehensive comparative-genomic analysis of genes which form clusters of paralogs in all or most eukaryotic lineages but not in prokaryotes. Here, we describe the results of such an analysis which are compatible with the notion of an extensive early paralogization in eukaryotic evolution and allow us to identify the prevalent functional features of the ancestral eukaryotic paralogs (to which we will also refer to as stem paralogs, to emphasize their origin prior to the divergence of the principal eukaryotic lineages).
MATERIALS AND METHODS
The set of orthologous eukaryotic proteins
The starting data set included the latest release of the database of eukaryotic clusters of orthologous groups of proteins (KOGs) [http://www.ncbi.nlm.nih.gov/COG/new/shokog.cgi (41)]. The complete (or nearly complete) genomes of Plasmodium falciparum, Giardia lamblia, Magnaporthe grisea and Orysa sativa were obtained from ftp://ftp.ncbi.nih.gov/genbank/genomes/Plasmodium_falciparum/; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein&cmd=Search&dopt=DocSum&term=txid184922; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein&cmd=Search&dopt=DocSum&term=txid242507; and http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml, respectively. The sequences of the predicted proteins encoded in these genomes were assigned to KOGs using a combination of two approaches, namely, the COGNITOR method (42) and RPS-BLAST against the KOG-derived profiles in CDD database (43), followed by manual verification of the assignments. The COGNITOR program runs BLASTP searches against a KOG sequence database (composition-based statistics and low complexity filtering were turned on to determine the list of homologous sequences with a E-value threshold of 0.01; scores of unfiltered searches were used to rank the hits) and identifies the KOG with the highest similarity to the query. To ensure robust KOG assignments, all weak predictions, as well as conflicts between COGNITOR and CDD results, were examined manually. Spurious hits (mostly due to compositional bias) were eliminated; ambiguous cases were resolved using additional PSI-BLAST searches.
Reconstruction of the ancestral eukaryotic KOG set
We used simple, phyletic-pattern-based rules to infer a set of KOGs that, most likely, were present in the last eukaryotic common ancestor (LECA). All the KOGs which had at least one representative in two or more of the following four major lineages were considered to be ancestral and were assigned to LECA: plants (Arabidopsis thaliana, O.sativa), animals (Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens), fungi-microsporidia (Saccharomyces cerevisiae, Schizosaccharomyces pombe, M.grisea, Encephalitozoon cuniculi) and apicomplexa-diplomonadida (P.falciparum, G.lamblia). While the former three groups comprise distinct branches of the eukaryotic phylogenetic tree supported by different methods of phylogenetic analysis (44–46), the species in the fourth group belong to separate lineages. However, both Giardia and Plasmodium are parasites which are prone to gene loss and, therefore, if genes were lost differentially in these two lineages, their grouping could compensate for the effect of such gene loss. Similarly, though the animal-fungi clade (Opisthokonta) is well-established (45,46), we took a liberal approach to the reconstruction of LECA by including KOGs shared by animals and fungi, again, in order to compensate for potential multiple gene losses in other lineages. The reconstruction was also repeated with the Opisthokont clade taken into account, with qualitatively identical results, as discussed below.
Identification of clusters of paralogous eukaryotic KOGs
Ancestral eukaryotic KOGs can be divided into two groups: (i) those that have identifiable prokaryotic homologs and (ii) those that are, apparently, eukaryote-specific (Figure 1A). To cluster the former, we employed the data on the KOG-to-COG (eukaryotic to prokaryotic) correspondence (47) which were obtained by using RPS-BLAST search with KOG queries against the COG-derived PSSMs in the CDD database (43): the KOGs that mapped to the same COG were considered paralogous. A case-by-case examination of the CDD search results was performed for the KOGs with below-the-threshold hits (0.01 < E < 1) to CDD profiles in order to identify additional prokaryotic homologs; the pattern of conserved residues and structural and functional data were taken into account whenever available. For the eukaryote-specific KOGs, we first used the results of a RPS-BLAST search of selected representatives of each KOG against the complete CDD database. Those KOGs that hit the same position-specific scoring matrix (PSSM) (E < 0.01) were clustered and considered paralogous. Since CDD database contains many redundant profiles, (e.g. two PSSMs for related variants of methionine aminopeptidase domain, cd01086 and cd01088), application of a formal cross-hit criterion might lead to underclustering (if, e.g. member of one KOG are recognized only by the cd01086 profile and of the other one only by the cd01088 profile). Thus, CDD hits for all KOGs were examined for biologically relevant connections between different profiles. The remaining KOGs, which were not recognized by any PSSM from the CDD database, were subject to single-linkage clustering by sequence similarity. Specifically, for all proteins from these KOGs, an all-against-all BLAST search was run (E-value threshold of 0.001) and a pair of KOGs was linked if at least one-third of the proteins from one KOG had proteins from the other KOG as their best hits. The results of these comparisons were manually checked for spurious hits in compositionally biased sequence segments.
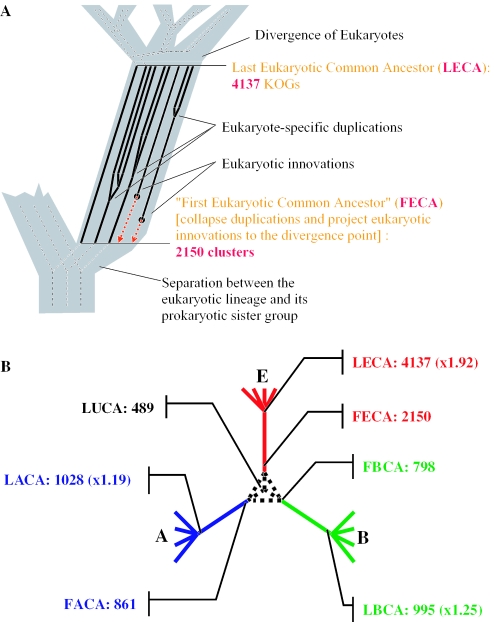
Figure 1.
Last and ‘first’ common ancestors. (A) A scheme of the procedure used to derive the gene sets in the last and ‘first’ common ancestors of eukaryotes. (B) The gene sets of ‘first common ancestors’ of eukaryotes, archaea and bacteria derived from the gene repertoires of the respective last common ancestors and identification of ancestral duplications. Abbreviations: A, archaea; B, bacteria; E, eukaryotes; LECA, last eukaryotic common ancestor; FECA, first eukaryotic common ancestor; LACA, last archaeal common ancestor; FACA, first archaeal common ancestor; LBCA, last bacterial common ancestor; FBCA, first bacterial common ancestor; LUCA, last universal common ancestor.
Inferring origins of ancestral eukaryotic KOGs
We inferred the likely origin of each ancestral eukaryotic KOG by identifying their closest prokaryotic homologs using the data on KOG-to-COG correspondence (see above). The origin of the prokaryotic COGs was, again, inferred on the basis of the pattern of their distribution across bacterial and archaeal phyla (see Supplementary Material for details). KOGs that did not have identifiable prokaryotic homologs and those whose prokaryotic orthologs inferred were not to be of ancient origin, (i.e. probably have been horizontally acquired from eukaryotes) were considered as eukaryote-specific. The KOGs homologous to genes assigned to the LUCA (48) were regarded as inherited from LUCA. The KOGs related to ancient archaeal or bacterial protein families (not assigned to LUCA) were regarded to be of archaeal or bacterial origin, respectively (see Supplementary Material for details). Presumably, genes of archaeal origin were inherited from the common ancestor of Archaea and Eukaryota; those of bacterial origin were acquired by eukaryotes via organellar symbiogenesis and, possibly, other HGT events.
Additionally, for each KOG with orthologs in both prokaryotic kingdoms, it was determined whether the KOG members were likely to have a closer affinity to the bacterial or to the archaeal orthologs. This was done by running BLAST (49) comparisons of eukaryotic proteins against their prokaryotic counterparts, ranking prokaryotic proteins according to their average rank in the BLAST hit lists for different eukaryotic queries and then by using Wilcoxon–Mann–Whitney U-test (P-value threshold of 0.05) to determine if bacterial or archaeal proteins have a tendency to be more closely related to their eukaryotic homologs. Inferences made with this approach were validated by examination of phylogenetic trees as described in the next section.
Evolutionary history of clusters of paralogous eukaryotic KOGs: distinguishing duplication from pseudoparalogs by phylogenetic analysis
Clusters of ancestral eukaryotic KOGs that shared a common closest prokaryotic homolog COG were subjected to further phylogenetic analysis. Alignments of sequences of the KOG and COG members belonging to the same cluster were produced using the MUSCLE (50) program; abnormally short sequences and alignment sites with >33% of gap characters were removed. The large (up to several hundred) number of sequences precluded the use of computationally expensive phylogenetic reconstruction techniques, such as maximum likelihood, for the large-scale analysis. Neighbor-Joining trees were constructed using the PROTDIST and NEIGHBOR programs of PHYLIP package (51). The trees were examined in order to determine whether the respective KOGs were related to each other by duplication or by (supposed) independent acquisitions from a prokaryotic source(s). The latter scenario was accepted if the eukaryotic subtrees formed distinct clades and joined the different prokaryotic clades (e.g. one KOG in a cluster was related to the bacterial and another one to the archaeal clade in the COG tree). The alignments for a number of selected clusters were manually refined (taking into account structural and functional information whenever available), the Neighbor-Joining trees were further optimized by local rearrangements using the MolPhy package and RELL bootstrap values were calculated (52).
Identification of ancestral duplications in bacteria and archaea
For this purpose, we employed the sets of COGs for the LUCA, last archaeal common ancestor (LACA), and the last bacterial common ancestor (LBCA) which were inferred using the described previously weighted parsimony approach (48). Two approaches were combined to estimate the number of duplications along the ‘trunk’ of the bacterial tree (branch leading to LBCA; Figure 1). COGs present in LBCA but absent in LUCA and displaying significant similarity to each other as determined by RPS-BLAST search with the COG-specific PSSMs in the CDD database were projected to a single entity at the base of the common bacterial branch. COGs present in LUCA, for which the median number of paralogs was ≤1 for archaea and >1 for bacteria, were inferred to have experienced a duplication or paralogization via HGT along the branch leading to LBCA. The number of duplications along the ‘trunk’ of the archaeal tree (branch leading to LACA; Figure 1) was inferred in the same manner.
Comparison of cluster distributions
Mapping orthologous sets (C/KOGs) from a last common ancestor of a group to the base of the respective branch yields paralogous clusters which, presumably, arose via duplication(s) of a single ancestral gene or via HGT yielding pseudoparalogs. The paralogy quotient (PQ) is the ratio of the number of orthologous sets to the number of (pseudo)paralogous clusters, which is equal to the average size of the cluster. The distributions of the cluster sizes were statistically compared to detect trends in the extent and pattern of paralogization. To compare two distributions of clusters, the observed frequencies of cluster sizes were binned, with each bin containing at least eight clusters. Typically, bins corresponding to small clusters include a single size class, (e.g. all single-KOG clusters, double-KOG clusters, etc.), whereas bins corresponding to larger families may span many size classes, most of them empty. Binned distributions were compared using the χ2-statistics.
RESULTS AND DISCUSSION
The extent of ancestral paralogy in the three domains of life
We identified 4137 orthologous protein clusters KOGs which, as could be inferred from their phyletic-patterns, were probably inherited by modern eukaryotes from the LECA. Allowing for the possibility of lineage-specific loss of ancestral genes, we used a liberal approach to the reconstruction of the gene set of LECA such that genes shared by any two of the major eukaryotic lineages were assigned to LECA (see Materials and Methods for the details of ancestral gene set reconstruction and the Makarova_Paralogous_KOGs spreadsheet in the Supplementary Material for the complete list of ancestral KOGs). These KOGs form 2150 clusters of paralogs (including those that consist of a single-KOG; see Materials and Methods for details of identification of paralogous clusters). Since each of the 4137 ancestral KOGs was inferred to have been present in LECA, the duplications (or HGT) leading to the emergence of these ancient (pseudo)paralogous clusters, by definition, predate LECA. Accordingly, we assigned the progenitor of each of the 2150 paralogous clusters to a hypothetical ancestral entity which we designated the first eukaryotic common ancestor (FECA) (Figure 1A and the Makarova_Paralogous_KOGs spreadsheet in the Supplementary Material). By definition, FECA did not have any eukaryote-specific paralogs although, it had a considerable number of ancestral paralogs inherited from archaea and bacteria. The ratio of the inferred numbers of genes in LECA and FECA, which we designated the PQ, appears to be a useful parameter for quantitative assessment of the contribution of duplications to the evolution of the given lineage. The above estimates gives PQ = 1.92 for eukaryotes.
To assess potential biases in these estimates, we implemented two modifications to the above procedure. Firstly, we split the LECA KOGs into individual domains identified by CDD search before clustering; secondly, we considered animals and fungi-microsporidia as a single clade when determining the set of LECA KOGs. These modifications resulted in PQ values of ∼1.97 and ∼1.90, respectively, with the distributions of cluster sizes statistically undistinguishable from that obtained with the original procedure (Supplementary Table 1S). Another possible source of error involves ancestral paralogy hidden in over-clustered KOGs. Among the KOGs assigned to LECA, approximately one-third had a potential lumping problem, with the median number of paralogs of two or greater. Case-by-case inspection of these KOGs using similarity-based clustering and phylogenetic tree analysis suggested that the great majority include genuine lineage-specific expansions, rather than ancestral duplications (data not shown). Finally, some of the ancestral paralogs, probably, were missed in the present work due to limitations of sequence-based approaches, even those that include careful analysis of profile search results. Generally, we believe that the catalogue of ancestral eukaryotic paralogs presented here is reasonably complete, even if conservative.
The origin of the eukaryotic cell involved an enormous increase in organizational complexity compared to its prokaryotic progenitors, and it seems plausible that ancestral duplications (and, possibly, HGT leading to pseudoparalogy) substantially contributed to this evolutionary transition. We were interested to determine whether or not extensive paralogization at the base of a major lineage was unique to eukaryotes. Although the reconstruction of the last common ancestors of bacteria and archaea involves more assumptions than the reconstruction of LECA, because of extensive HGT in prokaryotes, we estimated the approximate number of ancestral paralogs for the bacterial and archaeal branches using weighted parsimony [Figure 1B; (48)]. These estimates yielded PQ values of 1.19 for archaea and 1.25 for bacteria, notably lower than the above value for eukaryotes.
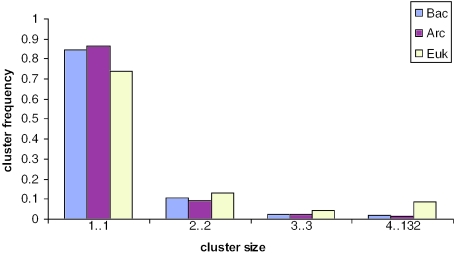
Comparison of the size distributions of the clusters of ancestral paralogs in eukaryotes, archaea and bacteria further illustrates the differences. Although, in the inferred ancestral gene sets for all three kingdoms, a significant majority of the genes did not have paralogs, the tail of the distribution was marked heavier in eukaryotes, with the excess of large clusters of paralogs being particularly notable (Figure 2). The differences between the eukaryotic distribution and those for archaea and bacteria were highly statistically significant (P-value of 4 × 10−15 for the archaea-eukaryote comparison and 3 × 10−11 for the bacteria-eukaryote comparison, according to the χ2-test); the archaeal and bacterial distributions were statistically indistinguishable. These observations show that early evolution of eukaryotes involved exceptionally extensive paralogization compared to the similar stages in the evolution of bacteria and archaea and suggest a major contribution of ancestral paralogs to the emergence of eukaryotic complexity.
Figure 2.
Size distributions of ancestral paralogous clusters in eukaryotes, archaea and bacteria. Relative frequencies of clusters of different size are shown for the three divisions of life.
Ancestral paralogy among LECA genes of different origins
The genes of LECA can be roughly divided into four classes according to their origin: (i) inherited from LUCA, (ii) inherited from archaea (or from the common archaeal-eukaryotic ancestor), (iii) those of bacterial origin (derived, in large part, from the mitochondrial endosymbiont and, possibly, via other routes) and (iv) eukaryotic innovations. We inferred the most likely origin of each ancestral eukaryotic KOG from the correspondence between KOGs and the prokaryotic COGs which were established using RPS-BLAST searches as described previously (47) and additional, case-by-case analyses. The provenance (ancestral, i.e. traced back to LUCA, archaeal or bacterial) of each of the prokaryotic COG with a eukaryotic ortholog(s) was then inferred by phyletic-pattern analysis (see Materials and Methods and Supplementary Material for details). The KOGs without prokaryotic homologs (or with few homologs that were not considered to be ancient prokaryotic genes) were taken to be eukaryotic innovations. We compared the size distributions of paralogous clusters in the four classes to each other and to the general distribution among LECA genes. Perhaps counter-intuitively, the results show a pronounced excess of stem eukaryotic paralogs in the set of KOGs inherited from LUCA and a deficit of duplications among proteins that were considered as eukaryotic innovations; the levels of paralogy among the KOGs of archaeal and bacterial origin were intermediate and statistically indistinguishable from each other and from the overall distribution (Table 1). The lower level of paralogy among eukaryote-specific genes could be trivially explained, at least in part, by relatively late emergence of some of these genes along the branch leading to LECA, because of which these genes simply had less time to duplicate than the genes inherited from prokaryotes. The overabundance of paralogs among LUCA-derived eukaryotic genes is of greater interest. We hypothesize that the eukaryotic protein core, especially, information-processing systems, was largely formed by duplication of the components of already well-coordinated and adapted systems inherited from LUCA and subsequent diversification of the emergent paralogs (see also below).
Table 1.
Ancestral duplications among eukaryotic genes of different inferred origins
| Inferred origin | Number of KOGs | Number of clusters | P(χ2) | Comment |
|---|---|---|---|---|
| Archaeal | 280 | 153 | 0.49 | No significant difference from the general distribution |
| Bacterial | 923 | 415 | 0.67 | No significant difference from the general distribution |
| Archaeal or bacterial | 239 | 117 | 0.02 | No significant difference from the general distribution |
| LUCA | 1003 | 407 | 1.8 × 10−21 | Excess of duplications |
| Eukaryotic | 1692 | 1058 | 6 × 10−12 | Deficit of duplications |
Ancestral duplication and pseudoparalogs
As mentioned above, eukaryotes have acquired a substantial number of genes from the mitochondrial endosymbiont and, possibly, from other endosymbionts because of which some of the apparent paralogous clusters actually represent pseudoparalogy. Among the 420 clusters comprised of the 1804 LECA KOGs of inferred prokaryotic origin, 171 clusters (41%) consist of two or more (up to seven) subclusters with discordant phylogenetic affinities as determined by sequence similarity analysis and phylogenetic tree analysis (see Materials and Methods for details). These subclusters were inferred to be pseudoparalogs whereas the KOGs within each subcluster appeared to be a bona fide paralogs, i.e. related by duplication (see the Makarova_Paralogous_KOGs spreadsheet in the Supplementary Material for the complete list of paralogous and pseudo-paralogous clusters). The phylogenetic trees in Figure 3 (see also Supplementary Material for details) exemplify the detected evolutionary patterns of clusters of paralogs and pseudoparalogs. The five KOGs in Figure 3A (IMP4 domain-containing RNA-binding proteins involved in splicing and ribosomal biogenesis) clearly are of archaeal origin and evolved via serial duplication at the onset of eukaryotic evolution. The set of paralogous KOGs in Figure 3B, which all consist of GTPases with diverse functions, shows a more complex pattern suggestive of a combination of ancient duplications with pseudoparalogy. Specifically, KOG1424, KOG2484 and KOG2423 are of obvious archaeal descent and have evolved via two consecutive duplications in eukaryotes (the placement of one of the proteins from KOG2484 within the archaeal cluster is, probably, a long-branch attraction artifact). In contrast, both KOG1249 and KOG2485 show strong affinities with distinct bacterial branches suggesting that at least two HGT events were involved in the evolution of this cluster of KOGs which map to the same prokaryotic COG. Further complexity is added to the evolutionary scenario of this cluster by the observation that KOG2484 shows unexpected heterogeneity, with general archaeal affinity but with one of the members (At4g02790, labeled ‘KOG2484Ath’ in Figure 3B) clearly grouping within the cyanobacterial branch. This KOG includes two members from Arabidopsis, one of which is of archaeal origin whereas the other one clearly originated by gene transfer from the chloroplast; thus, these genes, although belonging to the same KOG, are typical pseudoparalogs. The cluster in Figure 3C is even more complex, with five KOGs including functionally diverse FAD-binding proteins apparently originating from five different bacterial taxa. In this case, the archaeal members of the family do not form a clade such that the entire history of the family appears to be dominated by HGT from bacteria. Once again, KOG1231 is a ‘mixed bag’, with members from different eukaryotes showing affinity to distinct bacterial lineages.
Figure 3.
Phylogenetic trees of clusters of homologous KOGs illustrating ancestral eukaryotic duplications and pseudoparalogy. (A) A case of multiple ancestral duplications of a gene of archaeal origin. IMP4 domain-containing proteins. (B) A cluster with a mixed history of duplication of pseudoparalogy. Predicted GTPases. (C) A cluster of multiple pseudoparalogs. FAD/FMN-containing dehydrogenases. Eukaryotic branches are shown in red, archaeal branches are shown in blue, and bacterial branches are shown in black. Only the numbers of (pseudo)paralogous KOG, the numbers of the homologous COG (a single one for each tree) and, where relevant, major bacterial taxa are indicated. Trees with all species names indicated are given in the Supplementary Material. The maximum likelihood trees were constructed using ProtML program (52) to perform local rearrangements on the Neighbour-Joining tree as described previously (80). Nodes with RELL bootstrap support >70% are boldfaced.
Of the 171 clusters that showed evidence of pseudoparalogy, 54 (13% of the clusters with prokaryotic homologs) consist of KOGs of apparent archaeal and bacterial origin (Supplementary Table 2S). These clusters represent the dominant theme in pseudoparalogy whereby acquisition of a bacterial gene via HGT, most likely, from an endosymbiont, adds a pseudoparalog to an ancestral eukaryotic gene. Indeed, among these 54 clusters of mixed archaeal and bacterial origin, 39 (72%) include proteins involved in translation, mostly aminoacyl-tRNA synthetases and ribosomal proteins, which are often represented by cytosolic and mitochondrial versions. Some of the other pseudo-paralogous clusters, e.g. those including molecular chaperones, are also related to the translation system albeit less directly (Supplementary Table 2S). Among the rest of the pseudoparalogy cases, it was hard to identify specific patterns, with the phylogenetic affinities of pseudoparalogs scattered among bacterial taxa. This is likely to reflect both obliteration of specific phylogenetic signal and the genuine diversity of the HGT sources.
The structural and functional gamut of ancient eukaryotic paralogs
The availability of the catalogue of ancestral eukaryotic (pseudo)paralogs allows us to examine in detail the structural and functional repertoire of the proteins that were propagated by duplication (and, to some extent, also by HGT) during evolution from FECA to LECA. Supplementary Tables 2 and 3S summarize the principal features of the largest clusters of paralogs of different origins. Remarkably, the majority of these clusters seem to center at two related functional (and, in part, structural) themes: (i) protein–protein interactions and superstructure formation mediated primarily by repetitive protein domains (WD-40, HEAT/ARM, TPR) and (ii) regulation of protein folding, trafficking and degradation (RINGs, DNAJ, SAR1/G GTPases, mitochondrial carrier proteins). The striking abundance of WD-40 repeat proteins among the conserved eukaryotic KOGs that are represented by a single gene in each species has been noticed previously (47). These proteins are subunits of major, eukaryote-specific protein complexes, such as the rRNA processosome (53), and the presence of numerous paralogs in LECA indicates that (nearly) the entire architecture of these complexes, with the unique functions of individual subunits, evolved at a very early stage of eukaryotic evolution via multiple duplications of genes for superstructure-forming proteins (see also below). Similarly, the HEAT/ARM repeat-containing proteins seem to perform unique structural roles in various chromatin-associated complexes and in the nuclear pore; the numerous karyopherins, which are directly responsible for transporting cargo through the nuclear pore, are, mostly, paralogous, HEAT-repeat-containing proteins (54,55).
Notably, almost all large clusters of (pseudo)paralogous KOGs of archaeal descent consist of proteins involved in information-processing systems, such as the chromatin and the replication machinery, the basal protein degradation system, the proteasome, and the RNA degradation machine, the exosome. This reflects the well-known vertical relationships between archaeal and eukaryotic informational systems (28,56–60). While maintaining the functional continuity of these systems with their archaeal progenitors, eukaryotes have evolved extensive complexity of specificities and regulatory interactions—to a large extent, by virtue of massive paralogization. There seem to be no dominant, unifying themes among the top paralogous clusters of LUCA and bacterial origins whereas the eukaryotic innovations are dominated by proteins involved in specific protein–protein interactions and protein fate (Supplementary Tables 2 and 3S).
Ancestral paralogy in different functional classes of eukaryotic genes
We compared the distributions of paralogous cluster sizes among all functional categories of KOGs, which contained >150 members, to the overall distribution (Table 2). For most of the categories, the distributions were statistically indistinguishable from each other and the overall distribution; however, three major deviations were detected. A significant excess of stem paralogs compared to the general background was detected in only one functional category, namely, molecular chaperones and other proteins involved in protein fate determination. Numerous large and small clusters of different origins were detected among these proteins (Table 3). Indeed, it appears plausible with the emerging cell compartmentalization on the outset of eukaryotic evolution triggered selection for diversification and specialization of the molecular machines involved in protein folding, trafficking and degradation. Many notable duplications in this group, such as the proteasome subunits, molecular chaperones of the HSP40, HSP60, HSP70 and HSP90 families, and ubiquitin system components, have been discovered and discussed previously (33,34,61–64).
Table 2.
Ancestral duplications in different functional categories of eukaryotic genesa
| Functional class | P(χ2) details | Number of KOGs in the largest cluster | Largest cluster |
|---|---|---|---|
| Translation | 7 × 10−4 (excess of size two clusters; deficit of larger clusters) | 4 | EF2 |
| Replication and repair | 0.1 (no difference) | 6 | Cdc46/Mcm |
| Transcription | 0.2 (no difference) | 16 | HOX |
| Cytoskeleton | 0.2 (no difference) | 22 | Profilin superfamily |
| Chaperones and related proteins involved in protein fate determination | 2.7 × 10−3 (excess of duplications) | 30 | RINGs |
| Signal transduction | 0.99 (no difference) | 39 | S/T kinases |
| Energy metabolism | 3 × 10−4 (deficit of duplications) | 21 | Mitochondrial carrier protein |
| Secretion | 0.2 (no difference) | 22 | Profilin-like proteins |
| RNA processing and modification | 0.26 (no difference) | 31 | RRM |
aThe rough functional classification of eukaryotic genes was adopted from the KOG database (47).
Table 3.
Ancestral paralogous clusters among genes involved in protein fate determination
| Cluster description | Number of KOGs | Inferred origin |
|---|---|---|
| RINGs in E3 ubiquitin ligases | 28 | Eukaryotic |
| Ubiquitin-specific protease | 18 | Eukaryotic |
| E2 ubiquitin protein ligase | 18 | Eukaryotic |
| DNAJ-like | 17 | LUCA |
| 20S proteasome α/β subunits | 14 | Archaeal |
| AAA+-type ATPase (COG0464) | 11 | Archaeal or LUCA |
| PINT domains | 10 | Eukaryotic |
| HSP60-like | 9 | LUCA |
| Cyclophilin family | 9 | Bacterial |
| Ubiquitin-like proteins | 9 | Eukaryotic |
| E3 ubiquitin protein ligase (HECT domain) | 8 | Eukaryotic |
By contrast, genes coding for proteins involved in energy production and conversion show a significant deficit of ancestral duplications (Table 2). Conceivably, most of these systems were acquired more or less ready-made from the mitochondrial endosymbiont, on many occasions, probably, with displacement of ancestral versions.
Finally, the set of KOGs involved in translation is enriched for clusters of size two, whereas larger clusters are rare in this group. As discussed above, these doublets are, mostly, pseudoparalogs brought about by the mitochondrial endosymbiosis such that many proteins involved in translation exist in two versions, cytosolic and mitochondrial, as discussed above. The lack of larger clusters in this functional category could be due to the selection against imbalance in multisubunit complexes and other tightly coordinated systems (65).
Generally, although ancestral paralogy spans all functional spheres of the eukaryotic cell, the excess of structural subunits of eukaryote-specific complexes and of proteins with broadly defined chaperone-like functions is the most remarkable manifestation of the extensive early paralogization in eukaryotic evolution. These seem to be the types of protein functions which are most directly linked to the increased complexity of the eukaryotic cell, which simultaneously creates niches and demands for versatile mechanisms of protein and RNA processing and topogenesis.
‘Frozen’ clusters of paralogs
The gene duplication process is inherent to genome evolution and never stops, hence numerous lineage-specific duplications, including major lineage-specific expansions of paralogous families (24,26,66); for much of the evolution of life (with the likely exception of multicellular eukaryotes), HGT seems to have been equally pervasive (38–40), leading to the emergence of pseudoparalogs. However, not all (pseudo)paralogous gene clusters belong to such expansions—many can be traced to a unique event in the trunk of a taxon tree, with very few or no subsequent additions. We dubbed these evolutionarily stable paralogous clusters ‘frozen duplications’ (with the understanding that some of these clusters may include pseudoparalogs). It appears likely that further proliferation of these clusters was prevented by purifying selection eliminating additional duplications which become deleterious because they disrupt the balance between the expression levels of interacting proteins (65,67). Several cases of ‘frozen duplications’ in eukaryotes have been detected and discussed previously (28,34,64). We identified ‘frozen duplications’ within the set of ancestral paralogous KOGs as those that had no pronounced lineage-specific expansions (median number of paralogs within each of the paralogous KOGs <2.5 proteins per species). The list of the most prominent ‘frozen duplications’ (Table 4) conspicuously differs from the overall list of top paralogous clusters (Table 5) in that the former does not include serine/threonine kinases and SAR1/G GTPases which are prominent in the overall list. Apparently, the elaboration and diversification of the regulatory pathways in multicellular organisms drove numerous lineage-specific duplications of kinases and G-type GTPases, whereas the functions of many other eukaryotic complexes were already fully evolved and fixed in LECA. Strikingly, the list of the most prominent ‘frozen duplications’ is dominated by repeat-containing, superstructure-forming proteins; many of these proteins have been shown to be essential for survival in yeast S.cerevisiae and/or the nematode C.elegans (47,68,69). Together, these observations emphasize the fundamental importance of these structural proteins for the emergence of the eukaryotic cell complexity and the role of selection for balance in their evolution.
Table 4.
The top 10 ‘frozen’ ancestral paralogous clusters in eukaryotes
| Cluster description | Number of KOGs in cluster | Inferred origin |
|---|---|---|
| WD-40 | 93 | Bacterial |
| HEAT/ARM | 34 | Bacterial |
| TPR | 28 | LUCA |
| RRM (RNA-binding) | 26 | Bacterial |
| RINGs | 21 | Eukaryotic |
| Helicases | 17 | LUCA |
| snRNP-like | 15 | LUCA |
| SNARE-like proteins | 14 | Eukaryotic |
| PINT domains | 14 | Eukaryotic |
| C2H2-type Zn-fingers | 14 | Eukaryotic |
Table 5.
The top 10 ancestral paralogous clusters in eukaryotes
| Cluster description | Number of KOGs in cluster | Inferred origin |
|---|---|---|
| WD-40 | 132 | Bacterial |
| Serine/threonine kinases | 69 | LUCA |
| RRM (RNA-binding) | 60 | Bacterial |
| HEAT/ARM | 48 | Bacterial |
| RINGs | 47 | Eukaryotic |
| TPR | 34 | LUCA |
| GTPases (SAR1/G) | 30 | Archaeal or bacterial |
| Helicases | 26 | LUCA |
| Mitochondrial carrier protein | 24 | Eukaryotic |
| DNAJ-like domains | 20 | LUCA |
Highly diverged and previously undetected ancestral eukaryotic paralogs
Functionally uncharacterized ancestral paralogs are of special interest with regard to the possibility of prediction of yet unknown essential functions. However, the number of unexpected findings of such uncharacterized ancient paralogs in the present study was surprisingly small. In most cases, there is either direct functional information or clear indication of the probable function from the domain composition of the proteins in question, e.g. the confident prediction of the ubiquitin ligase function for the numerous uncharacterized RING-finger-containing proteins. It appears that, although a wealth of details remains to be filled in, the general functional census of the ancestral eukaryotic paralogs is nearly complete (see the full results at ftp://ftp.ncbi.nih.gov/pub/koonin/euk_origin).
However, on many occasions, identification of ancestral paralogs required detection of subtle similarity and ‘cryptic’ domains through the use of sensitive, iterative database searches. Most of the genes in such KOGs remain annotated as ‘hypothetical proteins’ in GenBank and other databases although, for many of them, the domain architecture has been described in specialized publications (Supplementary Table 4S).
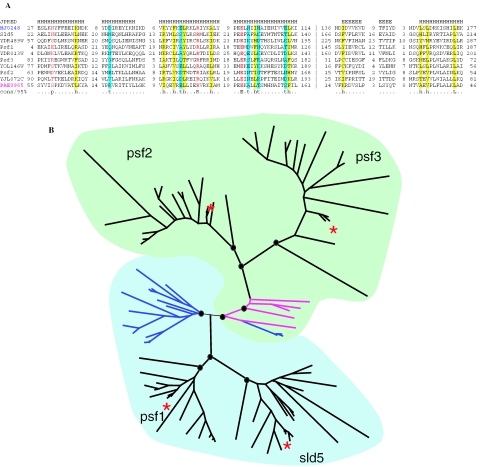
The most unusual ancestral paralogous cluster analyzed here consisted of four eukaryotic KOGs which are distantly related to uncharacterized archaeal proteins from COG1711. Recently, it has been shown that these eukaryotic proteins (Sld5 and Psf1,2,3) form the hetero-tetrameric GINS complex involved in DNA replication initiation (70,71). Notably, the function of these eukaryotic proteins has been accurately predicted on the basis of the conservation of genomic context, i.e. adjacency of the COG1711 gene to the DNA polymerase sliding clamp (PCNA ortholog) gene in several archaeal genomes (72). Here, we analyzed this protein family (hereinafter GINS family) in greater detail. We found that archaea encode two forms of the GINS proteins, one of which appears to have been derived from the other by circular permutation of a small domain (Figure 4A). One of these forms is most typical of Crenarchaeota, whereas the second one is found, largely, in Euryarchaeota (Figure 4B). Eukaryotes also have both forms and, despite the low sequence conservation, specific relationships appear to exist between Psf2/Psf3 and the crenarchaeal homologs, on the one hand, and Psf1/Sld5 and euryarchaeal homologs, on the other hand. This conclusion is supported both by the shared permutation points and by the phylogenetic tree topology (Figure 4B). The heteromeric structure of the eukaryotic GINS complex and the fact that most of the archaeal genome encode a single gene of this family suggest that the eukaryotic complex evolved from a homo-tetramer to the hetero-tetramer via two rounds of duplication and a permutation after the first round. However, the early stages in the evolution of the GINS family remain murky. One possibility is that the common ancestor of archaea and eukaryotes already encoded both permuted forms, which were subsequently differentially lost. Another scenario involves eukaryotes inheriting a single gene from their last common ancestor with archaea, permutation in one of the archaeal lineages, and acquisition of the permuted form by an early eukaryote (thus, pseudoparalogy would enter the history of this family). Subsequently, both forms were duplicated in the eukaryotic lineage, similarly to other eukaryotic genes for proteins that form multimeric complexes, such as the proteasome, the DNA replication licensing MCM complex, the chaperonin TCP complex, and others (31–34) (33,34). This example emphasizes that at least some of the ancestral eukaryotic duplications evolved through complex and not always readily decipherable chains of events which might combine duplication and pseudoparalogy.
Figure 4.
Evolution of the GINS family. (A) Multiple alignment of the selected GINS proteins. Sequences are denoted by gene names: Sld5, Psf1, Psf3, Psf2—experimentally characterized GINS proteins from Xenopus laevis (70); YDR489W, YDR013W, YOL146W, YJL072C—orthologous proteins from S.cerevisiae (71); MJ0248—homolog from the euryarchaeon Methanocaldococcus jannaschii; PAE0965—homolog from the crenarchaeon Pyrobaculum aerophilum. The positions of the first and the last residue of the aligned region in the corresponding protein are indicated for each sequence. The numbers within the alignment represent poorly conserved inserts that are not shown. The vertical dashed line separates the permuted region. The colouring is based on the consensus (calculated for all sequences in the alignment) shown underneath the alignment; h/yellow indicates hydrophobic residues (ACFILMVWYHRK), t/cyan indicates turn-forming residues (ASTDNVGPERK), p/red indicates charged residues (STEDKRNQH), positions with identical amino acids are boldfaced. The secondary structure was predicted using the JPRED program (81). H indicates α-helix, E indicates extended conformation (β-strand). (B) Schematic representation of the phylogenetic tree of the GINS family. The representation is based on a maximum likelihood tree of 97 sequences of GINS family reconstructed using ProtML program. Nodes with bootstrap support >70% are marked by circles. Euryarchaeal branches are shown in blue, and the Crenarchaeal branches are shown in magenta. The two coloured areas denote the two permuted forms of the protein. Branches corresponding to the Sld5, Psf1, Psf3, Psf2 proteins from X.laevis are marked by red asterisks.
GENERAL DISCUSSION AND CONCLUSIONS
The pivotal role of gene duplications in the evolution of eukaryotes had been obvious for a long time, at least since the publication of Ohno's classical book (5). In the genomic era, it became clear that lineage-specific expansion of paralogous gene families is one of the principal paths taken by eukaryotes to adapt to their specific environments and life styles (26,61). Here, we quantitatively and qualitatively characterize a particularly interesting set of eukaryotic paralogs, those that were inferred to predate the last common ancestor of the known eukaryotic lineages but are either represented by a non-duplicated ancestral form or absent in prokaryotes. By definition, these paralogous clusters evolved concomitantly with or shortly after the emergence of the eukaryotic cell, and it seems likely that extensive paralogization made an important contribution to this momentous evolutionary transition. We found that the extent of paralogy traced to the onset of eukaryotic evolution is substantially (and highly statistically significantly) greater than that at the comparable stages of evolution of bacteria and archaea, supporting the notion of a burst of paralogization, primarily via gene duplication, but also via HGT, as a hallmark of early eukaryotic evolution. Conceivably, this increase in the fixation rate of (pseudo)paralogous genes was precipitated by a cataclysmic event leading to a sharp drop in the population size of the proto-eukaryote and the ensuing weakening of purifying selection, which in turn led to an increase in the survival time of duplications and genes acquired via HGT and an increased probability of their fixation in evolution (73,74). An interesting candidate for such a catastrophe could be the acquisition of the proto-mitochondrial endosymbiont, which might have had the effect of starting off eukaryotic evolution from a miniscule chimeric population.
Comparative-genomic analysis of plants, fungi and animals strongly suggests that, on many independent occasions during evolution, whole-genome duplication (polyploidization) took place, with subsequent differential loss of paralogs in lineages descendant from the one with the genomic duplication (75–79). We cannot rule out that whole-genome duplication occurred also at the onset of eukaryotic evolution although, given the amount of evolutionary change that transpired since these events, it is hard, if not impossible, to distinguish between this scenario and a burst of paralogization resulting from a greatly increased probability of fixation of the individual gene duplications and genes acquired via HGT.
Structural, functional and evolutionary survey of the ancestral eukaryotic paralogs revealed four notable trends: (i) while gene duplication is, undoubtedly, the main path to paralogization, apparent HGT from bacteria yielding pseudoparalogy also played an important role, contributing to nearly half of the clusters with prokaryotic homologs, (ii) the most ancient genes, apparently inherited from LUCA, made greater contribution to the set of stem eukaryotic paralogs than genes of more recent origin, (iii) the set of stem paralogs, particularly, the ‘frozen’ ones (those that have undergone minimal or no lineage-specific expansion), is dominated by proteins involved in superstructure formation and containing repetitive domains, such as WD-40, HEAT/ARM, and TPR, and (iv) the only functional category of eukaryotic genes that is substantially enriched in stem duplications are the molecular chaperones and other proteins involved in protein fate determination, including post-translational modification, targeting, trafficking and regulated degradation.
The quantitative preponderance of the LUCA heritage, rather than eukaryote-specific genes, among the stem paralogs came as a surprise although, anecdotally, it had been well-appreciated previously that certain ubiquitous genes, e.g. RNA polymerase subunits, have multiple paralogs in all eukaryotes. Apparently, diversification of the ancestral gene set was one of the principal sources of early eukaryotic innovation. Equally, if not more unexpected seems to be the prevalence of repeat-containing proteins among the stem paralogs [in part, an observation that has come to light during the previous analysis of highly conserved orthologous genes in eukaryotes (47)]. These proteins are usually considered to be ‘mere’ building blocks in multisubunit complexes, e.g. HEAT/RM repeats in chromatin-associated complexes, and WD-40 in the rRNA processosome. However, the remarkable early diversification of these proteins, as well as the ‘freeze’ imparted on many of them afterwards, indicate that these functions are unique and fundamentally important for the eukaryotic cell. Given the prevalence of these repeat-containing, structural proteins among the stem duplications, it would not be a gross exaggeration to suggest that, to a large extent, their proliferation ‘made the eukaryotes’. The excess of stem duplications among chaperones, ubiquitin system components, and other proteins involved in protein fate determination, compared with the other functional classes of eukaryotic genes, is compatible with this notion in as much as chaperone functions are required for multisubunit complex assembly. Generally, the proliferation of chaperones and functionally related proteins probably should have been expected. Indeed, the sheer size of the eukaryotic cell and its extensive internal compartmentalization seem to necessitate diversification of various chaperone-type functions.
For nearly all stem duplications, there is either direct experimental information on the protein functions or, at least, a clear functional prediction based on diagnostic domain architecture. Thus, all numerous paralogous proteins containing WD-40 repeats can be confidently predicted to function as structural components of multisubunit complexes, whereas all RING-finger proteins are most likely to be ubiquitin ligases. At this level, it may be claimed that the set of stem paralogs had been functionally characterized. However, many of these are extremely general predictions. A full understanding of the functional repertoire of the eukaryotic stem duplications requires much additional experimentation, which undoubtedly will reveal crucial functional distinctions between ancient paralogs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Eva Czabarka and Liran Carmel for the helpful discussions of statistical approaches used in this work, Vladimir Slesarev for technical assistance, and Martin Lercher and Igor Rogozin for useful discussions. This work was supported by the Intramural Research Program of the NIH, National Library of Medicine. Funding to pay the Open Access publication charges for this article was provided by The Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fisher R.A. The possible modification of the response of the wild type to recurrent mutations. Am. Nat. 1928;62:115–126. [Google Scholar]
- 2.Haldane J.B.S. The part played by recurrent mutation in evolution. Am. Nat. 1933;67:5–19. [Google Scholar]
- 3.Muller H.J. The origination of chromatin deficiencies as minute deletions subject to insertion elsewhere. Genetics. 1935;17:237–252. [Google Scholar]
- 4.Bridges C.A. Salivary chromosome maps. J. Hered. 1935;26:60–64. [Google Scholar]
- 5.Ohno S. Evolution by Gene Duplication. Berlin-Heidelberg-NY: Springer-Verlag; 1970. [Google Scholar]
- 6.Hughes M.K., Hughes A.L. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol. Biol. Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- 7.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch M., Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 10.Kondrashov F.A., Rogozin I.B., Wolf Y.I., Koonin E.V. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch M., Conery J.S. The evolutionary demography of duplicate genes. J. Struct. Funct. Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- 12.He X., Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gogarten J.P., Kibak H., Dittrich P., Taiz L., Bowman E.J., Bowman B.J., Manolson M.F., Poole R.J., Date T., Oshima T., et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc. Natl Acad. Sci. USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad. Sci. USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho G., Doolittle R.F. Intron distribution in ancient paralogs supports random insertion and not random loss. J. Mol. Evol. 1997;44:573–584. doi: 10.1007/pl00006180. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L., Anantharaman V., Koonin E.V. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L., Mazumder R., Vasudevan S., Koonin E.V. Trends in protein evolution inferred from sequence and structure analysis. Curr. Opin. Struct. Biol. 2002;12:392–399. doi: 10.1016/s0959-440x(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 18.Friedman R., Hughes A.L. Pattern and timing of gene duplication in animal genomes. Genome Res. 2001;11:1842–1847. doi: 10.1101/gr.200601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman R., Hughes A.L. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol. Biol. Evol. 2003;20:154–161. doi: 10.1093/molbev/msg017. [DOI] [PubMed] [Google Scholar]
- 20.Romero D., Palacios R. Gene amplification and genomic plasticity in prokaryotes. Annu. Rev. Genet. 1997;31:91–111. doi: 10.1146/annurev.genet.31.1.91. [DOI] [PubMed] [Google Scholar]
- 21.Brown C.J., Todd K.M., Rosenzweig R.F. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol. Biol. Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 22.Stark G.R., Wahl G.M. Gene amplification. Annu. Rev. Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- 23.Schwab M. Amplification of oncogenes in human cancer cells. Bioessays. 1998;20:473–479. doi: 10.1002/(SICI)1521-1878(199806)20:6<473::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 24.Jordan I.K., Makarova K.S., Spouge J.L., Wolf Y.I., Koonin E.V. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 2001;11:555–565. doi: 10.1101/gr.166001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 26.Lespinet O., Wolf Y.I., Koonin E.V., Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maynard Smith J., Szathmary E. The Major Transitions in Evolution. Oxford: Oxford University Press; 1997. [Google Scholar]
- 28.Dacks J.B., Doolittle W.F. Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell. 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 29.Archambault J., Friesen J.D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol. Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filee J., Forterre P., Sen-Lin T., Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- 31.Kearsey S.E., Labib K. MCM proteins: evolution, properties, and role in DNA replication. Biochim. Biophys Acta. 1998;1398:113–136. doi: 10.1016/s0167-4781(98)00033-5. [DOI] [PubMed] [Google Scholar]
- 32.Gupta R.S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 33.Hughes A.L. Evolution of the proteasome components. Immunogenetics. 1997;46:82–92. doi: 10.1007/s002510050245. [DOI] [PubMed] [Google Scholar]
- 34.Gille C., Goede A., Schloetelburg C., Preissner R., Kloetzel P.M., Gobel U.B., Frommel C. A comprehensive view on proteasomal sequences: implications for the evolution of the proteasome. J. Mol. Biol. 2003;326:1437–1448. doi: 10.1016/s0022-2836(02)01470-5. [DOI] [PubMed] [Google Scholar]
- 35.Koonin E.V., Wolf Y.I., Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 2001;11:240–252. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koonin E.V., Makarova K.S., Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koonin E.V. Orthology, paralogy, and evolutionary genomics. Annu. Rev. Genet. 2005 doi: 10.1146/annurev.genet.39.073003.114725. in press. [DOI] [PubMed] [Google Scholar]
- 38.Doolittle W.F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 39.Doolittle W.F., Boucher Y., Nesbo C.L., Douady C.J., Andersson J.O., Roger A.J. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc. Lond B. Biol. Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. discussion 57–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersson J.O., Sjogren A.M., Davis L.A., Embley T.M., Roger A.J. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr. Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- 41.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A., Panchenko A.R., Shoemaker B.A., Thiessen P.A., Geer L.Y., Bryant S.H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roger A.J. Reconstructing early events in eukaryotic evolution. Am. Nat. 1999;154:S146–S163. doi: 10.1086/303290. [DOI] [PubMed] [Google Scholar]
- 45.Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 46.Baldauf S.L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 47.Koonin E.V., Fedorova N.D., Jackson J.D., Jacobs A.R., Krylov D.M., Makarova K.S., Mazumder R., Mekhedov S.L., Nikolskaya A.N., Rao B.S., et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirkin B.G., Fenner T.I., Galperin M.Y., Koonin E.V. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol. Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 52.Adachi J., Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 53.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malik H.S., Eickbush T.H., Goldfarb D.S. Evolutionary specialization of the nuclear targeting apparatus. Proc. Natl Acad. Sci. USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neuwald A.F., Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown J.R., Doolittle W.F. Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makarova K.S., Aravind L., Galperin M.Y., Grishin N.V., Tatusov R.L., Wolf Y.I., Koonin E.V. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 58.Forterre P., Philippe H. Where is the root of the universal tree of life? Bioessays. 1999;21:871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 59.Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- 60.Forterre P., Brochier C., Philippe H. Evolution of the Archaea. Theor. Popul. Biol. 2002;61:409–422. doi: 10.1006/tpbi.2002.1592. [DOI] [PubMed] [Google Scholar]
- 61.Chervitz S.A., Aravind L., Sherlock G., Ball C.A., Koonin E.V., Dwight S.S., Harris M.A., Dolinski K., Mohr S., Smith T., et al. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bochtler M., Ditzel L., Groll M., Hartmann C., Huber R. The proteasome. Annu. Rev. Biophys. Biomol. Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- 63.Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nature Cell. Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 64.Archibald J.M., Blouin C., Doolittle W.F. Gene duplication and the evolution of group II chaperonins: implications for structure and function. J. Struct. Biol. 2001;135:157–169. doi: 10.1006/jsbi.2001.4353. [DOI] [PubMed] [Google Scholar]
- 65.Papp B., Pal C., Hurst L.D. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 66.Friedman R., Hughes A.L. Gene duplication and the structure of eukaryotic genomes. Genome Res. 2001;11:373–381. doi: 10.1101/gr.155801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veitia R.A. Nonlinear effects in macromolecular assembly and dosage sensitivity. J. Theor. Biol. 2003;220:19–25. doi: 10.1006/jtbi.2003.3105. [DOI] [PubMed] [Google Scholar]
- 68.Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 69.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 70.Kubota Y., Takase Y., Komori Y., Hashimoto Y., Arata T., Kamimura Y., Araki H., Takisawa H. A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev. 2003;17:1141–1152. doi: 10.1101/gad.1070003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Makarova K.S., Koonin E.V. Comparative genomics of Archaea: how much have we learned in six years, and what's next? Genome Biol. 2003;4:115. doi: 10.1186/gb-2003-4-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lynch M., Conery J.S. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 74.Koonin E.V. A non-adaptationist perspective on evolution of genomic complexity or the continued dethroning of man. Cell Cycle. 2004;3:280–285. [PubMed] [Google Scholar]
- 75.McLysaght A., Hokamp K., Wolfe K.H. Extensive genomic duplication during early chordate evolution. Nature Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 76.Skrabanek L., Wolfe K.H. Eukaryote genome duplication—where's the evidence? Curr. Opin. Genet. Dev. 1998;8:694–700. doi: 10.1016/s0959-437x(98)80039-7. [DOI] [PubMed] [Google Scholar]
- 77.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 78.Wolfe K. Evolutionary genomics: yeasts accelerate beyond BLAST. Curr. Biol. 2004;14:R392–394. doi: 10.1016/j.cub.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 79.Kellis M., Birren B.W., Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 80.Wolf Y.I., Rogozin I.B., Koonin E.V. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cuff J.A., Clamp M.E., Siddiqui A.S., Finlay M., Barton G.J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.