Abstract
We identified a truncated allele of dam1 as a multicopy suppressor of the sensitivity of cdc13-117 (cyclin B) and mal3-1 (EB-1) cells to thiabendazole, a microtubule poison. We find that Dam1 binds to the plus end of spindle microtubules and kinetochores as cells enter mitosis and this is dependent on other components of the fission yeast DASH complex, including Ask1, Duo1, Spc34 and Dad1. By contrast, Dad1 remains bound to kinetochores throughout the cell cycle and its association is dependent on the Mis6 and Mal2, but not Mis12, Nuf2 or Cnp1, kinetochore proteins. In cells lacking Dam1, or other components of the DASH complex, anaphase is delayed due to activation of the spindle assembly checkpoint and lagging sister chromatids are frequently observed and occasionally sister chromatid pairs segregate to the same spindle pole. We find that the mitotic centromere-associated Klp5/Klp6 kinesin complex is essential in cells lacking components of the DASH complex. Cells lacking both Dam1 and Klp5 undergo a first cell cycle arrest in mitosis due to a failure to establish bipolar chromosome attachment.
Keywords: Dam1, DASH, kinetochore, microtubule, Schizosaccharomyces pombe
Introduction
In order for each set of sister chromatids to be separated to opposite spindle poles, each kinetochore must be bound to microtubule(s) from one pole and its sister kinetochore to microtubule(s) from the opposite pole, a process known as chromosome biorientation. The mechanism governing the establishment of chromosome biorientation has been examined in some detail in budding yeast, in which each kinetochore is bound by only a single microtubule (Winey et al, 1995). As chromosomes are replicated, both sister chromatids are frequently captured by spindle microtubules from the same pole, a configuration known as syntelic attachment. In this situation, tension cannot be applied across the mitotic spindle (Dewar et al, 2004). As cells enter mitosis, the lack of spindle tension is sensed at the kinetochore by the Aurora-like Ipl1 kinase, which triggers microtubule detachment so that each sister chromatid pair can be attached to spindle microtubules from opposite poles (amphitelic attachment) (Tanaka et al, 2002; Dewar et al, 2004). Ipl1 phosphorylates a number of microtubule binding proteins and kinetochore-associated proteins in vitro (Cheeseman et al, 2002; Tanaka et al, 2002; Shang et al, 2003). One of these, Dam1, is particularly important for correcting improper chromosome attachment because a dam1 allele that mimics its constitutive phosphorylation partially suppresses the lethality of a temperature-sensitive ipl1 mutant (Cheeseman et al, 2002). Saccharomyces cerevisiae Dam1 is a subunit of a 10-component DASH complex, which forms both rings and spiral structures on microtubules in vitro (Miranda et al, 2005; Westermann et al, 2005). Components of the budding yeast DASH complex, including Dam1, Duo1, Spc34, Dad1 and Ask1, are essential and connect the centromere to the plus end of spindle microtubules throughout the cell cycle (Hofmann et al, 1998; Cheeseman et al, 2001a, 2001b; Enquist-Newman et al, 2001; Janke et al, 2002; Li et al, 2002). The activity of the Ipl1 kinase is counteracted by the Glc7 type 1 phosphatase, which is in turn regulated by the type 1 phosphase inhibitor, Glc8 (Francisco et al, 1994; Tung et al, 1995). The mechanism by which the lack of spindle tension activates Ipl1 is, however, not known.
Although a structural homologue of Ask1 has been identified in fruitflies, it is not known whether the DASH complex is conserved in other organisms (Li et al, 2002). Moreover malorientation of spindle microtubules in human cells appears to be resolved by microtubule disassembly rather than detachment of spindle microtubules from kinetochores (Lampson et al, 2004). Intriguingly, Aurora B controls the localisation and activity of MCAK, a mitotic centromere-associated kinesin of the kinesin-13 family, which stimulates microtubule disassembly (Andrews et al, 2004; Lan et al, 2004; Lawrence et al, 2004). The reason for this difference is unclear but may lie in the fact that budding yeast and human centromeres differ markedly in structure. The budding yeast centromeres are short (150 bp) and provide a binding site for a single spindle microtubule, which can be either syntelically or amphitelically attached. By contrast, human centromeres are larger (∼1 Mb) and attract 20–30 microtubules per kinetochore and can encounter merotelic attachments, in which a sister chromatid is attached to microtubules from both poles, as well as syntelic and amphitelic attachments.
Several lines of evidence suggest that the fission yeast, Schizosaccharomyces pombe, is a useful model system in which to examine the mechanisms governing the establishment of chromosome biorientation. Firstly, fission yeast centromeres closely resemble those in animal cells in that they are large (∼35–110 kb) and contain multiple inverted repeat units, which are important for establishing centromeric cohesion (Pidoux and Allshire, 2004). Secondly, each kinetochore is bound to 2–4 microtubules and thus sister chromatid pairs theoretically can become both syntelically and merotelically attached (Ding et al, 1993). Thirdly, spindle microtubules are only nucleated during mitosis and only attach to kinetochores during this phase of the cell cycle (Ding et al, 1993). Proteins that bind fission yeast centromeres can be classified into two groups: those that bind throughout the cell cycle, such as the histone H3 variant Cnp1, Ndc80, Nuf2, Mal2, Mis6 and Mis12, and those that only bind during mitosis, such as the Klp5 and Klp6 kinesins and Alp14 and Dis1 TOG-like proteins (Nabeshima et al, 1998; Goshima et al, 1999; Takahashi et al, 2000; Garcia et al, 2001, 2002a, 2002b; Nakaseko et al, 2001; Wigge and Kilmartin, 2001; West et al, 2002). In this paper, we have identified a truncated allele of dam1 in screens for multicopy suppressors of the sensitivity of both cdc13-117 (cyclin B) and mal3-1 (EB-1 homologue) cells to thiabendazole (TBZ). We show that Dam1 is a component of a nonessential DASH complex, which binds to spindle microtubule tips and kinetochores as cells enter mitosis. Components of the DASH complex are nonessential for viability but play an overlapping role with the Klp5/Klp6 kinesin in establishing bipolar chromosome attachment during mitosis. Surprisingly, one of the components of the DASH complex, Dad1, is also a constitutive component of the kinetochore. We discuss the implications of these findings with respect to how amphitelic attachment of kinetochores to spindle microtubules is established in fission yeast.
Results
Identification of Dam1 as a multicopy suppressor of cdc13-117 and mal3-1 mutants
The Cdc2/Cdc13 (Cdk1/cyclin B) complex binds spindle poles and spindles in mitosis. Consistent with a role in controlling spindle microtubule stability, cdc13-117 cells are sensitive to growth in the presence of 12.5 μg/ml TBZ (Supplementary Figure 1A; Booher and Beach, 1988). We screened for multicopy suppressors of the inability of cdc13-117 cells to proliferate in the presence of 12.5 μg/ml TBZ and identified four suppressors including cdc13+ (five clones), a truncated allele of dis2+ type 1 phosphatase (eight clones) and a small heat-stable inhibitor of type 1 phosphatase (one clone). The latter two gene products are homologues of the S. cerevisiae Glc7 and Glc8 proteins, respectively (Francisco et al, 1994; Tung et al, 1995). In addition, we identified a truncated allele of a gene (SPAC589.08c) that encodes a protein of 155 amino acids that shares a short region of sequence homology to the S. cerevisiae Dam1 microtubule binding protein (three clones) (Supplementary Figure 1B and Supplementary Table I). We identified the same truncated dam1(1–127) allele in an independent screen for multicopy suppressors of the TBZ sensitivity and chromosome loss defect of mal3-1 cells (Supplementary Figure 1C and D). The Mal3 protein (homologue of human EB-1) binds the plus ends of both interphase and spindle microtubules (Beinhauer et al, 1997; Kerres et al, 2004). In this study, we have examined the role of Dam1 in fission yeast mitosis.
Dam1 regulates mitotic progression and resistance to environmental stress
To investigate the function of S. pombe. (S.p.) Dam1, one allele of dam1 was deleted with the kanamycin resistance gene in an h−/h+ diploid (Bähler et al, 1998). Tetrad dissection of Δdam1/dam1+ heterozygous diploids gave rise to four viable spores on germination that showed a 2:2 segregation of kanamycin resistance, indicating that Dam1 is nonessential for viability. Despite this, a proportion of Δdam1 cells display mitotic abnormalities that include hypercondensed or fragmented chromosomes, chromosome mis-segregation (unequal chromatin staining) or ectopic septation in the absence of chromosome segregation (cut phenotype), as judged by staining with DAPI and calcofluor (Figure 1A and B). Notably, cells lacking Dam1 are more sensitive to TBZ than wild-type cells, whereas dam1(1–127):kanR cells, the C-terminal 28 amino acids of which have been removed, are more resistant to TBZ, suggesting that the C-terminal 28 amino acids of S.p. Dam1 are important for controlling microtubule stability (Figure 1C). Additionally, Δdam1 cells are unable to proliferate at high temperature (37°C) or in the presence of 0.6 M KCl, an osmotic stress, phenotypes not shared by dam1(1–127):kanR cells (Figure 1D). These results indicate that Dam1 is required for microtubule stability and resistance to environmental stress.
Figure 1.
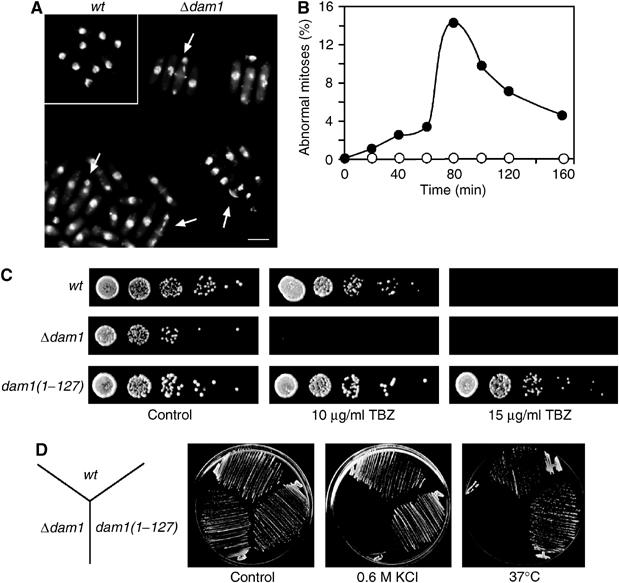
Dam1 controls mitotic progression and resistance to environmental stress. (A) Log phase wild-type or Δdam1 cells were fixed and stained with DAPI and calcofluor. Scale bar=10 μm. Arrows indicate abnormal mitotic cells. (B) Log phase wild-type (open circles) or Δdam1 cells (closed circles) were synchronised in early G2 and incubated in fresh medium. At the times indicated, cells were fixed and the percentage of mitotic abnormalities expressed as a percentage of the total number of cells (n=300). (C) Serial dilutions (10-fold) of wild-type, Δdam1 or dam1(1–127):kanR cells were spotted onto YE medium either in the absence (control) or presence of either 10 μg/ml TBZ or 15 μg/ml TBZ and incubated for 3 days at 30°C. (D) Wild-type (wt), Δdam1 or dam1(1–127):kanR cells were streaked onto YE medium in the absence (control) or presence of 0.6 M KCl at 30°C or on YE medium at 37°C for 3 days.
Dam1 associates to spindle microtubule tips and kinetochores during mitosis
The subcellular localisation of Dam1 was first examined in fixed dam1-gfp ndc80-cfp cells. Dam1 does not localise to any identifiable cellular structure in interphase, but forms a single spot in the nucleus in early mitosis and multiple spots during prometaphase and metaphase, suggesting that it is recruited to kinetochores as cells enter mitosis (Figure 2A). Higher magnification images of fixed dam1-gfp ndc80-cfp cells show that each of the Dam1 signals is associated closely, but not coincidentally, with Ndc80 during metaphase and that the extent of overlap between the Dam1 and Ndc80 signals increases after sister chromatid separation (Figure 2B). In movies of dam1-gfp cells, we observe, in addition to up to six stable Dam1 dots, other Dam1 dots that rapidly appear and disappear during prometaphase and metaphase (see arrows in Figure 3A). By analysing fixed dam1-gfp ndc80-cfp cells, we find that these Dam1 dots do not colocalise with Ndc80 (see arrow in Figure 3B). We suspected that these may represent Dam1 on intranuclear spindle microtubules, which are highly dynamic and can normally only be visualised by live imaging (Zimmerman et al, 2004). However, plasmid-borne overexpression of dam1 from the thiamine-repressible nmt1 promoter inhibits cell proliferation and causes cells to arrest in a preanaphase state with abnormally elongated intranuclear spindle microtubules, which can be observed after fixation (Supplementary Figure 2). In fixed nmt1-GFP-dam1 cells, Dam1 is observed both on the mitotic spindle and at the plus end of an intranuclear microtubule, which is nucleated at an angle to the spindle (Figure 3C). Consistent with this, we find that Dam1 frequently colocalises with Mal3 in early mitosis, although their localisation is mutually independent (Supplementary Figure 3A–C). To determine whether the extra Dam1 dots are inside or outside the nucleus, we visualised dam1-gfp cut11-cfp cells in mitosis. Cut11 is associated with the nuclear envelope during interphase and with the nuclear envelope and spindle pole bodies during mitosis (West et al, 1998). The image in Figure 3D shows at least seven Dam1 dots, six of which lie in a line between the spindle pole bodies whereas the seventh is located on the inner face of the nuclear envelope (see arrow in Figure 3D). In late anaphase, Dam1 is concentrated at the separated spindle poles with fainter signals along the spindle inside the nucleus, although no Dam1 could be seen at the spindle midzone (Figure 3E). Taken together, these data strongly suggest that Dam1 binds to intranuclear spindle microtubules and kinetochores during early mitosis.
Figure 2.
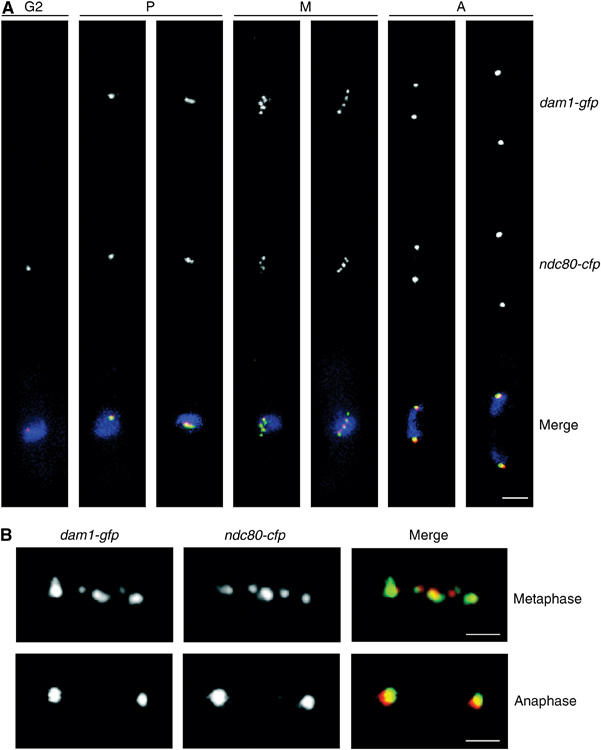
Dam1 binds kinetochores during mitosis. (A) Log phase dam1-gfp ndc80-cfp cells were fixed and analysed by fluorescence microscopy. Cells were categorised for cell cycle distribution based on cell size and distribution of kinetochores as either interphase (G2), prometaphase (P), metaphase (M) or anaphase (A). Scale bar=2 μm. (B) Higher magnification images of fixed dam1-gfp ndc80-cfp cells before and after anaphase. Dam1 is shown in green and Ndc80 is shown in red. Scale bar=1 μm.
Figure 3.
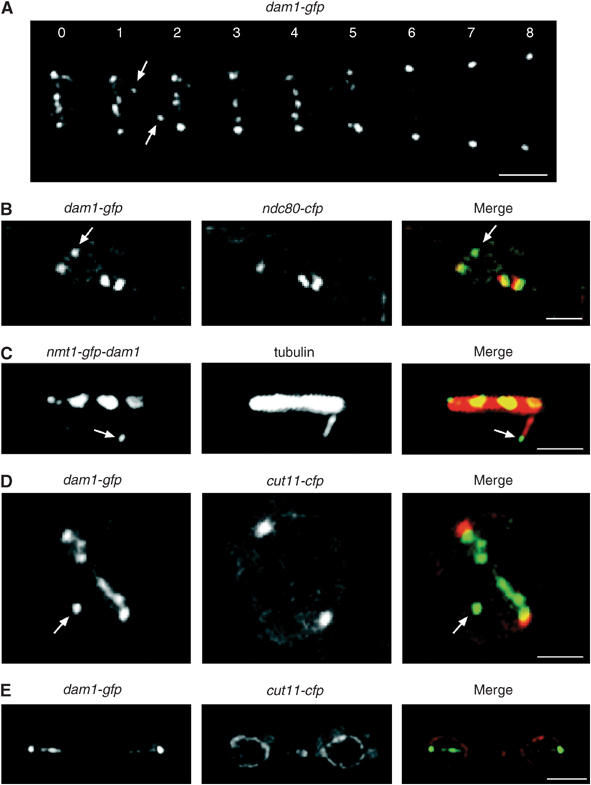
Dam1 binds non-kinetochore-associated spindle microtubule tips in mitosis. (A) Images from a movie of dam1-gfp cells in mitosis taken at 60 s intervals. Scale bar=2 μm. (B) High-magnification image of fixed dam1-gfp ndc80-cfp cells in mitosis. The arrow indicates non-kinetochore-associated Dam1. Scale bar=1 μm. (C) Log phase nmt1-GFP-dam1 cells were grown in the absence of thiamine for 24 h and fixed and stained with anti-tubulin antibody (tubulin). Scale bar=1 μm. The arrow shows Dam1 (green) on the plus end of an intranuclear spindle microtubule (red). (D) High-magnification image of dam1-gfp cut11-cfp cells before anaphase onset. Six Dam1 dots are observed in a line between the spindle poles. The arrow indicates a seventh Dam1 dot (green), which is inside the nuclear envelope (red). Scale bar=1 μm. (E) Image of dam1-gfp cut11-cfp cells after anaphase. Scale bar=3 μm.
Identification of components of the fission yeast DASH complex
We screened the S. pombe protein database for homologues of the budding yeast DASH complex and identified Duo1, Spc34, Ask1, Dad1 and Dad3 (Supplementary Table I). In addition, we found two previously identified gene products, Hos2 and Hos3, that are structurally homologous to S.c. Dad2 and S.c. Hsk3, respectively (Supplementary Table I). The hos2+ and hos3+ mutants were identified by their inability to proliferate under conditions of high osmotic stress, a phenotype also associated with loss of Dam1 (Aoyama et al, 2000; Nakamichi et al, 2000). By analysing the S. pombe DNA sequence database, we also identified homologues of the budding yeast Spc19 and Dad4 proteins (Supplementary Table I). We deleted the duo1, spc34, ask1 and dad1 genes and found that none are essential for viability (Figure 4A; data not shown). However, Δduo1, Δspc34, Δask1 and Δdad1 cells are sensitive to growth on 10 μg/ml TBZ or 0.6 M KCl or growth at high temperature, as observed with Δdam1 cells (Figure 4A; data not shown). Double mutant combinations between any of these strains showed no additive phenotype. Moreover, Duo1, Spc34, Ask1 and Dad1 are required for localisation of Dam1 in mitosis (Figure 4B). In addition, we find that Hos2 and Hos3 are required for Dam1 association to kinetochores and that Duo1-GFP, Spc34-GFP and Ask1-GFP proteins display an identical localisation pattern to Dam1-GFP (data not shown). Together, these results suggest that the DASH complex is structurally conserved in fission yeast.
Figure 4.
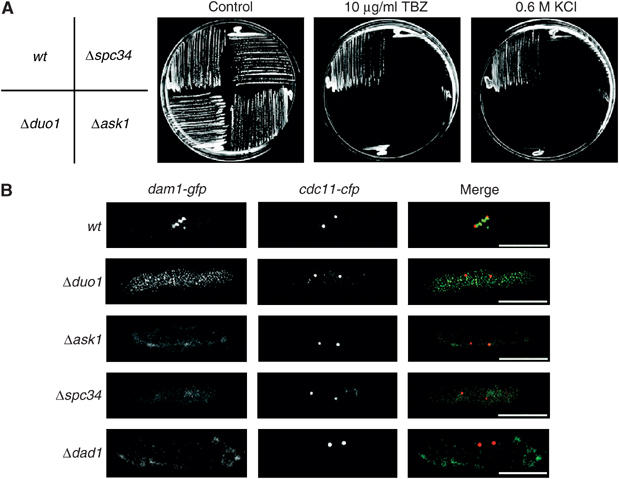
Localisation of Dam1 is dependent on Spc34, Ask1, Duo1 and Dad1. (A) Wild-type, Δspc34, Δduo1 and Δask1 cells were streaked either onto rich medium at 30°C containing either no addition (control), 10 μg/ml TBZ, 0.6 M KCl or on rich medium at 37°C and incubated for 3 days. (B) Log phase dam1-gfp cdc11-cfp, Δduo1 dam1-gfp cdc11-cfp, Δspc34 dam1-gfp cdc11-cfp, Δask1 dam1-gfp cdc11-cfp and Δdad1 dam1-gfp cdc11-cfp cells were fixed and stained with DAPI and calcofluor and visualised by fluorescence microscopy. Scale bar=5 μm.
Dad1 binds the fission yeast kinetochore throughout the cell cycle
Surprisingly, one of the components of the fission yeast DASH complex, Dad1, associates to kinetochores throughout the cell cycle, as judged by analysis of dad1-gfp ndc80-cfp cells (Figure 5A). Localisation of Dad1 to kinetochores is independent of Dam1, although its association to the late anaphase B spindle is dependent on Dam1 (Figure 5B; data not shown). Localisation of Dad1 to the kinetochore is not observed in mis6-302 or mal2-1 mutants at the restrictive temperature, but is unaffected in mis12-537, cnp1-1 or nuf2-4 mutants under the same conditions, indicating that association of Dad1 to the kinetochore is dependent on the Mis6 and Mal2 kinetochore proteins (Figure 5C; data not shown). In addition, kinetochore localisation of Dam1 was abolished in mis6-302 and mal2-1 mutants, but not in mis12-537 or cnp1-1 mutants, suggesting that the Mis6 complex provides the docking site for the DASH complex at kinetochores (data not shown). We were unable to construct double mutants between Δdad1, Δdam1 or Δask1 and either mis6-302 or mal2-1, although double mutants between Δdad1, Δdam1 or Δask1 and mis12-537 were viable (Table I). This suggests that the DASH complex is required for the stability of the Mis6 complex in mitosis. Notably, however, whereas double mutants between either Δdam1 or Δask1 cells and cnp1-1 are viable, Δdad1 cnp1-1 cells are inviable at all temperatures, indicating that Dad1 has an additional role not shared by other components of the DASH complex (Table I). Together, these results suggest that fission yeast Dad1 is both a component of the fission DASH complex and a constitutive component of the fission yeast kinetochore.
Figure 5.
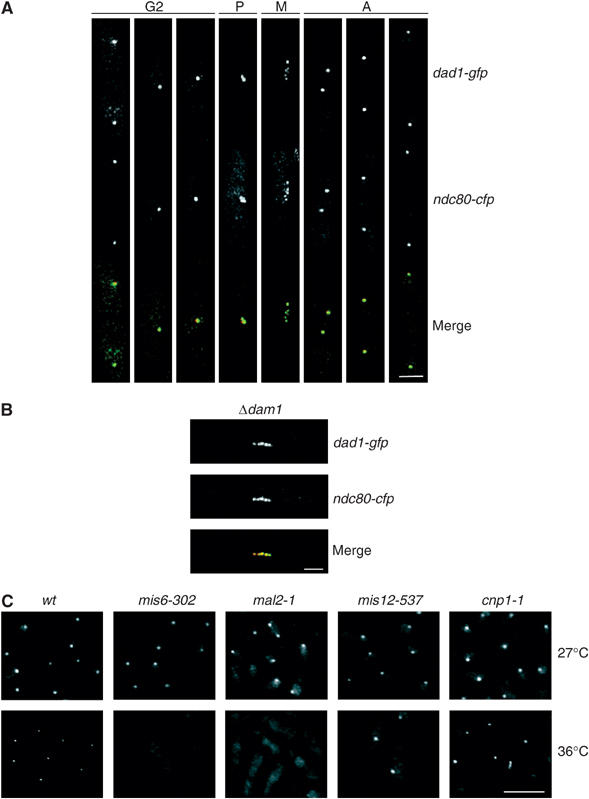
Dad1 is a constitutive component of the fission yeast kinetochore. (A) Log phase dad1-gfp ndc80-cfp cells were stained with DAPI and visualised by fluorescence microscopy. Cells at different cell cycle stages are shown. Scale bar=2 μm. (B) Mitotic Δdam1 dad1-gfp ndc80-cfp cells were visualised by fluorescence microscopy. Scale bar=2 μm. Dad1 is shown in green and Ndc80 is shown in red. (C) Log phase dad1-gfp, mis6-302 dad1-gfp, mal2-1 dad1-gfp, mis12-537 dad1-gfp and cnp1-1 dad1-gfp cells were incubated for 6 h either at 27 or 36°C. Cells were fixed and visualised by fluorescence microscopy. Scale bar=10 μm.
Table 1.
Genetic interactions of the fission yeast DASH complex
| Mutants | Δdam1 | Δask1 | Δdad1 |
|---|---|---|---|
| Wild type | V | V | V |
| Δklp1/Δpkl1 | V | ND | V |
| Δklp2 | V | ND | V |
| Δklp3 | V | ND | V |
| Δklp4/Δtea2 | V | ND | V |
| Δklp5 | SL | SL | SL |
| Δklp6 | SL | SL | SL |
| Δmal3 | V | V | V |
| mal2-1 (27°C) | SL | SL | SL |
| mis6-302 (27°C) | SL | SL | ND |
| mis12-537 (27°C) | V | V | ND |
|
cnp1-1 (27°C) |
V |
V |
SL |
| All genetic interactions were tested at 30°C unless otherwise stated. V: viable; SL: synthetic lethal; ND: not determined. | |||
Dam1 is required for timely anaphase onset and accurate chromosome segregation
To examine the role of the DASH complex in mitosis, we monitored spindle microtubule dynamics in single cells. In wild-type cells, spindles remain at a constant length (phase 2) for 9±2 min at 30°C (Nabeshima et al, 1998; Figure 6A). By contrast, phase 2 lasts for 24±10 min in the absence of Dam1 at the same temperature (Figure 6A). To examine the effect of Dam1 on kinetochore and spindle pole dynamics, a Δdam1 ndc80-gfp cdc11-cfp strain was synchronised in early G2 and the percentage of cells in prometaphase and metaphase was analysed during the subsequent mitosis (Tournier et al, 2004). Consistent with the above result, cells lacking Dam1 remain in prometaphase or metaphase on average twice as long as control cells (as judged by the area under the curve) (Figure 6B). Filming of single cells also revealed considerable cell-to-cell variation in the time spent in prometaphase or metaphase. Images from representative movies of wild-type and Δdam1 cells undergoing sister chromosome separation is shown in Figure 6C. In the wild-type cell, kinetochores oscillate between the spindle poles and congress to the spindle midzone prior to anaphase onset, as previously observed (Figure 6C; Tournier et al, 2004). In this and other movies, kinetochores segregate equally to the spindle poles (3:3). In the cells lacking Dam1, the mitotic spindle was found to be 3.5 μm at anaphase onset compared to 2.6 μm in the wild-type cell, because the onset of anaphase A occurs just after the onset of phase 3 (spindle elongation) (Figure 6C). In the absence of Dam1, kinetochores failed to congress to the spindle midzone and one of the sister chromatids (marked with an arrow) lagged behind the other sister chromatid after anaphase, although it eventually segregated to the correct spindle pole (Figure 6C). Similar behaviour was observed in 14 out of 19 movies. Consistently, a linear Ch16 (ade6-M216) minichromosome is lost in 8±0.7% of cell divisions in Δdam1 cells, 800 times more frequently than in wild-type cells (Niwa et al, 1989). Together, these data indicate that Dam1 is required for proper chromosome segregation and timely anaphase onset in fission yeast.
Figure 6.
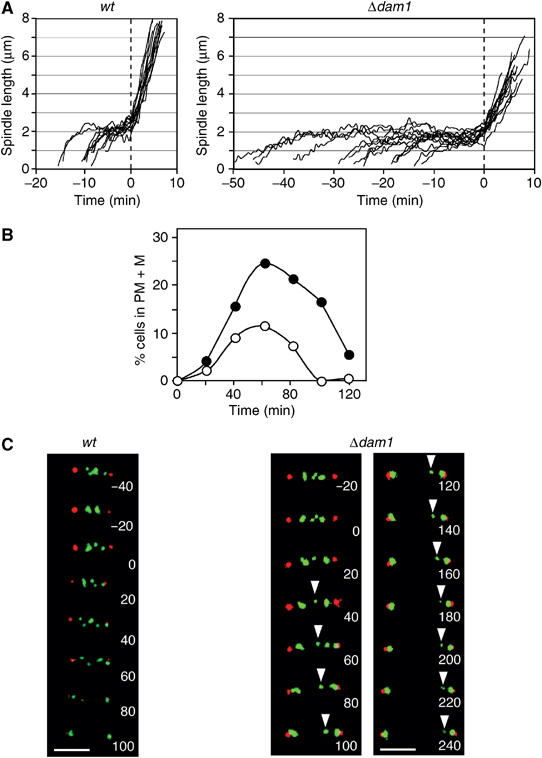
Dam1 is required for chromosome congression and timely anaphase onset. (A) Analysis of spindle length from multiple movies of nmt1-atb2-gfp (n=11) and Δdam1 nmt1-atb2-gfp (n=14) cells growing in rich medium containing thiamine at 30°C. Time points were taken every 30 s. (B) Log phase ndc80-gfp cdc11-cfp and Δdam1 ndc80-gfp cdc11-cfp cells were synchronised in early G2 and incubated in fresh medium. The percentage of cells in prometaphase and metaphase (PM+M) was calculated (n=250). (C) Images from a movie of an ndc80-gfp cdc11-cfp cell (left panels) or a Δdam1 ndc80-gfp cdc11-cfp cell (right panel) in mitosis. Ndc80 is shown in green and Cdc11 is shown in red. Time zero is taken as the onset of anaphase A. The arrow indicates a lagging sister chromatid. Scale bar=2 μm.
The spindle assembly checkpoint delays anaphase onset in the absence of Dam1
In wild-type cells, Cdc2/Cdc13 (Cdk/cyclin B) kinase binds spindle poles and short <2.5 μm preanaphase spindles, but is absent from spindles and spindle poles during anaphase B (Tatebe et al, 2001). In the absence of Dam1, Cdc13 binds to mitotic spindles for longer during prometaphase and metaphase, during which time the mitotic spindle elongates (Figure 7A). These results suggest that activation of the anaphase-promoting complex (APC) is delayed. To examine whether this is due to activation of the spindle assembly checkpoint (SAC), we monitored the localisation of Mad2 and Bub1, components of the SAC that bind to the kinetochore either when spindle microtubules are not attached or the spindle is not under tension (Tournier et al, 2004). We find that less than 1% of mitotic mad2-gfp cdc11-cfp cells display a non-SPB-associated Mad2 focus during mitosis, whereas these are observed in 12% of mitotic Δdam1 mad2-gfp cdc11-cfp cells, and in some cells multiple Mad2 foci are observed (Figure 7B). Additionally, Bub1 associates to kinetochores for longer during prometaphase and metaphase in Δdam1 bub1-gfp cdc11-cfp cells than in control cells (Supplementary Figure 4). Although both Δmad2 Δdam1 and Δbub1 Δdam1 double mutant cells are viable, the percentage of Δmad2 Δdam1 cells with a short (<3.5 μm) preanaphase mitotic spindle is less than in Δdam1 cells and similar to that observed in either wild-type or Δmad2 cells (Figure 7C). Importantly, however, both Δmad2 Δdam1 and Δbub1 Δdam1 strains display twice as many mitotic abnormalities as observed in Δdam1 cells (Figure 7D). These results suggest that the SAC delays anaphase to allow a proportion of Δdam1 cells to correctly establish bipolar chromosome attachment.
Figure 7.
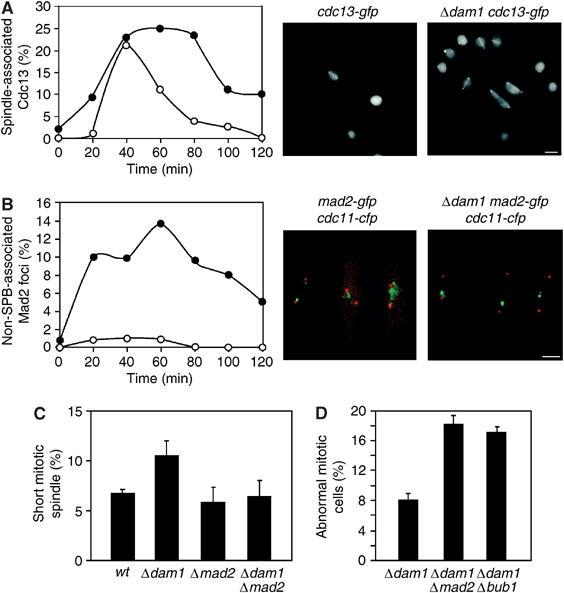
Loss of Dam1 causes activation of SAC. (A) Log phase cdc13-gfp and Δdam1 cdc13-gfp cells were synchronised in early G2 and incubated in fresh medium. At the times indicated, cells were fixed and visualised by fluorescence microscopy. The percentage cells with spindle or spindle pole-associated Cdc13 was calculated at each time point (n=250). Images of cdc13-gfp and Δdam1 cdc13-gfp cells are shown at the t=40 min time point. Scale bar=2 μm (B) Log phase mad2-gfp cdc11-cfp and Δdam1 mad2-gfp cdc11-cfp cells were synchronised in early G2 and incubated in fresh medium. At the times indicated, cells were fixed and visualised by fluorescence microscopy. Mad2 is shown in green and Cdc11 is shown in red. The percentage of cells with non-SPB-associated Mad2 foci was calculated at each time point (n=300). Images of mad2-gfp cdc11-cfp and Δdam1 mad2-gfp cdc11-cfp cells are shown at the t=40 min time point. Scale bar=2 μm. (C) Log phase nmt1-atb2-gfp, Δdam1 nmt1-atb2-gfp, Δmad2 nmt1-atb2-gfp and Δdam1 Δmad2 nmt1-atb2-gfp cells were grown in the presence of thiamine and the percentage of cells with short (<3 μm) preanaphase mitotic spindles determined (n=200). The results are the average of three independent experiments. (D) Log phase Δdam1, Δdam1 Δmad2 or Δdam1 Δbub1 cells were fixed and stained with DAPI and calcofluor. The percentage of mitotic abnormalities was expressed as a percentage of the total number of cells from three independent experiments (n=300).
The DASH complex and Klp5/Klp6 kinesin coordinately control bipolar chromosome attachment
The previous results persuaded us to examine the relationship between Dam1 and other proteins that function at the interface between spindle microtubules and the kinetochore. We find that Δmal3 Δdam1 double mutant cells are viable and only marginally more sensitive to TBZ than either Δmal3 or Δdam1 single mutants (Table I and Supplementary Figure 3D). By contrast, we were unable to construct a Δdam1 and either Δklp5 or Δklp6 mutant by tetrad analysis, indicating that Dam1 and Klp5/Klp6 perform an essential overlapping function (Figure 8A). Microscopic analysis revealed that spores lacking Dam1 and Klp5 germinated but arrested with a ‘cut' phenotype (Figure 8A). Double mutants between Δklp5 or Δklp6 and either Δduo1, Δspc34, Δask1 or Δdad1 also could not be constructed, indicating that other components of the DASH complex are essential in the absence of Klp5/Klp6 (Table I; data not shown). On the other hand, double mutants between Δdam1 or Δdad1 and either Δklp1 (Δpkl1), Δklp2, Δklp3 or Δklp4 (Δtea2) mutants, which are deleted for other fission yeast kinesins, are viable, indicating that the interaction between the DASH complex and Klp5/Klp6 is specific (Table I; Brazer et al, 2000; Browning et al, 2000; Troxell et al, 2001).
Figure 8.
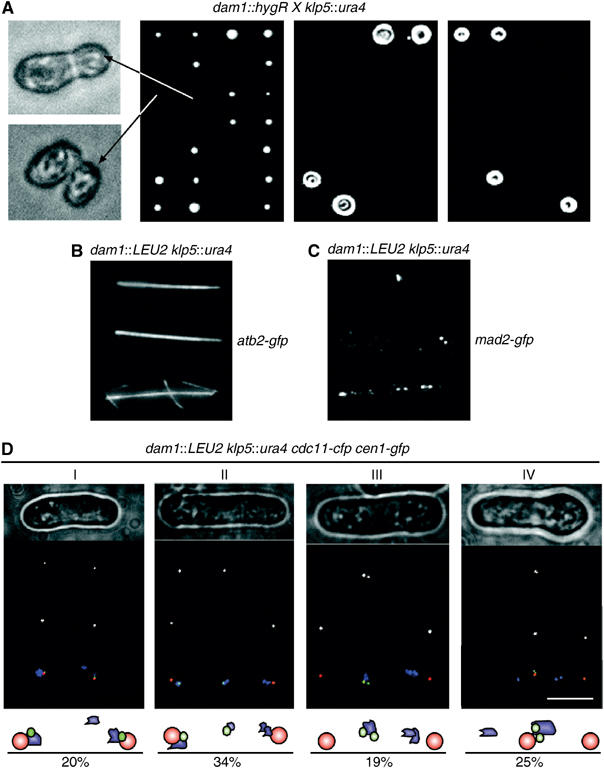
Dam1 and Klp5 are essential for establishing chromosome biorientation. (A) dam1∷hygR cells were crossed to klp5∷ura4 cells and the spores from asci were separated by tetrad dissection on rich medium (YES). Genotype was assessed by replica plating onto minimal medium lacking uracil (EMM−Ura) or rich medium containing hygromycin (YES+Hyg) and scored as either parental ditype (PD), nonparental ditype (NPD) or tetratype (TT). dam1∷hygR klp5∷ura4 cells from an NPD were visualised by light microscopy (left panels). (B) dam1∷LEU2 klp5∷ura4 nmt1-gfp-atb2 spores were germinated on minimal medium lacking uracil and leucine containing 10 μg/ml thiamine for 20 h at 30°C and then visualised by fluorescence microscopy. (C) dam1∷LEU2 klp5∷ura4 mad2-GFP:kanR spores were germinated on minimal medium lacking uracil and leucine for 20 h at 30°C. Cells were fixed and analysed by fluorescence microscopy. (D) dam1∷LEU2 klp5∷ura4 cen1-gfp:kanR cdc11-cfp:kanR spores were germinated on minimal medium lacking uracil and leucine for 20 h at 30°C. Germinated spores were analysed by phase contrast microscopy (phase) and by fluorescence microscopy for the localisation of centromere 1 (Cen1) or spindle poles (Cdc11). Merged images show the localisation of centromere 1 (green), spindle poles (red) and chromatin material (blue). Scale bar=5 μm. Four cells (I–IV) are shown that display a distinct terminal mitotic phenotype. A total of 150 germinated cells were scored for mitotic phenotype and expressed as a percentage of the total number of germinated cells.
To examine the nature of the essential function shared by Dam1 and Klp5, dam1∷LEU2 and klp5∷ura4 strains were crossed and the resulting ascospores incubated in medium without uracil or leucine so that only cells lacking both Dam1 and Klp5 could germinate. All germinated cells had an elongated mitotic spindle, indicating that Dam1 and Klp5 are not required for bipolar spindle formation (Figure 8B). However, multiple Mad2 foci were observed in most germinated dam1∷LEU2 klp5∷ura4 mad2-gfp cells, suggesting that bipolar chromosome attachment to microtubules is defective (Figure 8C). To assess the accuracy of sister chromatid separation, centromere 1 was marked with GFP (cen1-gfp) and spindle poles were marked with CFP (cdc11-cfp). In 20% of germinated dam1∷LEU2 klp5∷ura4 cen1-gfp cdc11-cfp cells, two sister chromatids of centromere 1 separated to opposite spindle poles but other chromatin material remained improperly segregated (Figure 8D, Cell I). In 34% of cells, one of the two sister chromatids failed to segregate to a spindle pole, either because it is unattached or merotelically attached to both poles (Figure 8D, Cell II). In 19% of cells, the sister chromatid pairs did not separate (Figure 8D, Cell III). Finally, in 25% of cells, both sister chromatids segregated to the same spindle pole, possibly because both sister chromatids are syntelically attached to microtubules from the same pole (Figure 8D, Cell IV). The remaining 2% of germinated cells showed no fluorescence signal and may represent double mutant cells that had died. To assess the individual contributions of Dam1 and Klp5, dam1∷LEU2 cen1-gfp cdc11-cfp and klp5∷ura4 cen1-gfp cdc11-cfp spores were incubated on agar pads and the segregation of chromosome 1 examined after the first mitosis following germination. In two out of 97 Δdam1 cells, both sister chromatids segregated to the same pole, although this was never observed in wild-type cells or in the absence of Klp5 (data not shown). To examine this more thoroughly, log phase dam1∷LEU2 cen1-gfp cdc11-cfp and klp5∷ura4 cen1-gfp cdc11-cfp cells were synchronised in early G2 and the segregation of chromosome 1 monitored after cells had passed through mitosis (Figure 9A and B). In cells lacking Dam1, either one of the two sister chromatids failed to segregate to a spindle pole or both sister chromatids segregated to the same pole (Figure 9B). By contrast, from analysis of 500 cells lacking Klp5, we observed some lagging sister chromatids during anaphase B but not unequal segregation of sister chromatids (Figure 9B). In other experiments, we found that sister chromatids segregate to the same spindle pole in 3.6±0.5% of log phase Δduo1 cells but not in the absence of Klp6 (data not shown). Together, these results indicate that the DASH complex and Klp5/6 kinesin play an overlapping role in establishing bipolar chromosome attachment.
Figure 9.
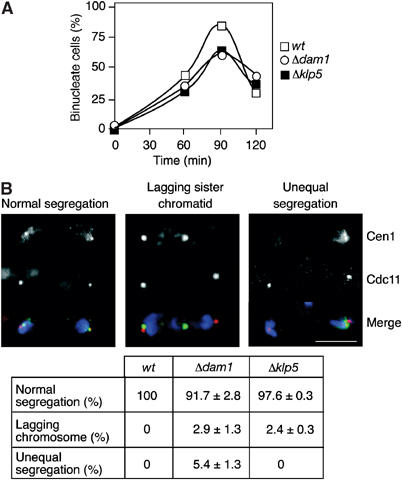
Dam1 and Klp5 have distinct effects on sister chromatid separation in rapidly dividing cells. (A) Log phase cen1-gfp cdc11-cfp:kanR cells (open squares), Δdam1 cen1-gfp cdc11-cfp:kanR cells (open circles) or Δklp5 cen1-gfp cdc11-cfp:kanR cells (closed squares) were synchronised in early G2 and incubated in fresh medium and the percentage of binucleate cells was expressed as a percentage of the total number of cells at the indicated time points (n=300). (B) The cells from (A) were fixed after 90 min and scored for either segregation of sister chromatids to opposite spindle poles (normal segregation), segregation of only one of the two sister chromatids to the spindle pole (lagging sister chromatid) or segregation of both sister chromatids to the same pole (unequal segregation). Scale bar=5 μm. The percentage of each segregation event was expressed as a percentage of the total number of cells (n=250).
Discussion
We have identified a truncated allele of dam1 in two independent genetic screens for multicopy suppressors of the TBZ sensitivity of cdc13-117 (cyclin B) and mal3-1 (EB-1) mutants. Fission yeast Dam1 is homologous to the budding yeast Dam1 protein but only associates to kinetochores during mitosis. We have identified homologues of all other components of the budding yeast DASH complex, including Duo1, Spc34, Ask1, Dad1, Hos2(Dad2) and Hos3(Hsk3) and found that these proteins are required for Dam1 localisation in mitosis. Most of these proteins, including Duo1, Spc34 and Ask1, show a similar localisation to Dam1, strongly suggesting that these proteins act in a complex. This conclusion is in agreement with the results of Liu et al (2005), who have purified a 10-subunit DASH complex from mitotically arrested fission yeast cells. Surprisingly, unlike budding yeast, none of the components of the fission yeast DASH complex are essential for viability. By careful time-lapse microscopy, we find that Dam1 also binds the plus end of intranuclear spindle microtubules. These microtubules are highly dynamic and may serve to search and capture unoccupied kinetochores during early mitosis. Although Dam1 frequently colocalises with the Mal3 plus end microtubule binding protein, localisation of Dam1 and Mal3 is mutually independent (Beinhauer et al, 1997; Kerres et al, 2004). Recently, the DASH complex has been found preferentially to bind GTP-tubulin in vitro, providing an explanation for its affinity for the plus end of microtubules (Westermann et al, 2005). These data are consistent with the findings in budding yeast that association of the DASH complex to kinetochores is dependent on interaction of spindle microtubules with the kinetochore (Li et al, 2002). We note that fission yeast Dam1 does not bind to the spindle midzone during anaphase B. It is not clear whether this is because the DASH complex is not loaded onto spindle midzone microtubules or that the DASH complex is only stabilised on the plus end of kinetochore-attached microtubules. Notably, the truncated dam1(1–127) allele isolated in our genetic screens confers TBZ resistance even when expressed from its own chromosomal locus. This suggests that the C-terminal 28 amino acids of Dam1 are important for regulation of microtubule stability and explains why this allele, and not full-length dam1, was isolated in our genetic screens.
Importantly, one of the components of the fission yeast DASH complex, Dad1, binds the kinetochore throughout the cell cycle and does so independently of Dam1, although association of Dad1 to the late anaphase B spindle is dependent on Dam1. Localisation of Dad1 to the fission yeast kinetochore is dependent on the Mis6 and Mal2, but not Mis12, Nuf2 or Cnp1, kinetochore proteins. These results, and those of the accompanying paper, suggest that Dad1 is both a component of the DASH complex and a component of a kinetochore subcomplex containing the Mis6 and Mal2 proteins (Liu et al, 2005). Notably, we find that Δdad1 cnp1-1 mutants are synthetically lethal whereas Δdam1 cnp1-1 and Δask1 cnp1-1 mutants are viable, indicating that Dad1 has an extra function that is not shared by other components of the DASH complex, possibly by contributing to the structural integrity of the kinetochore. We further find that Dam1 does not localise to kinetochores in mis6-302 or mal2-1 mutants, whereas its localisation is unaffected in mis12-537, nuf2-1 or cnp1-1 mutants, suggesting that the Mis6 complex provides the docking site for the DASH complex at kinetochores. We note that Dam1 and Ndc80 are seen adjacent to each other during prometaphase and metaphase, but the signals overlap more extensively after anaphase onset. This indicates that the binding site for the DASH complex on centromeres, or its higher order structure, may alter during mitosis. Intriguingly, the DASH complex has been shown to form both rings and spiral structures on microtubules in vitro (Miranda et al, 2005; Westermann et al, 2005). One exciting possibility is that kinetochore-associated Dad1 nucleates a change in DASH structure that stabilises amphitelic attachment of spindle microtubules to the kinetochore. To test this hypothesis, it will be necessary to construct Dad1 mutants that bind only the DASH complex but not the Mis6 complex.
In wild-type cells, kinetochores oscillate between the spindle poles during prometaphase and metaphase and congress to the spindle midzone just before anaphase onset. However, in most cells lacking Dam1, chromosome congression does not take place, anaphase is delayed and lagging sister chromatids are frequently observed during early mitosis. By analysis of kinetochore behaviour in both single cells and synchronised cell populations, we find that cells lacking Dam1 spend on average more than twice as long in prometaphase and metaphase than wild-type cells. Moreover, degradation of Cdc13 (cyclin B) is delayed in cells lacking Dam1, indicating that activation of APC is inhibited. Consistent with APC inhibition, in the absence of Dam1, we observe sustained recruitment of the Mad2 and Bub1 SAC proteins to kinetochores and show that the anaphase delay in Δdam1 cells is abolished by simultaneous inactivation of mad2. These results suggest that, in the absence of Dam1, some kinetochores are not fully attached to microtubules or that chromosome biorientation has not been established. Importantly, Δdam1 Δmad2 and Δdam1 Δbub1 cells display a higher percentage of mitotic abnormalities than Δdam1 cells, suggesting that in the absence of Dam1 the SAC delays anaphase onset to allow bipolar chromosome attachment to be established or corrected by another mechanism.
Components of the DASH complex (except Dad1) share a property of the Alp14, Dis1, Klp5 and Klp6 proteins, namely associating to kinetochores exclusively in M phase (Nabeshima et al, 1998; Garcia et al, 2001, 2002a, 2002b; Nakaseko et al, 2001; West et al, 2002). Previously, we have shown that inactivation of either Alp14 or Dis1 is lethal in the absence of Klp5 or Klp6 (Garcia et al, 2002a). By contrast, DASH mutants are inviable in the absence of Dis1, but not in the absence of Alp14, the reason for which is presently unknown (K Crawley and T Toda, unpublished data). In this study, we show that cells lacking both Dam1 and Klp5 are inviable due to chromosome mis-segregation during the first mitosis even though bipolar mitotic spindle assembly is not affected. In most cells lacking Dam1 and Klp5, one or both sister chromatids of chromosome 1 either fail to separate to opposite spindle poles or both sister chromatids segregate to the same spindle pole, indicating that the DASH complex and Klp5/6 kinesin coordinately control bipolar chromosome attachment in fission yeast. In some cells lacking Dam1 and Klp5, chromosome 1 is segregated properly to opposite spindle poles, indicating that other pathways mediate interaction of microtubules with kinetochores. This may be through interaction of microtubule-bound Mal3 (EB-1) with kinetochore-bound Spc7, or some other microtubule attachment pathway, but this process is either inefficient or not subject to a correction process (Garcia et al, 2002a; Kerres et al, 2004). In summary, our data suggest that the pathways that mediate bipolar chromosome attachment in fission yeast differ from those that operate in budding yeast. One reason for this may be because fission yeast kinetochores are bound to 2–4 microtubules, whereas budding yeast kinetochores are bound only to a single microtubule and do not encounter merotelic attachments. Further experiments are under way to address this and other possibilities.
Materials and methods
Cell culture and synchronisation
Media, growth and maintenance of strains were as described (Moreno et al, 1991). Strains used in this study are listed in Supplementary Table II. Cell cultures were grown at 28°C in YES unless otherwise stated. Cell synchrony was achieved by lactose gradient centrifugation, as previously described (Tournier et al, 2004). Cells were resuspended in fresh medium at 106 cells/ml and released at 28°C unless otherwise stated. The peak synchrony of septation was >50% in each experiment.
Strain construction
Gene deletions, carboxy-terminal GFP and CFP epitope tagging and amino-terminal GFP epitope tagging of genes were performed either by one- or two-step PCR-based gene targeting, as previously described (Bähler et al, 1998; Krawchuk and Wahls, 1999). In all gene deletions (Δ), the entire ORF was removed. The truncated dam1(1–127):kanR allele was constructed by inserting a stop codon at residue 128. A full list of oligonucleotides used is provided in Supplementary Table III. Compound tagged and mutant strains were constructed by standard genetic methods (Moreno et al, 1991).
Isolation of multicopy suppressors
Multicopy extragenic suppressors of the cdc13-117 and mal3-1 mutant phenotypes were isolated by transformation of the strains with an S. pombe genomic bank (Barbet et al, 1992). Cells were grown at 30°C. Ura+ transformants were replica-plated twice onto EMM plates containing either 12.5 μg/ml TBZ (for cdc13-117 cells) or 7 μg/ml TBZ (for mal3-1 cells). Plasmid-dependent suppressors were isolated by transformation into Escherichia coli. Multicopy suppressors of the mal3-1 mutant were retested for the ability to rescue the increased minichromosome loss phenotype of a mal3-1 (Ch16 (ade6-M210)) strain by visual screening for the suppression of colony sectoring (Niwa et al, 1989; Beinhauer et al, 1997).
Overexpression of dam1
The entire dam1 ORF was amplified from S. pombe genomic DNA using the 5′ primer GGCTCGAGATGGAGAAATACCAGAAAGC and 3′ primer ATCGGATCCTTATCTGGAAGCGGAATAGG and cloned into pREP3X after digestion with XhoI and BamHI to generate pREP3X-dam1. This plasmid was transformed into wild type and grown under selective conditions on EMM with 5 μg/ml thiamine to repress the nmt1 promoter. For high-level expression of dam1, cells were grown in liquid medium lacking thiamine for 20 h at 30°C.
Fixation and fluorescence microscopy
Cells expressing GFP- or CFP-tagged proteins were fixed in 3.7% formaldehyde at room temperature or in 100% cold methanol and washed in PBS. The length of fixation was optimised for the protein and tag and ranged from 5 to 15 min. Live analysis of cells expressing GFP- or CFP-tagged proteins was performed in an imaging chamber (CoverWell PCI-2.5, Grace Bio-labs) filled with 1 ml of 2% agarose in minimal medium and sealed with a 22 × 22 mm glass coverslip. For indirect immunofluorescence, nmt1-gfp-dam1 cells were incubated in 1.2 M sorbitol for 5 min and fixed for 1 h in 4% paraformaldehyde at room temperature before permeabilisation. Tubulin staining was performed with a monoclonal anti-Tat1 antibody (a kind gift from K Gull) and a rhodamine-conjugated anti-mouse secondary antibody. Fluorescence microscopy was performed on a Deltavision containing a photometrics CH350L liquid cooled CCD camera and Olympus IX70 inverted microscope with a × 100 objective equipped with Deltavision data collection system (Applied Precision, Issaquah, WA). Stacks of six Z-sections (0.5 μm apart) were taken at each time point with an exposure time of 1 s for both GFP and CFP.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Xiangwei He for communicating results prior to publication. We thank Hirofumi Aiba, Zac Cande, Karl Ekwall, Miguel Angel Garcia, Keith Gull, Kevin Hardwick, Yasushi Hiraoka, Tomohiro Matsumoto, Richard McIntosh, Paul Nurse, Corina Vietmeier-Decker, Kohta Takahashi, Ayumu Yamamoto and Mitsuhiro Yanagida for supplying strains and reagents. We also thank Kate Sullivan (Microscopy facility, NIMR) and Wai Han Yan (Photographics facility, NIMR) for technical assistance. TT is supported by Cancer Research UK. UF is supported by the Deutsche Forschungsgemeinschaft, Germany. JBAM is supported by grants from the Association for International Cancer Research, UK and the Medical Research Council, UK.
References
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR (2004) Aurora B regulates MCAK at the mitotic centromere. Dev Cell 6: 253–268 [DOI] [PubMed] [Google Scholar]
- Aoyama K, Kawaura R, Yamada H, Aiba H, Mizuno T (2000) Identification and characterization of a novel gene, hos3+, the function of which is necessary for growth under high osmotic stress in fission yeast. Biosci Biotechnol Biochem 64: 1099–1102 [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM (1992) Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114: 59–66 [DOI] [PubMed] [Google Scholar]
- Beinhauer JD, Hagan IM, Hegemann JH, Fleig U (1997) Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol 139: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher R, Beach D (1988) Involvement of cdc13+ in mitotic control in Schizosaccharomyces pombe: possible interaction of the gene product with microtubules. EMBO J 7: 2321–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazer SC, Williams HP, Chappell TG, Cande WZ (2000) A fission yeast kinesin affects Golgi membrane recycling. Yeast 16: 149–166 [DOI] [PubMed] [Google Scholar]
- Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR (2000) Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol 151: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G (2002) Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, Huffaker TC, Drubin DG, Barnes G (2001a) Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol 155: 1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G (2001b) Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol 152: 197–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU (2004) Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428: 93–97 [DOI] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR (1993) Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol 120: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman M, Cheeseman IM, Van Goor D, Drubin DG, Meluh PB, Barnes G (2001) Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol Biol Cell 12: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L, Wang W, Chan CS (1994) Type 1 protein phosphatase acts in opposition to Ipl1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol 14: 4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T (2002a) Spindle–kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J 21: 6015–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T (2002b) Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol 12: 610–621 [DOI] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J 20: 3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M (1999) Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev 13: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG (1998) Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol 143: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Tanaka TU, Lechner J, Schiebel E (2002) Four new subunits of the Dam1–Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J 21: 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A, Vietmeier-Decker C, Ortiz J, Karig I, Beuter C, Hegemann J, Lechner J, Fleig U (2004) The fission yeast kinetochore component Spc7 associates with the EB-1 family member Mal3 and is required for kinetochore–spindle association. Mol Biol Cell 15: 5255–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls W (1999) High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15: 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM (2004) Correcting improper chromosome–spindle attachments during cell division. Nat Cell Biol 6: 232–237 [DOI] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT (2004) Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol 14: 273–286 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L (2004) A standardized kinesin nomenclature. J Cell Biol 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bachant J, Alcasabas AA, Wang Y, Qin J, Elledge SJ (2002) The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev 16: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, Yates JR III, He X (2005) Molecular analysis of kinetochore architecture in fission yeast. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JL, De Wulf P, Sorger P, Harrison SC (2005) The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell 9: 3211–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Yamamoto E, Yamada H, Aiba H, Mizuno T (2000) Identification and characterization of a novel gene, hos2+, the function of which is necessary for growth under high osmotic stress in fission yeast. Biosci Biotechnol Biochem 64: 2493–2496 [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M (2001) M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol 11: 537–549 [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 8: 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 2: 521–534 [DOI] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, Aranda J, Fields S, Drubin DG, Barnes G (2003) Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell 14: 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219 [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K (2002) Evidence that the Ipl1–Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Tatebe H, Goshima G, Takeda K, Nakagawa T, Kinoshita K, Yanagida M (2001) Fission yeast living mitosis visualized by GFP-tagged gene products. Micron 32: 67–74 [DOI] [PubMed] [Google Scholar]
- Tournier S, Gachet Y, Buck V, Hyams JS, Millar JB (2004) Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol Biol Cell 15: 3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR (2001) pkl1(+)and klp2(+): two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell 12: 3476–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung HY, Wang W, Chan CS (1995) Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol Cell Biol 15: 6064–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR (2002) Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci 115: 931–940 [DOI] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR (1998) cut11(+): a gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelopee and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell 9: 2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G (2005) Formation of a dynamic kinetochore–microtubule interface through assembly of the Dam1 ring complex. Mol Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol 152: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH Jr, McDonald KL, McIntosh JR (1995) Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol 129: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S, Daga RR, Chang F (2004) Intra-nuclear microtubules and a mitotic spindle orientation checkpoint. Nat Cell Biol 6: 1245–1246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


