Abstract
Gene regulation at the post-transcriptional level is prevalent in all domains of life. In bacteria, ProQ-like proteins have emerged as important RNA chaperones facilitating RNA stability and RNA duplex formation. In the major human pathogen Vibrio cholerae, post-transcriptional gene regulation is key for virulence, biofilm formation, and antibiotic resistance, yet the role of ProQ has not been studied. Here, we show that ProQ interacts with hundreds of transcripts in V. cholerae, including the highly abundant FlaX small RNA (sRNA). Global analyses of RNA duplex formation using RIL-Seq (RNA interaction by ligation and sequencing) revealed a vast network of ProQ-assisted interactions and identified a role for FlaX in motility regulation. Specifically, FlaX base-pairs with multiple sites on the flaB flagellin mRNA, preventing 30S ribosome binding and translation initiation. V. cholerae cells lacking flaX display impaired motility gene expression, altered flagella composition and reduced swimming in liquid environments. Our results provide a global view on ProQ-associated RNA duplex formation and pinpoint the mechanistic and phenotypic consequences associated with ProQ-associated sRNAs in V. cholerae.
Graphical Abstract
Graphical Abstract.
Introduction
Vibrio cholerae is a major human pathogen causing millions of infections every year worldwide (1). Pathogenicity of V. cholerae critically depends on its motility, which is driven by a single polar flagellum (2–4). Flagella-mediated motility has been shown to be required for V. cholerae to cross the intestinal mucus barrier facilitating gut colonization and virulence gene expression (5–7). In addition, the flagellum has been implicated in various other phenotypes in V. cholerae including surface sensing, biofilm formation, quorum sensing and the secretion of auxiliary toxins (8–11).
Analogous to other Gram-negative pathogens, such as Salmonella enterica, the expression of motility-related genes in V. cholerae is controlled via a multi-tiered regulon that allows for the integration of extra- and intracellular signals (12,13). Specifically, coordination of flagella synthesis is achieved in a hierarchical four-step process involving several transcriptional regulators and alternative sigma factors. The master regulator of flagella synthesis is FlrA (class I), which together with RpoN, activates the expression of class II genes. Class II genes include flrBC and fliA, both encoding transcriptional regulators. FlrC is phosphorylated by FlrB and, in concert with RpoN, induces class III gene synthesis. There are more than a dozen class III transcripts known in V. cholerae including the flaA flagellin mRNA and the flaX sRNA, as well as several other genes encoding the structural components of the flagella apparatus (12,14). Additional structural components and sensory elements involved in flagella synthesis and function are grouped as class IV genes and their expression is activated by FliA (σ28). Notably, V. cholerae encodes five different flagellin genes of which flaA belongs to the class III genes, whereas flaB, flaC, flaD and flaE are class IV genes (15).
Besides transcriptional control, regulation of flagella assembly and motility has recently been reported to also involve post-transcriptional regulators, such as sRNAs (16–19). Bacterial sRNAs are a heterogeneous group of typically non-coding RNAs that mediate gene regulation by base-pairing with cis- or trans-encoded target mRNAs (20,21). The majority of characterized sRNAs interact with the ubiquitous RNA chaperone Hfq, promoting sRNA stability and RNA duplex formation with often multiple target transcripts (22). Besides Hfq, ProQ has been discovered to function as a second global RNA chaperone, interacting with hundreds of RNA species in model organisms, such as Escherichia coli and S. enterica (23–25). ProQ belongs to the family of FinO-like proteins, which are small (14–27 kDa), basic proteins that are encoded on proteobacterial genomes and mobile genetic elements, such as plasmids (26–29). FinO-like proteins from plasmids, for example, FinO from S. enterica and RocC from Legionella pneumophila, usually bind only few RNA species, whereas FinO proteins encoded on bacterial chromosomes, for example, ProQ from E. coli, S. enterica and Neisseria meningitidis, frequently show extensive RNA interactomes (30–34).
Mutation of the proQ gene has been associated with various phenotypes. For instance, S. enterica cells lacking proQ displayed increased sensitivity towards DNA-damaging agents, reduced virulence and biofilm gene expression, and produced higher numbers of antibiotic persister cells (35–39). The direct molecular underpinnings that cause these phenotypes are not fully understood as ProQ has diverse regulatory roles in the cell. For example, ProQ can not only promote the interaction of two trans-encoded transcripts (25,40) but also protect the 3′-end of mRNAs from exonucleolytic degradation (23). How ProQ switches between these functions is only incompletely understood and while there are several studies on ProQ in E. coli and S. enterica, only little is known about its regulatory roles in other organisms (32,41–43).
In this work, we studied the role of ProQ in V. cholerae. We discovered that ProQ interacts with hundreds of transcripts, including mRNAs and sRNAs. Among the latter, the FlaX sRNA is the main interaction partner of ProQ together with several previously uncharacterized sRNAs. We further show that FlaX is processed from the 3′-UTR (untranslated region) of the flaA mRNA by the major endoribonuclease RNase E and reveal the global ProQ-associated RNA interactome in V. cholerae using RIL-Seq analysis. The latter experiments also identified several interaction partners of FlaX, including the flaB mRNA, which encodes a secondary flagellin. Detailed analysis of the FlaX–flaB interaction revealed an unconventional regulatory mechanism involving base-pairing of FlaX at three independent sites in the 5′-UTR that act additively to downregulate protein synthesis. We propose a model in which FlaX antagonizes translation initiation of flaB through sequestration of the ribosome binding site (RBS) and two ribosome stand-by sites. In accordance with this model, we discovered that mutation of either proQ or flaX compromised the timing of motility gene expression, altered flagella composition and impaired motility. Taken together, our study shows that ProQ and its associated RNAs are important post-transcriptional regulators in V. cholerae and reveals the mechanistic underpinnings driving sRNA-mediated regulation of motility, a physiologically and ecologically crucial phenotype of this major human pathogen.
Materials and methods
Bacterial strains and growth conditions
All strains utilized in this study are detailed in the appendix in Supplementary Table S1. Detailed information on bacterial strains and their construction is listed in Appendix Supplementary Table S2. The wild-type strain used is V. cholerae O1 El Tor C6706. V. cholerae and E. coli cells were grown in LB medium at 37°C, unless stated otherwise. The growth medium was supplemented with antibiotics when necessary. The following concentrations were used: 50 U/ml polymyxin B, 100 μg/ml ampicillin, 50 μg/ml kanamycin, 5000 μg/ml streptomycin and 20 μg/ml chloramphenicol.
Strain construction
Plasmids were introduced from E. coli S17 λpir donor strains into V. cholerae via RK2/RP4-based conjugation. Transconjugants were subsequently selected using specific antibiotics and polymyxin B to inhibit the growth of E. coli. V. cholerae mutant strains were generated using the pKAS32 suicide vector. In summary, pKAS32 plasmids were conjugated into V. cholerae, and cells were selected for ampicillin resistance. Single colonies were then selected on streptomycin. The desired mutations were verified through PCR (Polymerase Chain Reaction) amplification and sequencing.
Plasmid construction
Plasmids and DNA oligonucleotides are listed in Appendix Supplementary Table S2 and S3, respectively. The proQ deletion eliminates both the ORF (Open Reading Frame) and the 5′-UTR (5' Untranslated Region) of the proQ gene. The flaX deletion removes 193 nucleotides from the middle of the flaX gene, leaving intact the 5′ overlap with flaA (which includes the flaA terminator) and the last 39 nucleotides at the 3′-end of the gene, forming a rho-independent terminator.
ProQ co-immunoprecipitation
Triplicates of V. cholerae WT and proQ::3xFLAG strains were grown in LB medium to reach low cell density (OD600 of 0.2) and high cell density (OD600 of 2). ProQ co-immunoprecipitation was performed as previously described (44). Complementary DNA (cDNA) libraries were prepared using the NEBNext Small RNA Library Prep Set for Illumina (NEB; E7300L).
RIP-seq analysis
The cDNA libraries were pooled and sequenced on a NextSeq1000/2000 system in paired-end mode with a read length of 100 nucleotides (45). Data analysis was conducted using the CLC Genomics Workbench (Qiagen). In summary, raw reads underwent adaptor trimming, and the resulting trimmed reads were mapped to the V. cholerae reference genome [National Center for Biotechnology Information (NCBI) accession numbers NC_002505.1 and NC_002506.1, https://www.ncbi.nlm.nih.gov/assembly/GCF_000006745.1] using default parameters. Fold enrichment in the proQ::3XFLAG-tagged samples compared to untagged control samples was calculated using the CLC ‘Differential Expression for RNA-Seq’ tool (Supplementary Table S4). To identify three enrichment patterns across the different parts of the mRNA (5′-UTR, coding sequence (CDS) and 3′-UTR), mapping was performed using CLC genomics with annotations of the 5′-UTR and 3′-UTR regions extracted from (62) and (49). Each mRNA region was quantified separately, and enrichment levels were calculated as described above.
RIL-seq experiment
RIL-seq experiment was conducted as described previously (46,47). Briefly, V. cholerae wild-type and proQ::3xFLAG strains were grown in duplicate in LB medium until OD600 of 0.2 (low cell density) and OD600 of 2.0 (high cell density). Cells equivalent to 40 OD600 units underwent protein-RNA cross-linking, cell lysis and co-immunoprecipitation. The co-immunoprecipitated RNA was then treated with RNase A/T1 and T4 RNA ligase. Following proteinase K digestion, RNA was extracted, fragmented and treated with TURBO DNase. Ribosomal RNA was depleted, and cDNA libraries were prepared and sequenced in paired-end mode using a HiSeq 2500 system.
RIL-seq analysis
Demultiplexed sequencing reads were analyzed with ChimericFragments (https://github.com/maltesie/ChimericFragments) (48).
RNA isolation, Northern blot analysis and quantitative real-time PCR
Culture aliquots (4 OD600 units) were collected at specified time points or cell densities and immediately mixed with 0.2 volumes of stop solution (95% ethanol and 5% phenol). Total RNA was extracted using Extrazol reagent. For northern blot analysis, RNA samples were separated on 6% polyacrylamide/7 M urea gels and then transferred to Nylon membranes (Sigma). The membranes were hybridized at 42°C in Roti-Hybri-Quick buffer (Roth) using [32P] end-labeled DNA oligonucleotides. The oligonucleotides used for probing are listed in Supplementary Table S4. The membranes were washed in three successive steps with SSC (5×, 1×, 0.5×)/0.1% SDS wash buffer. Signals were visualized using a Typhoon phosphorimager (GE Healthcare) and quantified with GelQuant software (BiochemLabSolutions). For probing multiple target sRNAs, membranes were stripped with 30 ml of ROTI®Hybri-Quick at 72°C for 30 min. For quantitative real-time PCR (qRT-PCR), total RNA was treated with TurboDNase at 37°C for 30 min. qRT-PCR was carried out using the Luna Universal One-Step RT-qPCR Kit (New England BioLabs) and a CFX96 Real-Time PCR System (Bio-Rad). 5S rRNA and recA served as reference genes. The oligonucleotides used for qRT-PCR are listed in Supplementary Table S4.
RNase E in vivocleavage assay
V. cholerae RNase E thermosensitive strain rne‐3071 TS (44,49) and an isogenic control strain were grown overnight in LB medium at 30°C, then diluted 1:500 into fresh LB medium and further grown at 30°C to OD600 of 1.0. Half of the replicates were then either immediately transferred into a 44°C or left to grow at 30°C for 30 min. L-arabinose was then added to 0.2% final concentration and samples were collected after 30 min induction. Total RNA was isolated and analyzed by northern blotting as described above.
Transcriptome analysis
Depletion of ribosomal RNA was done with custom-made biotinylated oligos as described in (50). Oligos used for depletion are indicated in Supplementary Table S4. Efficiency of the depletion was confirmed using an Agilent 2100 Bioanalyzer. cDNA libraries were constructed using NEBNext Multiplex Small RNA Library Prep Set for Illumina (#E7300) according to the manufacturer’s instructions. The quality was assessed on an Agilent 2100 Bioanalyzer. The libraries were sequenced using a Nextseq 1000/2000 platform (Illumina) in paired-read mode with 50 bp read length. Data analysis was performed in CLC Genomics Workbench (Qiagen) as described for RIP-seq.
End-labeling of DNA oligonucleotides
DNA oligonucleotides were 5′-end-labeled using T4 polynucleotide kinase (PNK; New England Biolabs) in the presence of [32P]-γATP. Briefly, 5 pmol of oligonucleotide was incubated in a 10 μl reaction containing 1 μl of 10X PNK buffer, 1 unit of T4 PNK and 1.5 μl of [32P]-γATP (15 μCi). The reaction was incubated at 37°C for 60 min. Labeled oligonucleotides were then purified using Sephadex G-25 columns (GE Healthcare) according to the manufacturer’s instructions.
Western blot analysis
Protein samples were separated using a 12.5% denaturing SDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis) gel. Electrophoresis was carried out at 30 mA per gel until the samples passed through the 4% polyacrylamide stacking gel, then increased to 50 mA per gel as they entered the resolving gel. Subsequently, samples were transferred to PVDF membranes via semi-dry blotting at 320 mA per gel for 1.5 h at 4°C. Anti-FLAG (Sigma; F1804) and anti-GFP (Sigma; 11 814 460 001) antibodies were used for detection. RNA polymerase (RNAP) was used as the loading control and detected with an anti-RNAP antibody (BioLegend; WP003). The signals were visualized with a Fusion FX EDGE imager (Vilber), and band intensities were quantified using BIO-1D software (Vilber).
RNA stability experiment
To monitor the stability of flaX and flaB, transcription was halted by adding rifampicin to a final concentration of 250 μg/ml. RNA samples were collected prior to induction, and at 2, 4, 8, 16 and 32 min following rifampicin addition. RNA levels were then measured using northern blot analysis or quantitative real-time PCR.
Motility assays
Plate-based motility assay was performed on semi-solid LB agar. Approximately 5 × 108 cells from overnight cultures grown at 30°C were harvested by centrifugation at room temperature (1 min, 16 000 rcf) and resuspended in 100 μl of LB broth supplemented with kanamycin (50 μg/ml). For inoculation, 1 μl of each culture was spotted onto 0.2% LB agar square plates (120 × 120 mm) supplemented with kanamycin. The plates were incubated at 30°C for 13 h, after which they were photographed from above. Colony areas were measured using ImageJ software, with the area serving as a proxy for motility.
Fluorescence measurements
GFP fusion reporter assays were conducted using E. coli Top 10 cells harboring a pXG-10 plasmid (51,52). This plasmid contained the 5′-UTR, and the first 20 amino acids of the predicted target mRNA fused in-frame with sfGFP, under the control of a PLtetO-1 promoter. In parallel, the strain also carried a pEVS143 plasmid that constitutively overexpressed the sRNA. Strains carrying an empty pEVS143 plasmid served as controls. To account for background fluorescence, samples not expressing fluorescent proteins were used for subtraction. Cell pellets corresponding to 1 OD600 unit were collected by centrifugation, washed twice with 1 × PBS (pH 7.4), and the fluorescence intensity was measured using a Spark 10M plate reader (Tecan, Männedorf, Switzerland).
RNA structure probing
RNA structure probing and mapping of flaB-FlaX binding sites was performed as previously described with minor modifications (53,54). In brief, 0.4 pmol of 5′-end-labeled RNA was denatured, chilled on ice before addition of 1 μg of yeast RNA (Invitrogen, #AM7118) and 1X structure buffer (0.01 M Tris, pH 7, 0.1 M KCl, 0.01 M MgCl2). Unlabeled RNA was then added in different molar ratios followed by incubation at 37°C for 10 min. The samples were then treated with either RNase T1 (0.1 U; Ambion, AM2283) for 3 min, lead (II) acetate (final concentration: 6.25 mM; Sigma-Aldrich, 316512–5G) for 1 min and 45 s or RNase V1 (2 × 10−5 U; Ambion, AM2275) for 1 min. Reactions were terminated by adding an equivalent volume of GLII loading buffer. RNase T1 and alkaline (OH) sequencing ladders were prepared with 0.8 pmol of 5′-end-labeled RNA and stopped by adding GLII loading buffer. Samples were separated by denaturing PAGE on 5% or 12% polyacrylamide / 7 M urea sequencing gels.
T7 transcription and 5′end-labeling of RNA
RNAs were synthesized via in vitro transcription following previously established protocols (53). Briefly, a DNA template containing the T7 promoter was amplified by PCR using specific primers (KPO-2770/KPO-2771 on pDD29 for FlaX; KPO-8899/KPO-8901 on pDD21 for flaB 5′-UTR + 20aa) and transcribed with the MEGAscript® transcription kit (AM1334, Thermo Fisher). The purified RNA (20 pmol) was dephosphorylated using calf intestinal alkaline phosphatase (NEB), followed by extraction with P:C:I and ethanol precipitation. The dephosphorylated RNA was subsequently labeled with [32P]-γATP (25 μCi) by incubation with 1 unit of PNK (NEB) for 1 h at 37°C. The RNA was then purified using a denaturing 6% polyacrylamide/7 M urea gel, eluted in RNA elution buffer (0.1 M sodium acetate, 0.1% SDS, 10 mM EDTA) overnight at 4°C, and recovered through P:C:I extraction.
In vitro translation experiments
Translation was carried out using the PURExpress In Vitro Protein Synthesis system (E6800S, New England BioLabs) (55). Invitro transcribed mRNA was generated by in-vitro transcription with the sense primer adding the T7 promoter to the + 1 site of the 5′-UTR and the antisense primer located downstream the gfp stop codon. Primers used are listed in Supplementary Table S4. Invitro synthesized mRNA was mixed with or without the sRNA in a 10 μl reaction volume following the manufacturer’s instructions. Reactions were incubated at 37°C and stopped by addition of equal volume of protein loading buffer. Detection of GFP-tagged FlaB was done by western blot analysis using a monoclonal GFP antibody (Sigma; 11814460001).
Toeprinting
The toeprinting assay was conducted using in vitro synthesized flaB mRNA. The mRNA was generated with a sense primer that incorporated a T7 promoter at the + 1 position of the 5′-UTR and an antisense primer positioned 60 nucleotides downstream of the start codon. For the assay, 0.2 pmol of mRNA was combined with 0.5 pmol of radiolabeled antisense primer (KPO-8901) in toeprint buffer A (50 mM Tris-acetate, pH 7.6, 5 mM Dithiothreitol (DTT), 500 mM potassium acetate). RNase-free water or sRNA was added to the annealing mixture, which was then denatured at 95°C for 1 min and chilled on ice for 5 min. Following this, the RNAs were refolded by incubating at 37°C for 5 min after the addition of dNTPs (final concentration of 0.5 mM) and toeprint buffer B (10 mM Tris-acetate, pH 7.6, 1 mM DTT, 100 mM potassium acetate). Next, 2 pmol of activated 30S ribosomal subunits (pre-incubated at 37°C for 15 min) were added, followed by a 5-min incubation at 37°C. Subsequently, 20 pmol of tRNAfMet (MP Biomedicals, 199 154) was introduced, and the formation of the 30S initiation complex (30S-IC) was allowed for 15 min at 37°C.
cDNA synthesis was then carried out by adding SuperScript™ II Reverse Transcriptase (Thermo, 18 064 014) and incubating the mixture at 37°C for 20 min. The reaction was terminated by adding stop buffer (50 mM Tris–HCl, pH 7.5, 0.1% SDS, 10 mM EDTA), followed by P:C:I extraction. The template RNA was subsequently hydrolyzed with 150 mM KOH at 95°C for 5 min, followed by neutralization with acetic acid. The resulting cDNA was precipitated using sodium acetate and ethanol, and then separated on a 10% sequencing gel.
Purifying flagella filaments for MS analysis
Purification of the flagella hook-basal body complex from V. cholerae was carried out following the method previously described by (8). Briefly, bacterial cells were collected at an OD600 of 1.0 and then resuspended in 30 ml of an ice-cold sucrose solution (0.5 M sucrose, 0.1 M Tris–HCl, pH 8.0) with 1 mg/ml lysozyme and 10 mM EDTA (pH 8.0). The mixture was incubated at 4°C for 30 min and lysed with 3 ml of 10% Triton X-100 followed by 3 ml of 0.1 M MgSO4. After gently stirring the mixture at 4°C for 2 h, 3 ml of 0.1 M EDTA (pH 11.0) was added. The lysate was then clarified by centrifugation at 5000 × g for 10 min, and the pH was adjusted to 11.0 using 5 N NaOH. Subsequently, the lysate was subjected to centrifugation at 100 000 × g for 60 min, after which the resulting pellet was resuspended in an alkaline sucrose solution (10% sucrose, 0.1 M KCl, 0.1% Triton X-100, pH 11.0) and centrifuged again at 100 000 × g for 60 min. The pellet was then resuspended in TET buffer (10 mM Tris-Cl, 0.1 M EDTA, 0.1% Triton X-100, pH 8.0), clarified by centrifugation first at 5000 × g for 10 min and then at 100 000 × g for 60 min. The final pellet was resuspended in 5 ml of an acidic solution (50 mM glycine, 0.1% Triton X-100, pH 2.5), incubated at room temperature for 30 min and then centrifuged at 100 000 × g for 60 min. The resulting pellets were washed twice with TE buffer (10 mM Tris-Cl, 0.1 M EDTA, pH 8.0) and finally centrifuged at 100 000 × g for 60 min to isolate the intact flagella hook-basal body complex.
Proteome analysis
Samples were prepared from SDS-PAGE gel bands according to the in-gel digestion protocol by (56). LC-MS/MS analysis was carried out on an Ultimate 3000 nano RSLC system coupled to a Q Exactive HF mass spectrometer (both Thermo Fisher Scientific, Waltham, MA, USA). Peptides were trapped (2 cm × 75 μm, 3 μm) and separated (50 cm × 75 μm, 2μm) on Acclaim Pep Map columns. Eluents A (0.1% [v/v] formic acid in water) and B (0.1% [v/v] formic acid in 90/10 acetonitrile/water [v/v]) were mixed as follows: 0 min at 4% B, 5 min at 8% B, 20 min at 12% B, 30 min at 18% B, 40 min at 25% B, 50 min at 35% B, 57 min at 50% B, 62–65 min at 96% B, 65.1–90 min at 4% B. Positively charged ions were generated at 2.2 kV using a Nanospray Flex Ion Source (Thermo Fisher Scientific) and monitored at m/z 300–1500, R = 60 000 full width at half maximum (FWHM) using a maximum injection time (ITmax) of 100 ms and an automatic gain control (AGC) target of 1 × 106. Precursor ions were selected for fragmentation in a data-dependent manner (Top15, z = 2–5) at an isolation width of m/z 2.0 amu for higher-energy collisional dissociation (HCD) fragmentation at 30% normalized collision energy (NCE). MS2 ions were scanned at R = 15 000 FWHM (ITmax = 80 ms, AGC = 2e5) using a dynamic exclusion of precursor ions for 25 s. The LC-MS/MS instrument was controlled by Chromeleon 7.2, QExactive HF Tune 2.8 and Xcalibur 4.0 software. Tandem mass spectra were searched against the UniProt reference proteome database (27 July 2020; https://www.uniprot.org/proteomes/UP000000584) of V. cholerae using Proteome Discoverer (PD) 2.4 (Thermo) and the algorithms of Sequest HT (version of PD2.4) and MS Amanda 2.0 and MS Fragger 2.4. Two missed cleavages were allowed for the tryptic digestion. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance was set to 0.02 Da. Modifications were defined as dynamic Met oxidation as well as static Cys carbamidomethylation. A strict false discovery rate (FDR) < 1% (peptide and protein level) and at least a search engine threshold >4 (Sequest HT), >300 (MS Amanda) and >8 (MS Fragger) were required for positive protein hits. The Percolator node of PD2.4 and a reverse decoy database was used for q-value validation of spectral matches. Only rank 1 proteins and peptides of the top scored proteins were counted. The Minora algorithm of PD2.2 was applied for relative protein abundance quantification (Supplementary Table S5).
Scanning electron microscopy
For scanning electron microscopy, droplets of a fresh culture were placed on a glass slide, covered with a coverslip and plunge frozen in liquid nitrogen. This was followed by an additional chemical fixation by putting skipping the coverslip off and putting the glass slides in the fixative, which consisted of 2.5% glutardialdehyde in 75 mM cacodylate buffer also containing 100 mM NaCl, 2 mM MgCl2 at pH 7.0 for at least 1h. After three washing steps with buffer, post-fixation was done with 1% osmium tetroxide for 30 min. This was followed by two washing steps with buffer and three washing steps with double-distilled water. Dehydration of the samples was done in a graded acetone series before critical-point-drying. The glass slides were then mounted on aluminum stubs and sputter-coated with 3 nm platinum. Scanning electron microscopy was carried out using a Zeiss Auriga 40 crossbeam workstation (Zeiss, Oberkochen, Germany) with an acceleration voltage of 2 kV and by using the SE-detector.
Bioinformatic tools
Sequence alignments were created using MultAlin ((57); http://multalin.toulouse.inra.fr/). RNA secondary structures were predicted with RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). RNA base-pairing interactions were predicted using the RNAhybrid ((58); https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) and IntaRNA ((59); http://rna.informatik.uni-freiburg.de/IntaRNA/).
Results
RIP-seq analysis detects ProQ RNA ligands in V. cholerae
ProQ is highly conserved in γ-proteobacteria, including V. cholerae (60,61). Analogous to the genomes of E. coli and S. enterica, the proQ gene of V. cholerae is located between the prc (encoding a protease) and msrC (encoding a methionine-(R)-sulfoxide reductase) genes and its transcription is driven by single promoter located 50 bps upstream of the translation start site (Supplementary Figure S1A; (62)). To study the expression of ProQ in V. cholerae, we next generated a chromosomally tagged ProQ::3XFLAG strain and monitored protein abundance at various stages of growth. We detected nearly constant ProQ production under all tested conditions with a mild increase in protein expression under stationary phase growth conditions (Supplementary Figure S1B) and similar results were obtained when we probed proQ mRNA levels using qRT-PCR (Supplementary Figure S1C). We also generated a proQ deletion mutant in V. cholerae and tested growth in rich media (LB; lysogeny broth). We found that growth of this mutant was comparable to V. cholerae wild-type cells and plasmid-borne overexpression of proQ did not affect growth either (Supplementary Figure S1D). Taken together, we conclude that ProQ is expressed in V. cholerae and that mutation of its corresponding gene does not cause any major changes in growth, at least under standard growth conditions.
ProQ of E. coli and S. enterica has been reported to interact with hundreds of mRNAs and dozens of sRNAs (23–25). To identify the binding partners of ProQ in V. cholerae, we performed RNA co-immunoprecipitation using chromosomally produced ProQ::3XFLAG as bait and determined the attached RNA ligands by high-throughput sequencing (Figure 1A). When compared to a control strain lacking the FLAG tag epitope, the relative amount of reads mapping to coding sequences and sRNAs increased in the ProQ co-immunoprecipitation, whereas reads corresponding to house-keeping RNAs, transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) were decreased (Figure 1B). We discovered 496 mRNA and 60 sRNA ligands at low cell density (OD600 of 0.2) and 463 mRNA and 75 sRNA ligands at high cell density (OD600 of 2.0) (Supplementary Table S4). In line with a previous report (23), we discovered that 3′-UTRs of mRNAs were preferentially bound by ProQ (Supplementary Figure S1E). We further classified the sRNAs based on their genomic location, determining whether they were transcribed from intergenic regions (IGRs) or encoded within mRNA 5′-UTRs or 3′-UTRs (Supplementary Figure S1F). In total, we detected 17 sRNAs from IGRs, 3 overlapping coding sequences, 6 from 5′-UTRs and 60 from 3′-UTRs. Quantification of the reads mapping to sRNAs genes also revealed that the FlaX sRNA (14) is highly abundant under both growth conditions followed by several uncharacterized sRNAs, for example, Vcr087, Vcr203, Vcr061, Vcr078, Vcr098 and Vcr221, as well as known sRNAs, such as FarS and OppZ (Figure 1A and C). Analogous to ProQ, mutation or overexpression of flaX did not affect growth of V. cholerae in rich media (Supplementary Figure S1G). In summary, ProQ interacts with a large number of RNA ligands in V. cholerae of which FlaX is the most abundant sRNA binding partner.
Figure 1.
RIP-seq analysis reveals sRNAs associated with ProQ in Vibrio choleae. V. cholerae expressing chromosomally FLAG-tagged ProQ (3xF; +) and wild-type cells (WT; -) were grown in LB medium to low (OD600 of 0.2) and high cell densities (OD600 of 2.0) and used for ProQ co-immunoprecipitation. (A) Stacked plot of the read distribution of significantly enriched sRNAs with the FLAG-tagged ProQ in comparison to the wild-type control (≥2-fold change and FDR adjusted P-value ≤ 0.05). Mapped reads to each sRNA were normalized to the total reads mapping of all enriched sRNAs. Plots for OD600 of 0.2 and OD600 of 2.0 are shown on left and right side, respectively. Top 20 sRNAs are shown in the insets on each side. Data were collected from three biological replicates. (B) The relative fraction of different RNA classes recovered from WT and FLAG-tagged ProQ (3xF) coIP at OD600 of 0.2 and OD600 of 2.0 are shown. (C) Protein and RNA samples were collected before (input) and after purification (coIP). Top panel: western blot (WB) to verify the expression and enrichment of the ProQ-3xFLAG protein, with RNAP serving as the loading control. Bottom panel: RNA samples were analyzed by northern blotting (NB) to validate the expression and enrichment of specific sRNAs. The 5S rRNA was used as a loading control.
The FlaX sRNA is processed from the 3′-UTR of flaA
The above experiments showed that the FlaX sRNA interacts with ProQ and occupies a major fraction of the protein at low and high cell densities (Figure 1A and C). The flaX gene is located in the 3′-UTR of flaA (encoding a major flagellin) in V. cholerae (Figure 2A and Supplementary Figure S2A) and was reported to accumulate as a ∼150 nt transcript (14). However, follow-up transcriptomic studies (62) and northern blot analysis (Figure 2B) showed that FlaX is ∼265 nts long and detected under all tested conditions, with strongest expression at late exponential growth conditions (OD600= 1.0). Mutation of proQ resulted in reduced FlaX levels in stationary phase, whereas plasmid-borne overexpression of proQ had the reverse effect (Figure 2B). To investigate whether differential expression of FlaX in the proQ mutant and the proQ overexpression strain resulted from altered transcription or rather relied on ProQ-mediated changes in FlaX stability, we treated V. cholerae cells with rifampicin and determined the stability of FlaX. Indeed, we discovered that ΔproQ cells displayed reduced FlaX stability, whereas elevated ProQ levels increased its stability (Figure 2C and Supplementary Figure S2B).
Figure 2.
FlaX is a ProQ-associated sRNA derived from the 3′-end of the flaA gene. (A) Schematic representation of flax genomic organization. The black arrow indicates the promoter that drives the expression of flaA-flaX. The flaA-flaX primary transcript (1504 nt) is processed thus releasing FlaX (265 nt). (B) V. cholerae wild-type and ΔproQ mutant cells, carrying either a control plasmid (pctrl) or a plasmid containing the proQ gene with its native promoter (pproQ), were grown in LB medium. Total RNA was extracted at various growth stages, and FlaX expression was examined using northern blot analysis. The 5S rRNA served as the loading control. (C) Stability of FlaX was monitored in wild-type cells and ΔproQ mutant cells, carrying either a control plasmid (pctrl) or a plasmid containing the proQ gene (pproQ). Cells were cultivated in LB medium to an OD600 of 1.0 and rifampicin was added to inhibit transcription. Northern blot analysis was performed to measure FlaX level at the specified time points. One representative gel is shown in Supplementary Figure S2B. The signal was quantified and plotted in exponential scale. The RNA half-life was determined by calculating the time point at which 50% of FlaX had degraded (horizontal line). Error bars represent the standard deviation from three biological replicates. (D) V. cholerae ΔflaX strains carrying either a wild-type (rne WT) or a temperature-sensitive (rne TS) RNase E allele, along with the pBAD-flaAX plasmid, were grown in LB medium at 30°C to OD600 of 1.0. Cultures were then divided into two halves, with one half maintained at the permissive temperature (30°C) and the other shifted to the nonpermissive temperature (44°C) for 30 min. flaA-flaX expression was next induced by adding L-arabinose (0.2% final concentration) for 30 min, and FlaX was monitored by northern blot. The solid triangle indicates full-length FlaX transcript, while the open triangles denote the processing intermediates. 5S rRNA served as a loading control.
Transcription of flaA and flaX is driven by a single promoter located upstream of the flaA gene (Figure 2A and (62)) and mutation of this promoter strongly reduced FlaX expression (Supplementary Figure S2C). Thus, we asked how FlaX is released from the 3′-UTR of flaA and which ribonuclease could be involved in the process. We hypothesized that the major ribonuclease E (RNase E) could be required as it has previously been demonstrated to aid the maturation of sRNAs that are processed from longer transcripts (63,64). To test this hypothesis, we employed a V. cholerae strain producing a temperature-sensitive RNase E protein (rneTS, (49)) and probed FlaX expression under permissive (30°C) and non-permissive temperatures (44°C) on northern blots. In line with our hypothesis, we found that FlaX expression was strongly reduced when RNase E was inactivated, whereas wild-type cells expressing intact RNase E, as well as rneTS cells at permissive temperature, did not show altered FlaX levels (Figure 2D). In accordance, with these results we discovered that mutation of the RNase E recognition motif (49,65) at the 5′ end of flaX inhibited accumulation of the full-length FlaX transcript (Supplementary Figure S2D). We also tested the effect of a related ribonuclease, RNase G, on FlaX accumulation in the cell. In contrast to RNase E, deletion of the gene encoding RNase G (rng) did not affect FlaX expression (Figure 2D). Taken together, we conclude that FlaX is a 3′-UTR-derived sRNA that depends on RNase E for maturation, while ProQ binding is required for full stability.
FlaX and ProQ regulate motility in V. cholerae
The 3′-UTR of mRNAs has recently been recognized as a rich source for sRNA regulators in various bacteria, which frequently foster or extend the biological functions of their preceding genes (64,66). Given that FlaX is produced from the 3′-UTR of flaA (Figure 2A), we thus speculated that the sRNA (together with ProQ) could be involved in motility regulation in V. cholerae. To test this hypothesis, we mutated the genes responsible for flagella regulation in V. cholerae (i.e. flrA, flrB, flrC, rpoN and fliA) and probed FlaX expression on northern blots (Figure 3A and B). We found that expression of FlaX depends on FlrA-C and RpoN, but is independent of FliA and thus belongs to the group of class III genes.
Figure 3.
FlaX is a class III gene of the flagella regulon and controls motility. (A) Transcriptional hierarchy of flagella synthesis in V. cholerae. (B) Role of different transcription factors of the flagella regulon on the transcription of FlaX was tested. V. cholerae ΔflrA, ΔflrB, ΔflrC, ΔrpoN and ΔfliA cells harboring either an empty control vector or an overexpression plasmid (pflrA, pflrB, pflrC, prpoN and pfliA) were cultivated in LB medium and RNA samples were collected at OD600 of 1.0. Northern blot analysis was performed to determine FlaX level. 5S rRNA served as loading control. The experiment was performed with three biological replicates. (C) Motility assay on semi-solid LB agar. Inoculum from wild-type cells, ΔproQ mutant cells and ΔflaX mutant cells carrying either a control plasmid (pctrl) or a plasmid containing the deleted gene (pproQ or pflaX) were spotted on 0.2% LB agar plates. After 13 h incubation at 30°C, plates were photographed (top panel). ΔmotX served as a negative control. Bottom panel: Colony areas were measured using ImageJ software and used as proxy for motility. Box plots represent the colony area in pixels. Data were collected from 10 biological replicates and colony areas were normalized to that of ΔmotX. Statistical significance was determined using ordinary one-way ANOVA (Analysis of Variance) with Dunnett’s multiple comparison (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). (D) Heatmap of transcriptome analysis of ΔflaX in comparison to wild-type cells both carrying a control plasmid (pctrl) and in ΔflaX cells carrying a plasmid containing the flaAX genes (pflaAX) in comparison to ΔflaX cells with a control plasmid (pctrl). RNA-seq analysis was performed at OD600 of 0.2 and OD600 of 2.0. Genes involved in flagella synthesis are shown.
We next asked if expression of FlaX and/or ProQ also affected the motility of V. cholerae. To this end, we tested the swimming motility of V. cholerae wild-type, ΔproQ, and ΔflaX cells in semi-solid agar (0.2%). Indeed, we discovered that both proQ- and flaX-deficient mutants displayed reduced motility when compared to isogenic wild-type cells (Figure 3C). Conversely, plasmid-borne overexpression of either proQ or flaX in their respective mutant backgrounds resulted in increased motility. These data are in line with a previous report showing that FlaX expression modulates motility in V. cholerae (14).
To better understand the molecular underpinnings that control motility regulation by FlaX in V. cholerae, we employed RNA-seq analysis to compare the transcriptomes of wild-type cells with a ΔflaX mutant and a plasmid-complemented strain overexpressing FlaX. Again, we performed these experiments at low and high cell density (OD600 of 0.2 and 2.0, respectively), which revealed dozens of differentially expressed genes at both cell densities in ΔflaX and FlaX overexpressing cells (Figure 3D and Supplementary Figure S3A). We noticed that several of these genes were associated with motility-related functions and in several cases ΔflaX and FlaX overexpressing cells displayed inverse expression patterns (Figure 3D). For example, flaX mutation resulted in increased expression of the flaB-flaG transcript at both cell densities, whereas this pattern was reversed when FlaX was overexpressed. Indeed, we discovered that FlaX overexpression reduced the stability of the flaB mRNA, when compared to a control sample (Supplementary Figure S3B). Taken together, these results established a role for FlaX (and ProQ) in motility control in V. cholerae and suggested that regulation of mRNAs encoding components of the flagella apparatus might be relevant in this context.
Global ProQ-associated RNA interactome studies reveal FlaX interaction partners
ProQ has been shown to mediate changes in gene expression by affecting stability of its RNA ligands (23) or by promoting base-pairing between trans-encoded RNAs (25,40). To investigate the latter, we used RIL-seq to globally map the ProQ-associated RNA interactome in V. cholerae. RIL-seq relies on ligation of protein-bound RNA pairs and thereby directly captures and identifies interaction partners (47). Analogous to the RIP-Seq experiments outlined above, we performed these analyses at low and high cell densities (OD600 of 0.2 and 2.0) allowing us to recover 299 484 and 117 561 ProQ-associated interactions, respectively (Figure 4A and B and Supplementary Figure S4A). We quantified the respective interactions according to their genomic origin, i.e. mRNAs or sRNAs, which revealed that the majority (89%) of ProQ-associated interactions involved mRNAs, whereas only 3% of all interactions included sRNAs (8% of the interactions involved IGRs, Supplementary Figure S4B). To facilitate the accessibility of these results, we employed the ChimericFragments bioinformatics pipeline (48) and generated an interactive web interface displaying all ProQ-associated interactions and putative RNA duplex formation (https://vch-interactome.uni-jena.de/).
Figure 4.
RIL-seq analysis uncovers the RNA interactome of ProQ in V. cholerae. (A and B) Circos plot displaying top 500 RIL-seq chimeras at low cell density (OD600 of 0.2) (A) and high cell density (OD600 of 2.0) (B). Interaction hubs for sRNAs with highest interactions are highlighted. The first and second chromosomes are represented in dark green and light green, respectively. The Circos plot was created using the Circos component of the Dash Bio package. FlaX is marked in red. (C) Relative distribution of FlaX targets identified in the ProQ RIL-seq based on the number of interactions at low cell density (left) and high cell density (right). The total number of mRNA-FlaX interactions was normalized to 100% (low cell density: 5605 interactions, high cell density: 8894 interactions). The top 20 interaction partners are listed, while the remaining partners are grouped under others. (D) Validation of FlaX targets predicted by RIL-seq. Translational GFP reporter fusions were co-transformed into E. coli Top10 cells along with either a constitutive FlaX expression plasmid or an empty control plasmid. GFP expression was quantified by fluorescence intensity measurements, with fluorophore levels in the control strains normalized to 1. GFP levels were calculated from three biological replicates, with error bars representing the standard deviation. Statistical significance was assessed using Welch’s t-test (ns not significant, *P ≤ 0.05, **P ≤ 0.01). (E) Effect of FlaX on FlaB production was monitored. V. cholerae wild-type and ΔflaX strains each carrying a chromosomal 3xFLAG epitope at the C-terminus of FlaB and harboring the indicated plasmids were cultivated in LB medium. Protein and RNA samples were collected at the indicated growth cell densities. FlaB::3XFLAG protein levels were analyzed by western blot, while FlaX RNA levels were assessed by northern blot. RNAP and 5S rRNA were used as loading controls for the western and northern blots, respectively. Quantification of data from two biological replicates is provided in Supplementary Figure S4C.
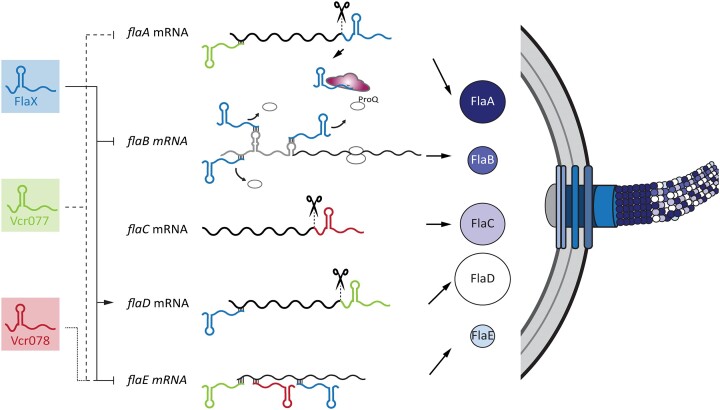
We next inspected the RIL-seq datasets for potential interaction partners of FlaX. Here, we discovered a total of 45 and 48 significant interactions (≥10 reads) at low and high cell densities, respectively (Figure 4C). The flaB flagellin mRNA was the most abundant FlaX partner transcript under both conditions followed by several other motility-related transcripts (fliD, flaG, flaD, flaE, flaC, flaA, flgL and flgE). To test if these transcripts are post-transcriptionally regulated by FlaX, we cloned the 5′-UTRs of these potential targets together with their initial coding sequence (expressed from a constitutive promoter) into a GFP-based reporter system and tested regulation by FlaX in E. coli (52). Five of the nine reporters showed significant regulation by FlaX: flaB, flaE, flaG and flaA were repressed, whereas flaD was induced (Figure 4D and Supplementary Figure S4D–G). Of note, flaB was most strongly inhibited by FlaX in these assays (∼ 500-fold), which prompted us to add a 3XFLAG epitope to the C-terminus of the chromosomal flaB gene and compare FlaB expression in wild-type, ΔflaX and flaX complemented cells by western blotting. Indeed, we discovered that FlaB levels were strongly increased under all growth conditions in the flaX mutant and that protein abundance was largely restored when FlaX was expressed from a plasmid (Figure 4E and Supplementary Figure S4C). These data show that FlaX acts post-transcriptionally to regulate genes involved in motility and that flaB is a main target of FlaX.
FlaX base-pairs with multiple target sites in the flaB mRNA
The mechanisms underlying gene regulation by base-pairing sRNAs in bacteria have been well-established for sRNAs interacting with Hfq (67). In contrast, how ProQ-binding sRNAs recognize and regulate trans-encoded target transcripts is less well understood (27,30,60). To address this question for FlaX in V. cholerae, we focused on flaB regulation as it was the most abundant interaction partner in our RIL-seq experiments (Figure 4C) and was strongly regulated by FlaX in our reporter assays (Figure 4D). Specifically, we performed structure probing analysis of the flaB 5′-UTR and added increasing concentrations of synthesized FlaX transcript to identify potential interactions sites in flaB (Figure 5A). We also performed the reverse experiments using radio-labeled FlaX and increasing concentrations of synthesized flaB 5′-UTR to determine the base-sequences in the sRNA (Supplementary Figure S5A). These experiments allowed us to determine the secondary structure of the flaB 5′-UTR and revealed three potential base-pairing sites in flaB and in FlaX (Figure 5B). Of note, all base-pairing sites in flaB (Supplementary Figure S5B) and FlaX (Supplementary Figure S2A) are highly conserved and analysis of the RIL-seq dataset using the ChimericFragments algorithm (48) predicted RNA duplex formation of FlaX with three separate binding sites in the flaB 5′-UTR, which we named flaB BS1-3 (Figure 5C–E). To confirm these predictions and to test the contribution of the individual base-pairing sites to regulation of flaB by FlaX, we mutated each of the corresponding sequences in flaB (as indicated in Figure 5C–E) and tested regulation by FlaX. We discovered that each mutation reduced repression of the flaB::GFP reporter by FlaX, however, no single mutation was sufficient to fully disrupt regulation (Figure 5F). Importantly, expression of FlaX variants carrying compensatory mutations for each of the sites restored regulation (Figure 5F and Supplementary Figure S5C). Therefore, we next generated flaB::GFP reporters carrying two (flaB1 + 2, flaB1 + 3 and flaB2 + 3) or three (flaB1 + 2 + 3) of the mutated sites. When compared to the single mutations, we observed that each additional mutation further reduced repression by FlaX and that mutation of all three sites fully abrogated regulation (Figure 5G and H). Again, expression of the relevant mutated FlaX variants (matching mutations flaB1 + 2, flaB1 + 3, flaB2 + 3 or flaB1 + 2 + 3) strongly enhanced repression of their respective reporters. Taken together, these results indicate that FlaX binds to three different base-pairing sites in the 5′-UTR of flaB that function together to post-transcriptionally control gene expression.
Figure 5.
FlaX base-pairs at three binding sites in the 5′-UTR of flaB mRNA. (A) In vitro structure probing of the 5′ end-labeled flaB 5′-UTR including the first 60 nucleotides of the CDS (0.4 pmol) was performed using RNase T1 (lanes 4–7), RNase V1 (lanes 8–11) and lead (II) (lanes 12–15) in the presence and absence of FlaX sRNA (0, 0.4 pmol, 4 pmol or 10 pmol, respectively). RNase T1 and alkaline (OH) ladders were used to map positions of individual nucleotides, with mapped G-residues indicated relative to the transcription start site (TSS). The potential binding sites for FlaX sRNA are indicated. (B) Secondary structure of the flaB 5′-UTR including the first 60 nucleotides of the CDS, as determined by bioinformatics predictions and revealed by chemical probing shown in (A). The FlaX binding sites are highlighted in blue. The start codon is highlighted in orange and the Shine-Dalgarno (SD) sequence is indicated by an orange line. (C–E) Predicted base-pairing interactions between FlaX and flaB mRNA. For flaB mRNA, positions are numbered relative to the start codon, while for FlaX, positions are numbered relative to its start site. The locations of nucleotide substitutions and the compensatory mutations in BS1 (C), BS2 (D) and BS3 (E) are indicated. (F–H) Fluorescence intensities were measured for E. coli strains carrying flaB translational GFP reporters, along with either a constitutive FlaX expression plasmid or an empty control plasmid. The fluorescence intensity of strains with the control plasmid was normalized to 1. Point mutations were individually introduced separately into each binding site in the 5′-UTR of flaB, with corresponding compensatory point mutations introduced in the FlaX sRNA (F). Fluorescence intensities for flaB translational GFP reporters with all combinations of double mutations (G) and triple mutations (H) were also measured. Data were collected from three biological replicates, and statistical analyses were conducted using Welch’s t-test (*P ≤ 0.05, **P ≤ 0.01).
Translation inhibition of flaB by FlaX involves sequestration of a ribosome standby site
The above data showed that FlaX acts post-transcriptionally on flaB using three base-pairing sequences (Figure 5C–E). Whereas base-pairing of FlaX with binding site 3 in flaB is likely to inhibit translation initiation by sequestration of the Shine-Dalgarno (SD) sequence, the role of binding sites 1 and 2 in this process remained unclear. Given that binding sites 1 and 2 are both located in single stranded regions and their high similarity with the SD consensus sequence (Figures 5B and 6A), we speculated that both sites could serve as ribosome standby-sites, known to increase translation efficiency of mRNAs (68). To test this idea, we deleted or mutated (exchanging BS1 and BS2 by consecutive cytosines; Figure 6A) each of the binding sites in the flaB::gfp reporter (Figure 6A) and monitored GFP levels. We discovered that all three deletions and mutations decreased GFP production. As expected, mutation of BS3 had the strongest effect (∼400-fold), as it disrupted the flaB SD sequence (Figure 6B). Deletion or mutation of BS2 reduced GFP expression by ∼8-fold followed by changes in BS1 (∼1.9 to 2.5-fold). In line with our previous data (Figure 5F–H), we found that all five mutated flaB::gfp variants were still repressed by FlaX, albeit to different levels (Supplementary Figure S6A). Taken together, these data suggest that binding sites 1 and 2 enhance translation of flaB and that base pairing of FlaX interferes with this process.
Figure 6.
FlaX inhibits ribosome binding in the 5′-UTR of the flaB mRNA. (A) The secondary structure of 5′-UTR flaB + first 60 nucleotides with FlaX binding sites highlighted in blue. Deletion of the different binding sites are marked in red. Residues that were substituted with cytosines (see panel B) are indicated by a red ‘C’ placed adjacent to each modified position. The start codon and the SD sequence are marked in orange. (B) Translational GFP reporter fusions with deletion mutations in each of the binding sites (as indicated in (A)) were transformed into E. coli Top10 cells. GFP production was measured, with fluorophore levels from the control strain normalized to 1. Error bars represent the standard deviation of three biological replicates. Statistical significance was determined using ordinary one-way ANOVA with Dunnett’s multiple comparison (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). (C and D) In vitro translation of the translational reporter fusion flaB::gfp mRNA. The protein was monitored by western blot at the indicated time points. Translation of single (C), double and triple mutants (D) (indicated in Figure 4C–E) was measured in the presence or absence of FlaX. Data were collected from two biological replicates. (E) Toeprint assay on flaB mRNA (0.2 pmol) in the presence or absence of FlaX. The presence of the 30S ribosomal subunit, the tRNAfMet and the flaB mRNA is indicated. Wild-type and mutant flaX are added in different ratios relative to the flaB mRNA (0.2 pmol, 0.6 pmol, 1 pmol and 2 pmol for flaX WT and 0.2 pmol and 0.6 pmol for flaX M1 + 2 + 3). Position of the 30S initiation complex is indicated, +15, relative to the start codon.
To better understand the mechanism underlying flaB translation and repression by FlaX, we employed in vitro translation assays using synthetically produced flaB::gfp and FlaX transcripts. Western blot analysis showed that FlaB:GFP protein was readily detectable after 10 min of incubation and gradually increased with time (Figure 6C and Supplementary Figure S6B and C). Addition of increasing concentrations of FlaX inhibited FlaB::GFP synthesis, further supporting the results from the in vivo assays (Figure 4D). We next synthesized the mutated variants of the flaB::gfp transcripts (carrying either mutations flaB BS1, 2 and 3 or the combined mutations; analogous to Figure 5F–H) and tested regulation by FlaX. Whereas none of the mutations impaired basal FlaB::GFP synthesis (Supplementary Figure S6C), we found that mutations 2 and 3 both reduced the effect of FlaX on flaB::gfp translation, whereas mutation of base-pairing site 1 had no significant impact (Figure 6C). Of note, mutation of binding site 2 had the strongest effect on the regulation of flaB::gfp by FlaX, which is in line with the corresponding in vivo data (Figure 5F). Combined mutation of base-pairing sites 1 and 2 almost fully abrogated translation repression of flaB::gfp by FlaX, and a similar pattern was observed when all three sites were mutated (Figure 6D).
Translation initiation in bacteria critically depends on binding of the 30S ribosome to the RBS and sRNAs often interfere with this process (69,70). Therefore, we employed toeprinting assays (71) to analyze the effect of FlaX on binding of the 30S ribosome to the flaB translation initiation region. In the presence of 30S ribosomes and fMet-tRNA, we detected a robust toeprint signal ∼15 nt upstream of the flaB start codon, demonstrating correct assembly of the translation initiation complex (Figure 6E). Addition of FlaX resulted in strong reduction of the toeprint signal, whereas the mutated FlaX variant carrying point mutations in the three flaB binding sites (FlaX-M1/2/3; see Figure 5E) failed to inhibit translation initiation (Figure 6E). We also tested the effect of the individual FlaX mutations in the toeprinting assays, and, vice versa, the impact of the flaB single point mutations on inhibition of by FlaX. Consistent with the experiments using the flaB::GFP reporter (Figure 5F–H) and the in vitro translation assays (Figure 6C and D), we discovered that mutation of binding site 2 in flaB (Supplementary Figure S6D) or FlaX (Supplementary Figure S6E) had the strongest impact on toeprint intensity, followed by binding site 3 and binding site 1. Based on these data, we propose that binding sites 1 and 2 could serve to recruit ribosomes to the flaB mRNA, which is inhibited by base pairing of FlaX.
FlaX affects flagella composition and the dynamics of flagella assembly
Flagella assembly in V. cholerae critically depends on the precise timing of flagella transcript synthesis that is organized into a four class regulon (Figure 3A and (12)). FlaX expression relies on RpoN and FlrA-C and thus belongs to the group of class III genes (Figure 3B). In contrast, flaB belongs to the class IV genes and thus is expressed later in the cascade together with other auxiliary flagellin genes, i.e.,flaC, flaD and flaE (72).
To examine if and how FlaX affects the dynamics of flagellin production in V. cholerae, we generated a flrA mutant and a flrA/flaX double mutant (blocking flagella gene induction and Figure 3A), which we complemented with a pBAD-flrA plasmid, allowing us to activate flagella gene expression through the addition of L-arabinose. We next followed the expression of all five flagellin genes (i.e. flaA, flaB, flaC, flaD and flaE) at various time-points after induction (0, 5, 15, 30 and 60 min) using real-time quantitative PCR. As expected, addition of L-arabinose to ΔflrA or ΔflrA/flaX cells carrying a control plasmid did not affect flaggelin mRNA expression (Figure 7A and B and Supplementary Figure S7A–D). Plasmid-borne production of FlrA in ΔflrA cells resulted in rapid activation of flaA mRNA reaching maximal expression already 15 min post induction (Figure 7A) and a similar pattern was observed for FlaX (Supplementary Figure S7A). In comparison, expression of flaB-E was delayed displaying highest levels 30 min (flaD, flaE) or 60 min after (flaB, flaC) after addition of the inducer (Figure 7B and Supplementary Figure S7B–D). In the flrA/flaX double mutant, the dynamics of flaA, flaC, flaD and flaE expression were similar to the patterns observed in ΔflrA cells; however, expression of flaB was significantly accelerated showing strong activation already after 15 min of induction and maximal expression after 30 min (Figure 7B).
Figure 7.
FlaX influences flagella assembly and composition. (A and B) V. cholerae ΔflrA and ΔflrA ΔflaX strains carrying either a control plasmid (pctrl) or pBAD-flrA plasmid with inducible promoter, were grown in LB medium at 37°C to OD600 of 1.0. pBAD-driven gene expression was next induced by adding L-arabinose (0.2% final concentration). mRNA levels of flaA (A) and flaB (B) were monitored by quantitative real-time PCR over time: 0, 5, 15, 30 and 60 min. All time points were compared to the pre-induction condition (0 min) and transcript abundance was set to 100. Data were collected from three biological replicates and normalized to the house-keeping gene, recA. (C) Mass spectrometry analysis of FlaA-E flagellin protein levels in wild-type, ΔflaX and ΔflaX pflaX cells. The relative abundances of each flagellin, based on spectral counts, are represented as stacked plot. (D) Motility assay on 0.2% LB agar was performed using inocula from wild-type, ΔflaX and ΔflaXΔflaB mutant cells carrying either a control plasmid (pctrl) or a plasmid with the sRNA (pflaX). The ΔmotX mutant was used as a negative control. Colony areas, measured in pixels via ImageJ, served as a motility proxy. Box plots show normalized colony areas from six replicates. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparisons (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
The above experiments suggested that FlaX mainly acts on flaB and that lack of flaX resulted in elevated flaB mRNA (Figures 3D and 7B) and FlaB protein levels (Figure 4E). Based on these results, we hypothesized that FlaX might modulate flagella filament composition, which could also explain the differences in motility observed for V. cholerae cells lacking or overexpressing FlaX (Figure 3C). To test this possibility, we isolated flagellin proteins from V. cholerae and quantified their relative abundances using mass spectrometry. We discovered that in wild-type cells, FlaD (∼41%), FlaA (∼30%) and FlaC (∼26%) were the most abundant flagellin proteins, whereas FlaB (∼3%) and FlaE (<1%) constituted only minor flagella components (Figure 7C). In contrast, in ΔflaX cells relative FlaB levels were strongly upregulated (constituting ∼30% of all flagellin proteins), whereas the abundance of FlaA was reduced to ∼14%. Plasmid-borne overexpression of FlaX in flaX-deficient cells restored and further reduced FlaB (∼1.5%) but also inhibited FlaA (11%) and increased FlaD (62%) levels (Figure 7C). The relative abundances of FlaC (∼25%) and FlaE (<1%) levels remained constant in all three strains. Taken together, these results suggest that FlaX not only affects the regulatory pattern associated with flagellin mRNA expression, but also the composition of the flagellum filament.
To correlate with these findings with the role of FlaX in motility of V. cholerae (Figure 3C), we next investigated the role of FlaB. Specifically, we speculated that increased FlaB levels (Figure 4E) and altered flagella composition (Figure 7C) in flaX-deficient cells might explain their reduced motility (Figure 3C). To test this idea, we deleted the flaB gene in ΔflaX cells and quantified motility in semi-solid agar (0.2%). Indeed, mutation of flaB strongly increased motility of the flaX mutant, which was comparable to cells over-expressing FlaX (Figure 7D). Likewise, plasmid-borne over-expression of FlaX no longer increased motility in cells lacking flaB. These results suggest that elevated FlaB levels inhibit motility of V. cholerae and that repression of flaB by FlaX can antagonize this effect.
Discussion
Flagella play crucial roles in bacterial motility and behavior (73,74). Flagella synthesis is a highly coordinated process involving multiple layers of regulation to ensure proper assembly and function. Over the past decades, we have learned a great deal about how flagella assembly is organized into hierarchical transcriptional programs that coordinate the order and timing of flagella synthesis (75,76). More recently, post-transcriptional gene regulation has also been implicated in flagella synthesis, including bacteria with peritrichous flagella, for example,E. coli (16–18), as well as amphitrichous bacteria, for example,Campylobacter jejuni (19). In this study, we discovered that post-transcriptional regulation is also relevant for motility and flagella synthesis in monotrichous bacteria, i.e.,V. cholerae.
Flagella synthesis in V. cholerae is organized into four classes of which flaA is the only class III flagellin gene, whereas flaB-E belong to class IV (Figure 3A). FlaA is also the only flagellin subunit that is required and sufficient for motility and this phenotype has been linked to a critical domain in FlaA that supports folding of the additional flagellin proteins (15,72,77). Thus, it is crucial that FlaA is produced prior to FlaB-E; however, previous data showed that the transition from class III to class IV gene expression takes place prior to the production and integration of FlaA into the filament (72). This organization demands for an additional regulatory element ensuring sufficient FlaA production before the other flagellin proteins are expressed. Our data suggest that FlaX could close this gap by inhibiting FlaB, which negatively affects motility (Figure 7D). We speculate that increased FlaB could reduce motility in two ways: first, a change in the ratio of FlaA versus FlaB proteins could impair or exceed the chaperone function of FlaA. Second, altered filament composition (Figure 7C) that impairs flagella stability and function. Indeed, using scanning electron microscopy, we discovered that ΔflaX mutants display elevated numbers of unflagellated cells, when compared to isogenic wild-type cells (Supplementary Figure S7E). In addition, timing of flagellin gene expression is likely crucial for correct filament assembly as mutation of flaX resulted in premature flaB expression when FlrA is induced (Figure 7C). It is currently unknown if V. cholerae employs differential filament composition to modify its motility in response to changing environmental conditions (12). Thus, it is noteworthy that FlaX overexpression resulted in enhanced motility (Figure 3C), suggesting that altered flagellin expression and filament composition (Figures 4D and 7D) can indeed modulate the motility behavior of V. cholerae. This regulatory concept might be relevant when V. cholerae transmits from aquatic to more viscous environments, such as the mucus-containing intestinal lining of the human host (78).
Other Vibrio strains also employ flagella consisting of several flagellins (12,79). The flaX gene is conserved in many Vibrios (Supplementary Figure S2A), suggesting that similar regulatory mechanisms for flagella synthesis could be relevant here as well. For example, in Vibrio parahaemolyticus and Vibrio campbellii flaX is located in the 3′-UTR of the mRNAs encoding the FlaC polar flagellin, which is homologous to FlaA of V. cholerae (80,81). The homolog of FlaB in V. parahaemolyticus and V. campbellii is FlaA and indeed the FlaX binding sites in the 5′-UTRs of their corresponding mRNAs are highly conserved (Supplementary Figure S5B). Of note, in both organisms, all flagellin genes are categorized as class IV genes, suggesting that post-transcriptional regulation could be the primary mechanism to establish ordered flagellin production.
Expression of flaX is inherently linked to flaA as both genes are transcribed from the same promoter (Figure 2A). Following transcription, accumulation of FlaX requires cleavage by RNase E (Figure 2D), a regulatory principle that has been documented for various Hfq-dependent sRNAs, as well as the ProQ-associated RaiZ sRNA (64). The recently discovered E. coli sRNAs UhpU, MotR, FliX and FlgO, whose expression is controlled by the flagella sigma factor σ28, are also encoded in the UTRs of their associated genes and also modulate motility (16). Our list of ProQ-associated sRNAs contains a large fraction of 3′-UTR-derived sRNAs (Figure 1D and Supplementary Figure S1E), including Vcr077 and Vcr078 (Figure 1C), which are located in the 3′-UTRs of the flaD and flaC flagellin genes, respectively. Both sRNAs share sequence similarity with FlaX (Supplementary Figure S7F) and are produced by ribonucleolytic cleavage (62). Inspection of their respective target spectra using our ProQ-dependent RIL-Seq datasets (Figure 4A and B) revealed dozens of putative target transcript including flagellin and other motility-associated genes. We tested several of these predictions using our GFP-based reporter system (Figure 4D). Indeed, we discovered that Vcr077 and Vcr078 both inhibited FlaE::GFP expression and Vcr077 also inhibited FlaA::GFP, whereas Vcr078 negatively affected FlaC::GFP levels (Supplementary Figure S7G). These findings might point to a more general regulatory concept in which flagellin mRNAs employ 3′-UTR-derived regulatory sRNAs to inhibit the expression of other flagellins (Figure 8). This process could benefit their own expression and might reinforce and extend transcriptional networks coordinating flagella synthesis.
Figure 8.
Model of FlaX-mediated regulation of flagella synthesis in V. cholerae. Flagella filaments in V. cholerae encompass five flagellins, of which only FlaA is a class III gene. FlaX is an sRNA processed by RNase E from the 3′-end of flaA and it is stabilized by ProQ. Its expression is controlled by FlrA, FlrB, FlrC and RpoN rendering it a class III gene as well. FlaX controls tightly the translation of flaB among other flagellins by base-pairing at three different positions in the 5′-UTR. By doing so, FlaX acts to reinforce the hierarchical order between class III and class IV genes. Thereby, FlaX controls motility by fine-tuning the composition of the flagella filament.
Altered expression of motility genes has been linked to ProQ-mediated regulation in other species as well (36) and mutation of proQ has been documented to cause various other phenotypes (27,30); however, with the exception of FlaX reported here, these have not yet been linked to individual sRNA regulators. This is contrary to the numerous phenotypes described for Hfq-dependent sRNAs from diverse microorganisms (20,67,82). Based on our data from V. cholerae, we speculate there could be two main reasons for this discrepancy. First, mutation of hfq in E. coli and related bacteria typically results in a complete loss of sRNA activity, whereas mutation of proQ also affects regulation, although to a minor extend (25,40). In accordance, we found that repression of FlaB::GFP by FlaX in a proQ mutant was only mildly reduced (∼2.5-fold) and a similar effect was detected for cells lacking the genes encoding both major RNA chaperones ProQ and Hfq (Supplementary Figure S7H). Thus, the phenotypes associated with ΔproQ cells, might not overlap with the phenotypes of individual sRNA mutants. Second, when comparing the fraction of recovered RNA duplexes according to their genomic origin, i.e., mRNAs and sRNAs, in our ProQ (Figure 4A and B) and Hfq (46) RIL-Seq datasets, we discovered that the vast majority (91%) of ProQ-associated interactions involved base-pairing between two mRNAs, whereas only 9% of all interactions included sRNAs (Supplementary Figure S7I). In contrast, the relative abundance of sRNAs in Hfq-mediated interactions was significantly higher (∼76%). Of note, the mRNA–mRNA pairs in the ProQ dataset frequently involved interactions within one transcript (e.g. between two mRNAs in same operon), suggesting that ProQ could play a role in intramolecular base pairing, whereas Hfq mainly facilitates interaction between trans-encoded transcripts.
At the mechanistic level, we speculate that strong translation of flaB is associated with two potential ribosome standby sites in the 5′-UTR of the mRNA that feed new ribosomes to the RBS. Ribosome standby sites are found in single-stranded regions of 5′-UTRs, where the 30S ribosomal subunit can bind and kinetically compete with RNA folding to reach the downstream RBS (83,84). This facilitates the recruitment of new ribosomes to mRNAs when the genuine start site is still occupied by the leading ribosome resulting in polysome formation and increased translation efficiency (68). Arguably one of the most thoroughly studied ribosome standby site is located in the distal 5′-UTR of the tisB toxin mRNA (85,86). In analogy to the FlaX–flaB interaction reported here, translation of tisB is facilitated by a ribosome standby site in the 5′-UTR, which in turn is repressed by the IstR antisense RNA (87–89). In contrast to FlaX, IstR is expressed from a genomic loci adjacent to the tisB gene and although IstR also binds to ProQ (24), the sRNA does not seem to regulate target mRNAs other than tisB.
The regulatory mechanism most frequently associated with sRNAs in bacteria involves base pairing with and sequestration of the target’s RBS resulting in reduced translation initiation, which is often followed by mRNA degradation (21). This mode of action has been documented for dozens of sRNA–target pairs requiring Hfq and has also been reported for ProQ (40). In few cases, full repression of an mRNA by an sRNA requires multiple target sites in the mRNA, as has been demonstrated for regulation of the manXYZ mRNA by SgrS in E. coli (90,91). However, in this case the individual target sites in manXYZ are separated by several hundred nucleotides and target two separate translation initiation sites of a polycistronic mRNA. In contrast, we discovered here that regulation of flaB by FlaX involves three consecutive binding sites in the mRNA’s 5′-UTR to reach full repression (Figure 5D–F). It is noteworthy that repression of FlaB::GFP by FlaX is unusually strong (∼500-fold; Figure 4D) when compared to other FlaX target genes (Figure 4D) and nearly all previously reported sRNA–target interactions using analogous experimental approaches. It is likely that the ribosome standby sites in the flaB 5′-UTR support efficient translation of mRNA, which could increase the dynamic range for post-transcriptional regulation. Importantly, this finding also sets the stage for follow-up genetic screens aiming to identify auxiliary regulatory factors involved in ProQ-mediated gene control.
Supplementary Material
Acknowledgements
We thank Andreas Starick and Yvonne Greiser for excellent technical support and the NICHD Molecular Genomics Core (Tianwei Li) for the sequencing of the RIL-seq libraries. We thank Gisela Storz and Jörg Vogel for comments on the manuscript and all members of the Papenfort lab for insightful discussions and suggestions.
Contributor Information
Rabea Ghandour, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany.
Daniel Devlitsarov, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany.
Phillip Popp, Humboldt-Universität zu Berlin, Institute for Biology, 10115 Berlin, Germany.
Sahar Melamed, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, Faculty of Medicine, The Hebrew University of Jerusalem, Jerusalem, Israel.
Michaela Huber, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany.
Malte Siemers, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany; Microverse Cluster, Friedrich Schiller University Jena, 07743 Jena, Germany.
Thomas Krüger, Department of Molecular and Applied Microbiology, Leibniz Institute for Natural Product Research and Infection Biology (Leibniz-HKI), 07745 Jena, Germany.
Olaf Kniemeyer, LMU Munich Biocenter, Ludwig-Maximilian-University of Munich, 82152 Munich, Germany.
Andreas Klingl, LMU Munich Biocenter, Ludwig-Maximilian-University of Munich, 82152 Munich, Germany.
Axel A Brakhage, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany; Microverse Cluster, Friedrich Schiller University Jena, 07743 Jena, Germany; Department of Molecular and Applied Microbiology, Leibniz Institute for Natural Product Research and Infection Biology (Leibniz-HKI), 07745 Jena, Germany.
Marc Erhardt, Humboldt-Universität zu Berlin, Institute for Biology, 10115 Berlin, Germany.
Kai Papenfort, Friedrich Schiller University, Institute of Microbiology, 07743 Jena, Germany; Microverse Cluster, Friedrich Schiller University Jena, 07743 Jena, Germany.
Data availability
The sequencing data from the RIL-seq experiment is deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession code ‘GSE275761’. The visualization for the V. cholerae ProQ RIL-seq dataset is accessible at https://vch-interactome.uni-jena.de/. The RIP-seq and RNA-seq sequencing data are available under the GEO accessions ‘GSE275750’ and ‘GSE275822’, respectively. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD055294. Further raw and processed data supporting the conclusions of this study are available upon request from the corresponding author.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
DFG [PA2820/7-1—Project‐ID 544846468, EXC 2051—Project‐ID 390713860 to K.P.]; Vallee Foundation (to K.P.); DFG [project Z2, project number 210879364 to A.B.]; European Research Council [CoG-101088027 to K.P.]. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
References
- 1. Kanungo S., Azman A.S., Ramamurthy T., Deen J., Dutta S.. Cholera. Lancet. 2022; 399:1429–1440. [DOI] [PubMed] [Google Scholar]
- 2. Guentzel M.N., Berry L.J.. Motility as a virulence factor for Vibrio cholerae. Infect. Immun. 1975; 11:890–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gardel C.L., Mekalanos J.J.. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 1996; 64:2246–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butler S.M., Camilli A.. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 2005; 3:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z., Miyashiro T., Tsou A., Hsiao A., Goulian M., Zhu J.. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl Acad. Sci. U.S.A. 2008; 105:9769–9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Millet Y.A., Alvarez D., Ringgaard S., von Andrian U.H., Davis B.M., Waldor M.K.. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014; 10:e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Syed K.A., Beyhan S., Correa N., Queen J., Liu J., Peng F., Satchell K.J., Yildiz F., Klose K.E.. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 2009; 191:6555–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dongre M., Singh B., Aung K.M., Larsson P., Miftakhova R., Persson K., Askarian F., Johannessen M., von Hofsten J., Persson J.L.et al.. Flagella-mediated secretion of a novel Vibrio cholerae cytotoxin affecting both vertebrate and invertebrate hosts. Commun. Biol. 2018; 1:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bennett R.R., Lee C.K., De Anda J., Nealson K.H., Yildiz F.H., O’Toole G.A., Wong G.C., Golestanian R.. Species-dependent hydrodynamics of flagellum-tethered bacteria in early biofilm development. J. R. Soc. Interface. 2016; 13:20150966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Utada A.S., Bennett R.R., Fong J.C.N., Gibiansky M.L., Yildiz F.H., Golestanian R., Wong G.C.L.. Vibrio cholerae use pili and flagella synergistically to effect motility switching and conditional surface attachment. Nat. Commun. 2014; 5:4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanchez S., Ng W.L.. Motility control as a possible link between quorum sensing to surface attachment in Vibrio species. Adv. Exp. Med. Biol. 2023; 1404:65–75. [DOI] [PubMed] [Google Scholar]
- 12. Echazarreta M.A., Klose K.E.. Vibrio flagellar synthesis. Front. Cell. Infect. Microbiol. 2019; 9:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lloyd C.J., Klose K.E.. The Vibrio polar flagellum: structure and regulation. Adv. Exp. Med. Biol. 2023; 1404:77–97. [DOI] [PubMed] [Google Scholar]
- 14. Dong T.G., Mekalanos J.J.. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 2012; 40:7766–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Echazarreta M.A., Kepple J.L., Yen L.H., Chen Y., Klose K.E.. A critical region in the FlaA Flagellin facilitates filament formation of the Vibrio choleraeflagellum. J. Bacteriol. 2018; 200:e00029-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Melamed S., Zhang A., Jarnik M., Mills J., Silverman A., Zhang H., Storz G.. Sigma(28)-dependent small RNA regulation of flagella biosynthesis. eLife. 2023; 12:RP87151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomason M.K., Fontaine F., De Lay N., Storz G.. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012; 84:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Lay N., Gottesman S.. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012; 86:524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. König F., Svensson S.L., Sharma C.M.. Interplay of two small RNAs fine-tunes hierarchical flagella gene expression in Campylobacter jejuni. Nat. Commun. 2024; 15:5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Papenfort K., Melamed S.. Small RNAs, large networks: posttranscriptional regulons in gram-negative bacteria. Annu. Rev. Microbiol. 2023; 77:23–43. [DOI] [PubMed] [Google Scholar]
- 21. Wagner E.G.H., Romby P.. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015; 90:133–208. [DOI] [PubMed] [Google Scholar]
- 22. Vogel J., Luisi B.F.. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011; 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Holmqvist E., Li L., Bischler T., Barquist L., Vogel J.. Global maps of ProQ bindinginvivo reveal target recognition via RNA structure and stability control at mRNA 3′ ends. Mol. Cell. 2018; 70:971–982. [DOI] [PubMed] [Google Scholar]
- 24. Smirnov A., Forstner K.U., Holmqvist E., Otto A., Gunster R., Becher D., Reinhardt R., Vogel J.. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melamed S., Adams P.P., Zhang A., Zhang H., Storz G.. RNA-RNA interactomes of ProQ and hfq reveal overlapping and competing roles. Mol. Cell. 2020; 77:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quendera A.P., Seixas A.F., Dos Santos R.F., Santos I., Silva J.P.N., Arraiano C.M., Andrade J.M.. RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front Mol. Biosci. 2020; 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Olejniczak M., Storz G.. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?. Mol. Microbiol. 2017; 104:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gonzalez G.M., Hardwick S.W., Maslen S.L., Skehel J.M., Holmqvist E., Vogel J., Bateman A., Luisi B.F., Broadhurst R.W.. Structure of the Escherichia coli ProQ RNA-binding protein. RNA. 2017; 23:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Attaiech L., Glover J.N.M., Charpentier X.. RNA chaperones step out of Hfq’s shadow. Trends Microbiol. 2017; 25:247–249. [DOI] [PubMed] [Google Scholar]
- 30. Holmqvist E., Berggren S., Rizvanovic A.. RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. Acta Gene Regul. Mech. 2020; 1863:194596. [DOI] [PubMed] [Google Scholar]
- 31. Kim H.J., Black M., Edwards R.A., Peillard-Fiorente F., Panigrahi R., Klingler D., Eidelpes R., Zeindl R., Peng S., Su J.et al.. Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones. Nat. Commun. 2022; 13:7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bauriedl S., Gerovac M., Heidrich N., Bischler T., Barquist L., Vogel J., Schoen C.. The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat. Commun. 2020; 11:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El Mouali Y., Gerovac M., Mineikaite R., Vogel J.. In vivo targets of Salmonella FinO include a FinP-like small RNA controlling copy number of a cohabitating plasmid. Nucleic Acids Res. 2021; 49:5319–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Durieux I., Ginevra C., Attaiech L., Picq K., Juan P.A., Jarraud S., Charpentier X.. Diverse conjugative elements silence natural transformation in Legionella species. Proc. Natl Acad. Sci. U.S.A. 2019; 116:18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rizvanovic A., Michaux C., Panza M., Iloglu Z., Helaine S., Wagner E.G.H., Holmqvist E.. The RNA-binding protein ProQ promotes antibiotic persistence in Salmonella. mBio. 2022; 13:e0289122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Westermann A.J., Venturini E., Sellin M.E., Forstner K.U., Hardt W.D., Vogel J.. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica Serovar typhimurium. mBio. 2019; 10:e02504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nichols R.J., Sen S., Choo Y.J., Beltrao P., Zietek M., Chaba R., Lee S., Kazmierczak K.M., Lee K.J., Wong A.et al.. Phenotypic landscape of a bacterial cell. Cell. 2011; 144:143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheidy D.T., Zielke R.A.. Analysis and expansion of the role of the Escherichia coliprotein ProQ. PLoS One. 2013; 8:e79656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bergman S., Andresen L., Kjellin J., Martinez Burgo Y., Geiser P., Baars S., Soderbom F., Sellin M.E., Holmqvist E.. ProQ-dependent activation of Salmonella virulence genes mediated by post-transcriptional control of PhoP synthesis. mSphere. 2024; 9:e0001824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smirnov A., Wang C., Drewry L.L., Vogel J.. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 2017; 36:1029–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yuan X., Eldred L.I., Kharadi R.R., Slack S.M., Sundin G.W.. The RNA-binding protein ProQ impacts exopolysaccharide biosynthesis and second messenger cyclic di-GMP signaling in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 2022; 88:e0023922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gulliver E.L., Sy B.M., Wong J.L., Deveson Lucas D.S., Powell D.R., Harper M., Tree J.J., Boyce J.D.. The role and targets of the RNA-binding protein ProQ in the gram-negative bacterial pathogen Pasteurella multocida. J. Bacteriol. 2022; 204:e0059221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leonard S., Villard C., Nasser W., Reverchon S., Hommais F.. RNA chaperones hfq and ProQ play a key role in the virulence of the plant pathogenic bacterium Dickeya dadantii. Front. Microbiol. 2021; 12:687484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huber M., Frohlich K.S., Radmer J., Papenfort K.. Switching fatty acid metabolism by an RNA-controlled feed forward loop. Proc. Natl Acad. Sci. U.S.A. 2020; 117:8044–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sprenger M., Siemers M., Krautwurst S., Papenfort K.. Small RNAs direct attack and defense mechanisms in a quorum sensing phage and its host. Cell Host Microbe. 2024; 32:727–738. [DOI] [PubMed] [Google Scholar]
- 46. Huber M., Lippegaus A., Melamed S., Siemers M., Wucher B.R., Hoyos M., Nadell C., Storz G., Papenfort K.. An RNA sponge controls quorum sensing dynamics and biofilm formation in Vibrio cholerae. Nat. Commun. 2022; 13:7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Melamed S., Peer A., Faigenbaum-Romm R., Gatt Y.E., Reiss N., Bar A., Altuvia Y., Argaman L., Margalit H.. Global mapping of small RNA-target interactions in bacteria. Mol. Cell. 2016; 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siemers M., Lippegaus A., Papenfort K.. ChimericFragments: computation, analysis and visualization of global RNA networks. NAR Genom. Bioinform. 2024; 6:lqae035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoyos M., Huber M., Forstner K.U., Papenfort K.. Gene autoregulation by 3′ UTR-derived bacterial small RNAs. eLife. 2020; 9:e58836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Culviner P.H., Guegler C.K., Laub M.T.. A simple, cost-effective, and robust method for rRNA depletion in RNA-sequencing studies. mBio. 2020; 11:e00010-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peschek N., Herzog R., Singh P.K., Sprenger M., Meyer F., Frohlich K.S., Schroger L., Bramkamp M., Drescher K., Papenfort K.. RNA-mediated control of cell shape modulates antibiotic resistance in Vibrio cholerae. Nat. Commun. 2020; 11:6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Herzog R., Peschek N., Frohlich K.S., Schumacher K., Papenfort K.. Three autoinducer molecules act in concert to control virulence gene expression in Vibrio cholerae. Nucleic Acids Res. 2019; 47:3171–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vogt L.N., Panis G., Schäpers A., Peschek N., Huber M., Papenfort K., Viollier P.H., Fröhlich K.S.. Genome-wide profiling of hfq-bound RNAs reveals the iron-responsive small RNA RusT in Caulobacter crescentus. mBio. 2024; 15:e0315323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruhland E., Siemers M., Gerst R., Spath F., Vogt L.N., Figge M.T., Papenfort K., Fröhlich K.S.. The global RNA-RNA interactome of Klebsiella pneumoniae unveils a small RNA regulator of cell division. Proc. Natl Acad. Sci. U.S.A. 2024; 121:e2317322121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Papenfort K., Podkaminski D., Hinton J.C., Vogel J.. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl Acad. Sci. U.S.A. 2012; 109:E757–E764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M.. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006; 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 57. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988; 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R.. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mann M., Wright P.R., Backofen R.. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017; 45:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liao Z., Smirnov A.. FinO/ProQ-family proteins: an evolutionary perspective. Biosci. Rep. 2023; 43:BSR20220313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J.et al.. Pfam: the protein families database. Nucleic Acids Res. 2014; 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Papenfort K., Forstner K.U., Cong J.P., Sharma C.M., Bassler B.L.. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl Acad. Sci. U.S.A. 2015; 112:E766–E775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Broglia L., Le Rhun A., Charpentier E.. Methodologies for bacterial ribonuclease characterization using RNA-seq. FEMS Microbiol. Rev. 2023; 47:fuad049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ponath F., Hör J., Vogel J.. An overview of gene regulation in bacteria by small RNAs derived from mRNA 3′ ends. FEMS Microbiol. Rev. 2022; 46:fuac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chao Y., Li L., Girodat D., Forstner K.U., Said N., Corcoran C., Smiga M., Papenfort K., Reinhardt R., Wieden H.J.et al.. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell. 2017; 65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Menendez-Gil P., Toledo-Arana A.. Bacterial 3′UTRs: a useful resource in post-transcriptional regulation. Front. Mol. Biosci. 2020; 7:617633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kavita K., de Mets F., Gottesman S.. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018; 42:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Andreeva I., Belardinelli R., Rodnina M.V.. Translation initiation in bacterial polysomes through ribosome loading on a standby site on a highly translated mRNA. Proc. Natl Acad. Sci. U.S.A. 2018; 115:4411–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodnina M.V. Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 2018; 10:a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Desnoyers G., Bouchard M.P., Masse E.. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013; 29:92–98. [DOI] [PubMed] [Google Scholar]
- 71. Yakhnin H., Babitzke P.. Toeprint assays for detecting RNA structure and protein-RNA interactions. Methods Mol. Biol. 2022; 2516:305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klose K.E., Mekalanos J.J.. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 1998; 180:303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grognot M., Taute K.M.. More than propellers: how flagella shape bacterial motility behaviors. Curr. Opin. Microbiol. 2021; 61:73–81. [DOI] [PubMed] [Google Scholar]
- 74. Wadhwa N., Berg H.C.. Bacterial motility: machinery and mechanisms. Nat. Rev. Micro. 2022; 20:161–173. [DOI] [PubMed] [Google Scholar]
- 75. Osterman I.A., Dikhtyar Y.Y., Bogdanov A.A., Dontsova O.A., Sergiev P.V.. Regulation of flagellar gene expression in bacteria. Biochemistry. 2015; 80:1447–1456. [DOI] [PubMed] [Google Scholar]
- 76. Subramanian S., Kearns D.B.. Functional regulators of bacterial flagella. Annu. Rev. Microbiol. 2019; 73:225–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Prouty M.G., Correa N.E., Klose K.E.. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 2001; 39:1595–1609. [DOI] [PubMed] [Google Scholar]
- 78. Grognot M., Mittal A., Mah’moud M., Taute K.M.. Vibrio cholerae motility in aquatic and mucus-mimicking environments. Appl. Environ. Microb. 2021; 87:e0129321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Khan F., Tabassum N., Anand R., Kim Y.M.. Motility of Vibrio spp.: regulation and controlling strategies. Appl. Microbiol. Biotechnol. 2020; 104:8187–8208. [DOI] [PubMed] [Google Scholar]
- 80. Kim Y.K., McCarter L.L.. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 2000; 182:3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Petersen B.D., Liu M.S., Podicheti R., Yang A.Y., Simpson C.A., Hemmerich C., Rusch D.B., van Kessel J.C.. The polar flagellar transcriptional regulatory network in Vibrio campbellii deviates from canonical Vibrio species. J. Bacteriol. 2021; 203:e0027621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Holmqvist E., Wagner E.G.H.. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017; 45:1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sterk M., Romilly C., Wagner E.G.H.. Unstructured 5′-tails act through ribosome standby to override inhibitory structure at ribosome binding sites. Nucleic Acids Res. 2018; 46:4188–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. de Smit M.H., van Duin J.. Translational standby sites: how ribosomes may deal with the rapid folding kinetics of mRNA. J. Mol. Biol. 2003; 331:737–743. [DOI] [PubMed] [Google Scholar]
- 85. Romilly C., Lippegaus A., Wagner E.G.H.. An RNA pseudoknot is essential for standby-mediated translation of the tisB toxin mRNA in Escherichia coli. Nucleic Acids Res. 2020; 48:12336–12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Romilly C., Deindl S., Wagner E.G.H.. The ribosomal protein S1-dependent standby site in tisB mRNA consists of a single-stranded region and a 5′ structure element. Proc. Natl Acad. Sci. U.S.A. 2019; 116:15901–15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Darfeuille F., Unoson C., Vogel J., Wagner E.G.. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007; 26:381–392. [DOI] [PubMed] [Google Scholar]
- 88. Edelmann D., Oberpaul M., Schaberle T.F., Berghoff B.A.. Post-transcriptional deregulation of the tisB/istR-1 toxin-antitoxin system promotes SOS-independent persister formation in Escherichia coli. Environ. Microbiol. Rep. 2021; 13:159–168. [DOI] [PubMed] [Google Scholar]
- 89. Vogel J., Argaman L., Wagner E.G., Altuvia S.. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004; 14:2271–2276. [DOI] [PubMed] [Google Scholar]
- 90. Rice J.B., Balasubramanian D., Vanderpool C.K.. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc. Natl Acad. Sci. U.S.A. 2012; 109:E2691–E2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Azam M.S., Vanderpool C.K.. Translation inhibition from a distance: the small RNA SgrS silences a ribosomal protein S1-dependent enhancer. Mol. Microbiol. 2020; 114:391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data from the RIL-seq experiment is deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession code ‘GSE275761’. The visualization for the V. cholerae ProQ RIL-seq dataset is accessible at https://vch-interactome.uni-jena.de/. The RIP-seq and RNA-seq sequencing data are available under the GEO accessions ‘GSE275750’ and ‘GSE275822’, respectively. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD055294. Further raw and processed data supporting the conclusions of this study are available upon request from the corresponding author.