Abstract
The genomic environments and the transcripts of the mitochondrial cox3 gene are investigated in three Arabidopsis thaliana ecotypes. While the proximate 5′ sequences up to nucleotide position −584, the coding regions and the 3′ flanking regions are identical in Columbia (Col), C24 and Landsberg erecta (Ler), genomic variation is detected in regions further upstream. In the mitochondrial DNA of Col, a 1790 bp fragment flanked by a nonanucleotide direct repeat is present beyond position −584 with respect to the ATG. While in Ler only part of this insertion is conserved, this sequence is completely absent in C24, except for a single copy of the nonanucleotide direct repeat. Northern hybridization reveals identical major transcripts in the three ecotypes, but identifies an additional abundant 60 nt larger mRNA species in C24. The extremities of the most abundant mRNA species are identical in the three ecotypes. In C24, an extra major 5′ end is abundant. This terminus and the other major 5′ ends are located in identical sequence regions. Inspection of Atcox3 transcripts in C24/Col hybrids revealed a female inheritance of the mRNA species with the extra 5′ terminus. Thus, a mitochondrially encoded factor determines the generation of an extra 5′ mRNA end.
INTRODUCTION
In seed plants, mitochondrial DNAs encode between 54 and 59 genes (1–4). The realization of this genetic information occurs in several consecutive steps, which are so far only partially understood. Transcription is initiated at various promoter sequences, some of which have been functionally characterized by in vitro analyses in different plant species (5,6). In dicots, an 18 bp sequence enclosing the transcription initiation site is necessary and sufficient to drive transcription in vitro (6,7). Other promoter motifs, summarized recently in Arabidopsis thaliana, diverge partially or completely from the CRTAGAGA nonanucleotide motif, but the functional requirements for these are so far unclear (8). After transcription, primary RNAs undergo a series of processing steps. For instance, many genes are transcribed into polycistronic precursor RNAs that become disassembled into smaller units (9). Apart from intron splicing, 5′ and 3′ ends are post-transcriptionally generated. The 3′ termini are at least in some instances polyadenylated, which appears to accelerate degradation of plant mitochondrial RNA (10–14). Recently, two exoribonucleases have been characterized and found to be important or essential components of the 3′ processing machinery. Both proteins are required for the generation of the correct atp9 3′ mRNA terminus, which is located just downstream of an inverted repeat. The knock-down of the AtmtPNPase provokes the accumulation of large 3′ extended atp9 transcripts. The knock-out of the AtmtRNaseII, a protein that is also involved in processing of ribosomal RNA in chloroplasts (15), causes a defect in the final 3′ trimming of atp9 mRNAs, which are a few nucleotides longer in the mutant plants than their wild-type counterparts (16).
In plant mitochondria, the 5′ ends of tRNAs and rRNAs are also post-transcriptionally generated. An RNaseP-like enzyme has been found to execute 5′ endonucleolytic processing on tRNA precursor molecules, a processing event which can similarly be expected to occur at the 5′ ends of rRNAs (17). Indeed, such an endonucleolytic generation of the 18S rRNA 5′ end has recently indirectly been substantiated by the identification of a 5′ leader sequence as a product of an endonucleolytic cut (18). 5′ processing has also been observed at plant mitochondrial mRNAs, but in contrast to 3′ processing the mode and mechanism of this process are unclear and might be manifold. For instance the sequence surrounding the 5′ end of the 26S RNA in Oenothera has been found conserved at a 5′ terminus of an atp9 mRNA and has thus been suggested to function as cis-element for the recognition and/or processing of the respective precursor molecules (19). But it can also be speculated that certain secondary structures are required for processing although no supporting experimental evidence has been found so far. The function of 5′ processing is also unclear. In analogy to such processing events in chloroplasts of Chlamydomonas reinhardtii, it can be speculated that the generation of certain 5′ termini might be a prerequisite of translation; however, no experimental data that would substantiate this hypothesis are yet available for plant mitochondria (20).
In the course of the characterization of the complete mitochondrial transcriptome in the model plant A.thaliana, we have investigated the gene arrangements and the steady state transcripts of the mitochondrially encoded cox3 gene in three different ecotypes. While no differences were observed in the 3′ region, within the gene and 5′ flanking sequence up to position −584 sequences further upstream differ between the three ecotypes. Our analyses suggest that these far upstream sequences influence the generation of 5′ ends located ∼140 nt downstream in sequences, which are identical in all three investigated ecotypes.
MATERIALS AND METHODS
Preparation of nucleic acids from A.thaliana
A.thaliana seedlings were grown under a 16 h/8 h light/dark regime (100 μmol/m2 s) at 23°C for 15 or 16 days. Total cellular DNA was isolated from above ground parts of the seedlings and cell suspension culture with PhytoPure kit according to the manufacturer's instructions (Amersham Bioscience). Total DNA from 28-day C24/Col hybrid old plants (21), which were gown under 16 h/8 h light/dark regime (60 μmol/m2 s), was isolated with DNeasy Plant Mini kits (Qiagen).
Total RNA was isolated from the same plants with an RNeasy Plant Mini kit (Qiagen). Extraction procedures followed the manufacturer's recommendations.
Mitochondria were isolated from cell suspension culture following a protocol published previously (22). mtDNA and mtRNA were extracted according to a protocol described previously (23). For the isolation of mtDNA, 3 M sodium acetate (pH 7.0) was used instead of 2 M sodium acetate, pH 4.0.
Northern and Southern hybridization
For Southern blot analysis, ∼10 μg of total DNA were digested with HindIII and size fractionated on 1% (w/v) agarose gels. Southern transfer and hybridization were performed with porablotNY+ membranes following a protocol given by the manufacturer (Macherey-Nagel). The DNA probe corresponding to the cox3 N-terminal region (positions 3–232 with respect to the ATG) was generated by PCR with primers cox3-5PS and cox3-3PS. Labeling of the probe was performed with Rediprime™ II Random Prime Labelling System and 50 μCi [α-32P]dCTP according to the manufacturer's protocol (Amersham Bioscience).
Southern blot analysis of 1–3 μg of mtDNA was performed under the same conditions as described above. 5′ end labeling of oligonucleotide probes was performed using polynucleotide kinase following the standard procedures (24). An mtDNA library established in pBluescript II was screened after colony transfer to porablotNY+ membranes according to the manufacturer's protocol.
For northern blot analysis, ∼10 μg of total RNA was sizes fractioned on agarose gels under denaturing conditions in the presence of glyoxale (24). Blotting and hybridization was performed using Duralon UV as recommended by the manufacturer (Stratagene).
PCR analyses
PCRs were performed with Taq DNA polymerase (Promega), BD Advantage™ 2 PCR Enzyme System (BD Bioscience) and Phusion™ High-Fidelity DNA polymerase (Finnzymes) according to conditions specified by the companies. PCR parameters for Taq and BD Advantage™ DNA polymerase were as follows: 3 min at 95°C, 35 cycles of 1 min at 94°C, 1 min at oligonucleotide-specific annealing temperature, 1 min (per kb product) at 72°C (Taq) or 68°C (BD Advantage DNA Pol) and a final elongation step of 3 min at the same temperature. Parameter for amplification reactions performed with Phusion™ High-Fidelity DNA polymerase were 30 s at 98°C, 35 cycles of 8 s at 98°C, 25 s at oligonucleotide-specific annealing temperature, 30 s (per kb product) at 72°C and a final elongation step of 5 min. Primers used in the individual reactions are indicated in the text. Primer sequences are available on request.
CR–RT–PCR was performed as described previously (25). Primers used are given in the text and indicated in Figure 5, respectively.
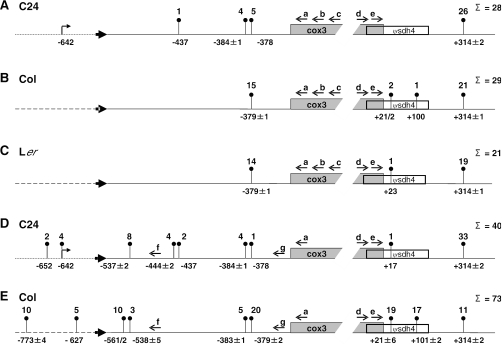
Figure 5.
CR–RT–PCR mapping of the 5′ and 3′ extremities of Atcox3 transcripts from ecotypes C24, Col and Ler. Analyses were performed with total RNA from green seedlings (A–C) and mtRNA from cell suspension cultures (D and E). The number of individual clones analyzed is given in the right margin. Prominent ends found in several clones are indicated by pins with number of clones above. The continuous line represents sequences identical in all investigated ecotypes; dashed lines are identical in Col and Ler; the dotted line is specific for C24 in this position. The locations of the ends are given in respect to the ATG or the stop codon of the Atcox3 gene. Primers (open arrows) used in the CR–RT–PCR are a: Atcox3-1; b: Atcox3-2; c: Atcox3-3PS; d: Atcox3-4; e: Atcox3-5; f: ATB2174-2; g: Atcox3-3.
Standard methods
Primer extension experiments, restriction analyses and other basic methods in molecular biology were carried out following the standard procedures (24) or protocols of the manufacturer. Sequence analysis was carried out with Thermo Sequenase sequencing chemicals and ALF express sequencers (Amersham Bioscience). In silico sequence analyses were carried out with different tools at the NCBI server (26,27).
RESULTS
A 1.8 kb insertion is present in the upstream region of the nuclear cox3 gene in A.thaliana ecotype Col
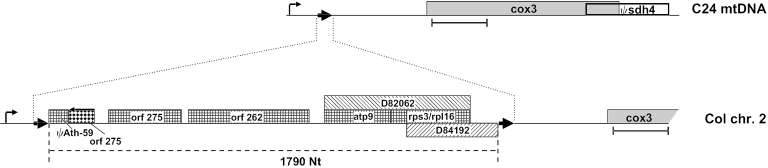
In the course of a systematic analysis of transcription in mitochondria of A.thaliana, mitochondrial sequences from ecotype C24 were compared with those of an mtDNA copy present in chromosome 2 in the nuclear DNA of ecotype Col (1,28). This nuclear mtDNA copy comprises ∼618 kb, of which ∼270 kb are sequenced. It is almost identical with the genuine mitochondrial DNA sequence determined in ecotype C24 (28,29). This comparison revealed the presence of an ∼1.8 kb insertion in the cox3 upstream region in the Col sequence (Figure 1). The insertion of 1790 bp is flanked by a 9 bp direct repeat, which is present in a single copy in the C24 mitochondrial sequence and which could potentially be involved in the insertion of this DNA fragment. The insertion is assembled from mtDNA fragments of various origins, a phenomenon frequently observed in these organelles (30). Major parts originate from rpl16/rps3, atp9, orf275 and orf262 sequences (1).
Figure 1.
Scheme of cox3 gene arrangements in the mitochondrial DNA of A.thaliana ecotype C24 (C24 mtDNA) and in the chromosome 2 mtDNA copy in the nuclear DNA of ecotype Col (Col chr. 2). A 1790 bp insertion flanked by nonanucleotide direct repeats (bold arrows) is present in Col. A putative cox3 promoter of the CNM-type (bent arrow) is found ∼640 (C24) and 2430 bp (Col) upstream of the cox3 reading frame (gray box), which overlaps with a Ψsdh4 gene (white box) in C24 and Col, respectively. Parts of this insertion are found in other genomic regions of the mtDNA of C24 (indicated by checkered boxes). An orf275 sequence in reverse orientation is given as rhomboid box with an arrow. rpl16/rps3 sequences found in the mtDNA of a maternal distorted leaf mutant are given by hatched boxes with the respective accession numbers (38). ΨAth-59 refers to a sequence with high similarity to a small non-coding RNA (44). The hybridization probe used in the Southern blot analysis of total DNA is given beneath the cox3 gene.
This insertion displaces a potential mitochondrial promoter present ∼640 bp upstream of the cox3 gene in ecotype C24 by 1.8 kb and thus could potentially influence transcription of this gene in ecotype Col. However, since this insertion was detected in the chromosomal copy the question arises whether the mtDNA of Col corresponds to the genomic arrangement observed in the nucleus or whether it corresponds to the mtDNA configuration present in C24 mitochondria.
Southern blot analysis reveals different Atcox3 environments in ecotypes Columbia (Col), C24 and Landsberg erecta (Ler)
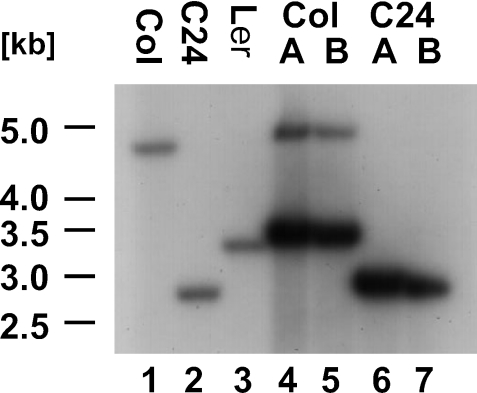
To investigate the genomic environment of the cox3 gene in these ecotypes, total DNA isolated from green seedlings was hybridized with a probe corresponding to the N-terminal part of the cox3 reading frame. The detection of HindIII fragments of 4.7 kb in Col, 2.9 kb in C24 and 3.4 kb in Ler identifies individual cox3 gene arrangements in the three ecotypes (Figure 2, lanes 1–3). Analogous results were obtained with EcoRV-digested DNAs (Col: 4.5 kb; C24 2.7 kb; Ler 7.2 kb) (data not shown). The fragment sizes detected in C24 and Col correspond to those expected from the sequences of mtDNA (C24) and chromosome 2 (Col), respectively (1,28). These hybridizations indicate that in green plants only single cox3 arrangements exist in the individual ecotypes, strongly suggesting that at least in Col the cox3 genomic environment is identical in mtDNA and in the mtDNA copy in chromosome 2. The presence of an mtDNA copy homologous to the one detected in ecotype Col was investigated by PCR. The results obtained strongly suggest that such homologous mtDNA copies do not exist in ecotypes C24 and Ler. This indicates that the insertion of the mtDNA copy in chromosome 2 in Col is an evolutionary recent event (Supplementary Figure S1).
Figure 2.
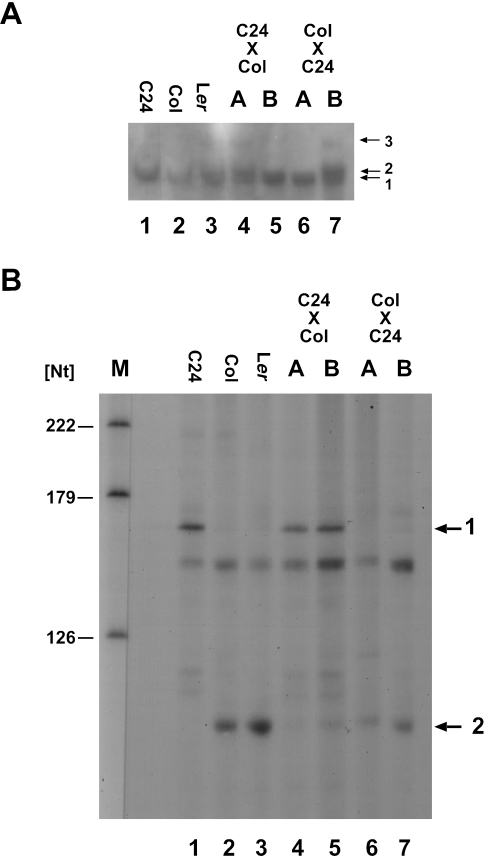
A cox3 probe detects different DNA fragments in A.thaliana ecotypes C24, Col and Ler. Total DNA isolated from green seedlings of ecotypes Col (lane 1), C24 (lane 2) and Ler (lane 3) as well as total DNA from two different cell suspension culture lines each (A and B) from Col (lanes 4 and 5) and C24 (lanes 6 and 7) were digested with HindIII (H) and hybridized with an Atcox3 probe.
A different hybridization pattern is observed in the Southern analysis of total DNA extracted from individually propagated cell suspension cultures. While again the expected fragments were detected in C24 DNA (Figure 2, 2.9 kb HindIII fragment in lanes 6 and 7; 2.7 kb EcoRV fragment, data not shown), each two fragments were visualized in the DNA of Col. Relatively weak signals correspond to the expected sizes observed in DNA from green plants (Figure 2, 4.7 kb HindIII fragments in lanes 4 and 5; 4.5 kb EcoRV fragments, data not shown). Additional strong signals are almost identical to those observed in green plants from Ler (Figure 2, 3.4 kb HindIII fragments in lanes 4 and 5; 7.2 kb EcoRV fragments, data not shown). An amplification of the 3.4 kb HindIII fragment by PCR followed by restriction digestion confirmed the identity of these fragments in Col cell suspension culture and Ler green plants (Supplementary Figure S2).
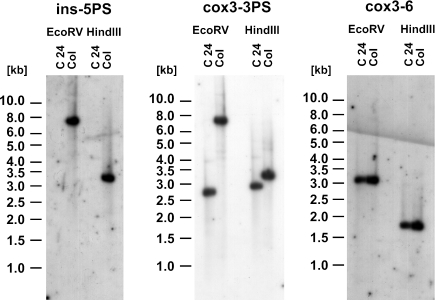
The different intensities of the signals in Col cell suspension culture could be due to the origin of the corresponding DNA fragments from the nucleus (weak signals) and the mtDNA (strong signals), which is present in much higher copy number in total DNA preparations. Thus, mtDNAs were isolated from cell suspension cultures (C24 and Col) and examined by Southern hybridization. An Atcox3-specific oligonucleotide probe (cox3-3PS) detects the expected DNA fragments in both HindIII- and EcoRV-digested DNAs from C24, which were identical with those found in all other hybridization analyses of this ecotype (Figure 3, center part; HindIII: 2.9 kb; EcoRV: 2.7 kb). In Col mtDNA also single fragments were detected (Figure 3, middle panel; HindIII: 3.4 kb; EcoRV: 7.2 kb). These correspond to the strong signals observed in the hybridization analysis of total DNA confirming the mitochondrial origin of these fragments (Figure 2, lanes 4 and 5). This also indirectly confirms that the relatively weak signals visualizing fragments with the expected size of 4.7 kb (HindIII) and 4.5 kb (EcoRV) are indeed of nuclear origin. An oligonucleotide probe specific for the insertion in Col (ins-5PS) only detects the predicted fragments in Col, confirming that this insertion or at least parts of it are also present in the mtDNA from this ecotype (Figure 3, left panel; HindIII: 3.4 kb; EcoRV: 7.2 kb). No hybridization of this probe was observed in the analysis of C24 mtDNA, suggesting that this insertion is completely absent from mtDNA of this ecotype. No differences were observed between the different ecotypes when hybridization probes correspond to Atcox3 3′ regions, suggesting that the different Atcox3 genomic environments detected in the investigated ecotypes as well as in different tissues of these (i.e. green plants and cell suspension cultures) are due to rearrangements in the 5′ regions (Figure 3, right panel; HindIII: 1.8 kb; EcoRV 3.1, data not shown).
Figure 3.
Southern blot analysis of mtDNA isolated from cell suspension cultures of ecotypes Col and C24. mtDNA was digested with HindIII and EcoRV, respectively, and hybridized with oligonucleotide probes specific for the 1790 bp insertion in Col (ins-5′PS, left panel), for the 5′ part of the cox3 reading frame (cox3-3PS, middle panel) and for the region downstream of the cox3 gene (cox3-6, right panel).
These experiments indicate that in Col cell suspension culture a cox3 arrangement becomes predominant that corresponds to the single detectable arrangement in green plants of ecotype Ler. This suggests that this arrangement pre-exists in Col most likely as a sublimon. Such substoichiometric molecules have previously been identified in maize mitochondria (31,32).
To exclude the possible contamination and/or mixtures of different ecotypes in the cell suspension culture, the ecotypes of the Col cell suspension cultures as well as of Col and Ler plants were verified using specific single nucleotide polymorphisms CER462304 and CER428393 (33) (data not shown).
The rearrangement in the mtDNA of Col suspension culture relocates a complete atp9 gene into the cox3 5′ region
For a detailed investigation of the cox3 genomic environment in the mtDNA of Col suspension cells, which might very similarly or identically also exist in ecotype Ler, a HindIII mtDNA genomic library was established and screened using the cox3-specific oligonucleotide probe (cox3-3PS). This identified several clones which contained 3.4 kb HindIII fragments, one of which was almost completely sequenced (data not shown).
The cox3 5′ region found in cell suspension culture is co-linear up to nucleotide position −1352 in respect to the ATG with the sequence of the mtDNA copy in chromosome 2 (Supplementary Figure S3), which is most likely identical to the original mtDNA in green plants of this ecotype (see above and Figures 2 and 3). This includes 65 bp of the 3′ terminal part of the atp9 gene and a 186 nt downstream region, which almost exactly corresponds to the 3′ non-translated region of the atp9 mRNA in both green plants and cell suspension culture [Forner and Binder, unpublished data, (16)]. In contrast to the sequence in chromosome 2 (Figure 1), a complete atp9 gene including a 5′ region containing two CNM-type promoters is present in the cell suspension specific cox3 arrangement in Col.
Different cox3 mRNAs are present in the three investigated A.thaliana ecotypes
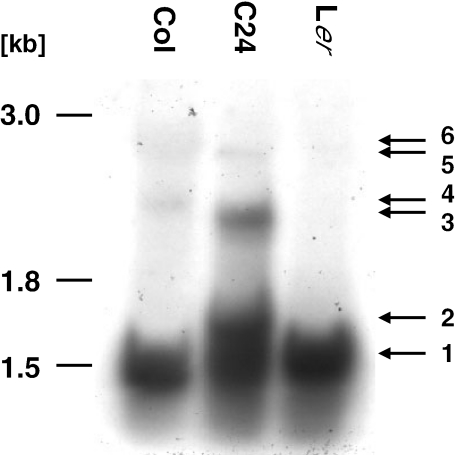
The analysis of the genomic environments of the cox3 gene in the three different ecotypes revealed substantial variations in the 5′ regions. To investigate the potential influence of these rearrangements on transcription, Atcox3 steady state transcripts from green seedlings were analyzed by northern hybridization. A 1.45 kb probe corresponding to the complete cox3 reading frame as well as 341 and 306 nt 5′ and 3′ flanking sequences detects mRNAs of ∼1500 nt from all three ecotypes (Figure 4). In C24, an additional major steady state mRNA species of ∼1560 nt and a minor RNA molecule of ∼2200 nt are detected. Additional transcripts are also detected in Col, but these are different from those found in C24. Thus, cox3 mRNAs of different sizes are present in the three ecotypes.
Figure 4.
Northern blot analysis of Atcox3 mRNA in total RNAs isolated from green seedlings of A.thaliana ecotypes C24, Col and Ler. A DNA probe representing the complete cox3 reading frame as well as 341 and 306 nt 5′ and 3′ flanking sequences detects transcript of ∼1500 nt (1) in all three ecotypes. A second prominent transcript of ∼1560 nt (2) is solely detectable in C24. In this ecotype, an ∼2.2 kb prominent (3) and an ∼2.8 kb low abundant (5) potential precursor RNA, respectively, are also observed. Such potential primary transcripts (4 and 6) with slightly different sizes are also present in Col. The individual RNAs are indicted by arrows and numbered in the right margin.
Different 5′ termini are found in identical cox3 upstream sequences in the three ecotypes
In order to determine the 5′ and 3′ extremities of the cox3 mRNA in the different ecotypes, CR–RT–PCR analyses were performed with total RNAs from green seedlings and mitochondrial RNA from cell suspension cultures. In all RNAs from all ecotypes and tissues identical major 3′ ends 314 (+/2) nucleotides downstream of the cox3 reading frame were found (Figure 5A–E). In Col cell suspension culture, additional major 3′ ends of shorter RNAs were present (Figure 5E). These ends were found 21 (±6) and 101 (±2) nucleotides, respectively, downstream of the cox3 stop codon and were likewise found in green seedlings of the same ecotype, however, at a much lower frequency (Figure 5B, 2 and 1 out of 29 clones).
In contrast to the 3′ termini, a complex situation is found for the 5′ ends. In green seedlings, major 5′ ends are found at position −379 (±1) in all investigated ecotypes (Figure 5A–C). These ends were detected after two PCRs with primer pairs b/d and a/e after initiating cDNA synthesis with primer c. In C24, two additional ends are detected at −384 (±2) and −437 (Figure 5A). With RNA from the cell suspension culture of C24 5′ ends were also found at −384 (±1) and −444/−437 (Figure 5D). With RNA from cell suspension culture from Col again a major 5′ end is detected at −379 (±2) but also a minor end at −383 (±1), which is not detected in green seedling of this ecotype. A CR–RT–PCR of cell suspension culture RNA was performed with 5′ primer g annealing 295–316 nt upstream of the oligonucleotide a used for the analysis of RNA from green seedlings. However, both primers essentially detect the same ends (Figure 5). In addition, in the analyses with primer a no ends were detected downstream of primer g. Taken together the CR–RT–PCR with these primer pairs detects four different 5′ ends. Two of these ends locate in close vicinity at −379 and −384 and can be considered as a single end, although the frequencies with which the individual ends were found differ between the investigated ecotypes. This 5′ end and the 3′ terminus at +314 define the major transcript of ∼1500 nt, detected in all three ecotypes (Figure 4). The other 5′ ends at −444 and −437, respectively, also represent probably a single terminus, which is unique to C24. This could be the 5′ end of the shorter of the two additional RNA species that are exclusively found in this ecotype (Figure 6, middle lane C24). To identify 5′ ends further upstream located, CR–RT–PCR with primer f located upstream of the detected major 5′ termini was performed with mtRNA from cell suspension culture from C24 and Col. These analyses detect identical 5′ ends at −537 (±2) and −538 (±5) (Figure 5D and E), respectively, in both ecotypes, and a Col-specific end at −561/−562. All the different 5′ ends found in the ecotypes investigated are located in a region in which the sequences are identical.
Figure 6.
Transcript analysis of C24 × Col and Col × C24 hybrids. (A) Northern hybridization of total RNA from ecotypes C24, Col, Ler (lanes 1–3) and each two plants (A and B) of C24 × Col and Col × C24 hybrids (lanes 4–7). The C24 specific RNAs of ∼1560 and 2800 nt are detected in the C24 parental line (lane 1) and in hybrids with C24 female parents (lanes 4 and 5). (B) Primer extension analysis of the same RNAs as described in (A) with primer Atcox3-3. Products of ∼160 nt (1) were generated on RNAs of C24 (lane 1) and of plants A and B with C24 as female parental line (lanes 4 and 5). These products correspond to 5′ ends at positions around −440, which are exclusively detected in the CR–RT–PCR of this ecotype (Figure 5A and D). A second prominent product (2) of ∼100 nt is detected in almost all plants, except in C24 in lane 1. This ends corresponds to 5′ termini around −380, which was found in the CR–RT–PCR in all ecotypes (Figure 5).
In steady state RNAs from cell suspension cultures from C24 and Col additional differing cox3 5′ ends were detected further upstream with 5′ primer f. However, these are located in a sequence region that differs between these ecotypes. In C24, these map to positions −642 and −652 directly at a predicted CNM-type promoter or 10 nt upstream, respectively. In Col 5′ termini at −627 and −773 (±4) are detected in another CR–RT–PCR and are situated in the 1790 bp insertion.
Another ecotype-specific 5′ end is detected at positions −561/−562 in Col cell suspension culture. Since this end has so far not been detected by the primer extension analysis, an experimental artifact can presently not be excluded.
Cox3 transcript analysis in C24/Col hybrids
Since the major Atcox3 5′ ends are located in identical sequence environments, two factors might be responsible for the generation of these varying termini: either a mitochondrially encoded factor which could act in trans or cis, or (a) trans factor(s) encoded in the different nuclear backgrounds of the ecotypes. We thus analyzed the Atcox3 5′ ends in individual plants of reciprocal crossings between C24 and Col. The identity of the nuclear background of the parent plants and these hybrids was confirmed using SNPs MASC03783 and MASC03308 (34) (data not shown). In addition, the configuration and thus origin of the mtDNA was tested by the presence or absence of the 1790 (+9 bp repeat) bp insertion by PCR (Supplementary Figure S4). In all plants tested, the configuration of the mtDNA corresponds to the female parent, which is in agreement with a strict maternal inheritance of mitochondria.
Total RNAs isolated from the hybrids and the parental ecotypes were investigated by northern hybridization. The cox3 probe used in these experiments has been described in the above mentioned northern analysis. In ecotypes C24 and Col the same mRNAs are detected as described above (Figure 4 and Figure 6A, lanes 1 and 2). In the hybrids, the pattern of cox3 mRNAs is always consistent with those found in the female parent, demonstrating a maternal inheritance of the C24-specific mRNA species of ∼1560 nt (Figure 6A, lanes 4–7).
Investigation of these mRNAs by primer extension analysis with oligonucleotide Atcox3-3 revealed a corresponding result. 5′ termini detected in the parental lines are consistent with those found in the CR–RT–PCR described above, except for C24. Here, primer extension signal 1 corresponds to the 5′ ends mapped at −444/−437; however, signal 2 corresponding to −384/−378 is not visible, although it has been clearly detected in the CR–RT–PCR and a corresponding mRNA is detected in the northern analysis (Figure 6B, lane 1; Figure 5A and D). In Col and Ler extension, signal 2 matches to the −378 end, while signal 1 corresponding to the 5′ end at −444/−437 is not observed. This is consistent with the results of the CR–RT–PCR analyses (Figure 6B, lanes 2 and 3; Figure 5B, C and E). The 5′ ends mapped in the hybrid plants are identical with those of the female parent. In hybrid plants with a C24 female parent, primer extension signal 1 is clearly detectable (Figure 6B, lanes 4 and 5) while this 5′ end is not found in hybrid plants with a Col female parent. Thus, the generation of the −444/−437 5′ end in C24 is independent from the ecotype-specific nuclear background but follows the maternal lineage, which is consistent with the northern analysis. This strongly suggests a cytoplasmic inheritance of this trait. Thus, a mitochondrial factor is responsible for the generation of the additional 5′ end located within the sequence identical in C24, Col and Ler.
DISCUSSION
A nonanucleotide direct repeat might be responsible for the integration or excision of the 1790 bp DNA fragment
Our analysis of the Atcox3 upstream sequence revealed three different genomic arrangements in the far upstream region of this gene. In Col, a 1790 bp insert is found to be flanked by a nonanucleotide direct repeat (5′-CTTTACGAG-3′), which is present as a single copy at this locus in C24 (Figure 1). These arrangements suggest that this nonanucleotide repeat is involved in the recombinatorial integration or excision of the insert. Such small repeats have been previously found to be involved in recombination events (35). For instance, in Oenothera a decanucleotide repeat is responsible for circularization and formation of a small subgenomic molecule, which contains the 3′ part of the 26S rRNA (36). Similarly, the 1790 bp insertion might be excised from the mtDNA genome by an intramolecular recombination of the nonanucleotide repeats in green plants of ecotype Col. However, no corresponding signal has been observed in the hybridization of total DNA of Col plants. This is most likely due to the low abundance of such a molecule, which would be present in substoichiometric amounts as described for sublimons (31). The identified nonanucleotide can be found in four other places within the mitochondrial genome of C24 (five places in total) and four times within the chromosome 2 sequence in Col at locations identical with those in C24, but there are so far no experimental data for the involvement of one of these repeats in a recombination event.
A third configuration is present in Ler
The 1790 bp insert in Col is composed of mtDNA fragments of different origins (Figure 1). Smaller sequence stretches with sizes between 32 and 71 bp are unique and are not found in the mitochondrial (C24), chloroplast (Col) or nuclear (Col) genome of A.thaliana. However, since the mitochondrial DNA copy within chromosome 2 of Col is not completely sequenced, it is possible that homologous sequences are present in another locations in the mtDNA (1,28,37). All other parts are duplicated sequences, whose origins are indicated in Figure 1. These are parts of other genes or ORFs. The insert is only partially conserved in the Ler configuration, which is also the predominant arrangement in the Col suspension culture (Supplementary Figure S3). Here, most likely a recombination has occurred within the atp9 sequence, which established a complete reading frame of this gene. The insert sequences downstream of the atp9 gene are conserved in fragments of a Maternal Distorted Leaf (MDL) mutant established in Ler (accession numbers: D82062 and D84192) (38). This supports that identical arrangements exist in Ler and Col. Most likely, the set up of the cell suspension culture has shifted the stoichiometry of the two pre-existing cox3 arrangements in Col, similar to the changes observed in the chm1-1 mutant (39,40), a phenomenon that has been observed in sublimons of other plant species and that contributes to the complexity of plant mitochondrial genomes (31,32,40–42). Interestingly, the size of the BamHI fragment deduced from the 3.4 kb HindIII fragment and the Ler sequence of data bank entry D82062 is 1.56 kb, which fits with the predominant fragment hybridizing with an atp9 in the chm1-1 mutant (40). Thus, the set up of the cell suspension culture might have somehow altered chm gene expression, which may have caused the shift in stoichiometry. However, detailed comparative studies of the atp9 arrangements in the different ecotypes are necessary to elucidate these complex genomic conditions. Differing mtDNA configurations have also been observed in other ecotypes and other genomic environments (43).
MtDNA copies in different ecotypes?
Three different 5′ arrangements are found in the mtDNA of the three investigated ecotypes. This raises the question whether potential nuclear mtDNA copies, if they exist, correspond to the individual mtDNA configurations or whether they all correspond to the configuration of the mtDNA copy in Col. In ecotypes C24 and Ler, our PCR analyses do not indicate the presence of an mtDNA copy homologous to the one in Col. This does not generally exclude other ecotypes to contain an mtDNA copy homologous to the one in Col; however, we assume that such a copy might only be present in ecotypes closely related to Col. Furthermore, the data suggest the insertion of the mtDNA in chromosome 2 of Col to be an evolutionary recent event. More studies with a large variety of different ecotypes are required to finally clarify this issue.
Different precursor RNAs are detectable in Col and C24
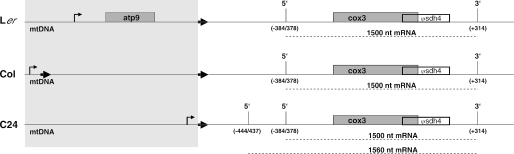
The northern analysis revealed different potential precursor RNAs in the different ecotypes. These might be attributed to transcription initiation at different promoters (Figure 7). In C24, a CNM-type promoter can be predicted with a potential transcription start point 642 bp upstream of the Atcox3 reading frame. Indeed, 5′ ends were mapped at this site confirming the functional importance of this promoter in this ecotype. However, other 5′ termini are mapped 10 nt upstream, suggesting a further 5′-located transcription initiation site. This promoter as well as the CNM-type sequence are also present in the Atcox3 arrangement in Col. Although the 1790 bp insertion (+9 bp repeat) increased the distance to the cox3 reading frame, these promoters are probably similarly active. However, it is also possible that transcription initiation sites are present within the insertion, which in addition could contribute to the transcription of this gene in Col. Several 5′ termini have been mapped around positions −773 and −627 within the insertion (Figure 5E); however, it is unclear whether they originate from processing or transcription initiation. Sequences at these 5′ ends do not show any striking similarity to new A.thaliana promoter sequences identified recently (8); however, considering their variability the presence of promoters at these ends can well be possible. The sequences at these ends are also present in the Ler arrangement. Thus, as in Col transcription initiation at these sequences may occur, although it is more likely that the cox3 is co-transcribed with the 5′-located atp9 gene, for which several promoters have been identified (8).
Figure 7.
Genomic arrangements and transcripts of the cox3 genes in three different A.thaliana ecotypes. The localization of the atp9, cox3 (gray boxes) and the Ψsdh4 genes (transparent box) are indicated in the mtDNAs of the investigated A.thaliana ecotypes here. The major 5′ and 3′ termini of the predominant mRNAs (dashed lines) are given including their position in respect to the ATG (5′ ends) and stop codon (3′ ends). Genomic regions that differ between the ecotypes are highlighted by a gray background. Mitochondrial promoters are given as bent arrows. Bold arrows represent the nonanucleotide direct repeat.
A mitochondrial factor is responsible for the generation of an alternative 5′ end
In all ecotypes, a major transcript of ∼1500 nt can be detected (Figure 4). The 3′ ends are consistently found 314 nt downstream of the cox3 stop codon (Figure 5). The generation of these ends either by transcription termination or by post-transcriptional processing thus seems to be independent from 5′ end formation, which is different in the investigated ecotypes. A major 5′ terminus is also consistently found in the three investigated ecotypes and crosses of them (Figures 4–6). Only in C24 the 5′ end at −384/−378 is not detectable in the primer extension analysis (Figure 6B, lane 1), although both the detection of the 1500 nt transcript and the CR–RT–PCR demonstrate the existence of this end (Figures 4, 5 and 6A). Thus, both cis and trans factors determining this end are identical in these ecotypes. While the latter are unknown, the cis elements are most likely located within the sequences identical in all three ecotypes ranging from 584 bp upstream of the cox3 reading frame to the 3′ ends.
In C24, an additional prominent 5′ end ∼60 nt upstream of the normal major 5′ terminus is detected (Figures 5 and 6B). This end is also located in a sequence region, which is identical in the investigated ecotypes (Figure 7). A nuclear encoded factor responsible for the generation of this end can be excluded, since the appearance of this end is maternally and thus mitochondrially inherited. Thus, a mitochondrially encoded cis element or trans factor specifies the formation of this 5′ end. Considering the coding capacity of all known mitochondrial genomes, a mitochondrially encoded trans factor required for specific transcription initiation or processing is highly unlikely. Instead, we assume that the differing sequences located at least 140 bp upstream are responsible for the appearance of this alternative 5′ end. These sequences most likely differently influence the folding of the precursor RNA, which in the case of C24 creates a secondary structure that allows a processing in this sequence region. Thus, there is influence across a distance of >140 nt. Generally two different scenarios are feasible: either the alternative folding allows base pairing at this −444/437 site and a processing by a double-strand-specific ribonuclease or in contrary the normally base paired region is made single stranded and thus accessible to a single-strand-specific ribonuclease.
Taken together, the detailed analysis of mitochondrial transcripts in different ecotypes is an additional useful tool to elucidate and characterize the requirements for transcript end formation in plant mitochondria.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Carmen Schilling-Kolle for taking care of the cell suspension cultures. This work is supported by DFG grant Bi 590/6-1 and a fellowship of the Studienstiftung des Deutschen Volkes to J.F. Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Unseld M., Marienfeld J.R., Brandt P., Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 2.Notsu Y., Masood S., Nishikawa T., Kubo N., Akiduki G., Nakazono M., Hirai A., Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 3.Kubo T., Nishizawa S., Sugawara A., Itchoda N., Estiati A., Mikami T. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA) Nucleic Acids Res. 2000;28:2571–2576. doi: 10.1093/nar/28.13.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapp W.D., Stern D.B. A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J. 1992;11:1065–1073. doi: 10.1002/j.1460-2075.1992.tb05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dombrowski S., Hoffmann M., Guha C., Binder S. Continuous primary sequence requirements in the 18-nucleotide promoter of dicot plant mitochondria. J. Biol. Chem. 1999;274:10094–10099. doi: 10.1074/jbc.274.15.10094. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann M., Binder S. Functional importance of nucleotide identities within the pea atp9 mitochondrial promoter sequence. J. Mol. Biol. 2002;320:943–950. doi: 10.1016/s0022-2836(02)00552-1. [DOI] [PubMed] [Google Scholar]
- 8.Kühn K., Weihe A., Borner T. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 2005;33:337–346. doi: 10.1093/nar/gki179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kuhn J., Däschner K., Binder S. The RNA world of plant mitochondria. Prog. Nucleic Acid Res. Mol. Biol. 2001;70:119–154. doi: 10.1016/s0079-6603(01)70015-3. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn J., Tengler U., Binder S. Transcript lifetime is balanced between stabilizing stem–loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol. 2001;21:731–742. doi: 10.1128/MCB.21.3.731-742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombrowski S., Brennicke A., Binder S. 3′-Inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J. 1997;16:5069–5076. doi: 10.1093/emboj/16.16.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagliardi D., Leaver C.J. Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J. 1999;18:3757–3766. doi: 10.1093/emboj/18.13.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagliardi D., Perrin R., Marechal-Drouard L., Grienenberger J.M., Leaver C.J. Plant mitochondrial polyadenylated mRNAs are degraded by a 3′- to 5′exoribonuclease activity, which proceeds unimpeded by stable secondary structures. J. Biol. Chem. 2001;276:43541–43547. doi: 10.1074/jbc.M106601200. [DOI] [PubMed] [Google Scholar]
- 14.Lupold D.S., Caoile A.G., Stern D.B. Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell. 1999;11:1565–1578. doi: 10.1105/tpc.11.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollenbach T.J., Lange H., Gutierrez R., Erhardt M., Stern D.B., Gagliardi D. RNR1, a 3′–5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrin R., Meyer E.H., Zaepfel M., Kim Y.J., Mache R., Grienenberger J.M., Gualberto J.M., Gagliardi D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004;279:25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- 17.Marchfelder A. Plant mitochondrial RNase P. Mol. Biol. Rep. 1995;22:151–156. doi: 10.1007/BF00988721. [DOI] [PubMed] [Google Scholar]
- 18.Perrin R., Lange H., Grienenberger J.M., Gagliardi D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004;32:5174–5482. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuster W., Brennicke A. Conserved sequence elements at putative processing sites in plant mitochondria. Curr. Genet. 1989;15:187–192. doi: 10.1007/BF00435505. [DOI] [PubMed] [Google Scholar]
- 20.Marchfelder A., Binder S. Plastid and plant mitochondrial RNA processing and RNA stability. In: Daniell H., Chase C., editors. Molecular Biology and Biotechnolgy of Plant Organelles. Dordrecht, Netherlands: Springer; 2004. pp. 261–294. [Google Scholar]
- 21.Meyer R.C., Torjek O., Becher M., Altmann T. Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 2004;134:1813–1823. doi: 10.1104/pp.103.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein M., Binder S., Brennicke A. Purification of mitochondria from Arabidopsis. Methods Mol. Biol. 1998;82:49–53. doi: 10.1385/0-89603-391-0:49. [DOI] [PubMed] [Google Scholar]
- 23.Binder S. Mitochondrial nucleic acid purification and analysis. Methods Mol. Biol. 1995;49:383–389. doi: 10.1385/0-89603-321-X:383. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Habror Laboratory Press; 1989. [Google Scholar]
- 25.Kuhn J., Binder S. RT–PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 2002;30:439–446. doi: 10.1093/nar/30.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusova T.A., Madden T.L. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Lin X., Kaul S., Rounsley S., Shea T.P., Benito M.I., Town C.D., Fujii C.Y., Mason T., Bowman C.L., Barnstead M., et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 29.Stupar R.M., Lilly J.W., Town C.D., Cheng Z., Kaul S., Buell C.R., Jiang J. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: implication of potential sequencing errors caused by large-unit repeats. Proc. Natl Acad. Sci. USA. 2001;98:5099–5103. doi: 10.1073/pnas.091110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 31.Small I., Isaac P.C., Leaver C.J. Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial diversity in maize. EMBO J. 1987;6:865–869. doi: 10.1002/j.1460-2075.1987.tb04832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small I., Suffolk R., Leaver C.J. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell. 1989;58:69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 33.Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 2002;129:440–450. doi: 10.1104/pp.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid K.J., Sorensen T.R., Stracke R., Torjek O., Altmann T., Mitchell-Olds T., Weisshaar B. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 2003;13:1250–1257. doi: 10.1101/gr.728603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andre C., Levy A., Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. doi: 10.1016/0168-9525(92)90370-J. [DOI] [PubMed] [Google Scholar]
- 36.Manna E., Brennicke A. Site-specific circularization at an intragenic sequence in Oenothera mitochondria. Mol. Gen. Genet. 1986;203:377–381. [Google Scholar]
- 37.Sato S., Nakamura Y., Kaneko T., Asamizu E., Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto W., Kondo H., Murata M., Motoyoshi F. Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell. 1996;8:1377–1390. doi: 10.1105/tpc.8.8.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Zapater J.M., Gil P., Capel J., Somerville C.R. Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell. 1992;4:889–899. doi: 10.1105/tpc.4.8.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelnoor R.V., Yule R., Elo A., Christensen A.C., Meyer-Gauen G., Mackenzie S.A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl Acad. Sci. USA. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janska H., Sarria R., Woloszynska M., Arrieta-Montiel M., Mackenzie S.A. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell. 1998;10:1163–1180. doi: 10.1105/tpc.10.7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackenzie S., McIntosh L. Higher plant mitochondria. Plant Cell. 1999;11:571–586. doi: 10.1105/tpc.11.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ullrich H., Lattig K., Brennicke A., Knoop V. Mitochondrial DNA variations and nuclear RFLPs reflect different genetic similarities among 23 Arabidopsis thaliana ecotypes. Plant Mol. Biol. 1997;33:37–45. doi: 10.1023/a:1005720910028. [DOI] [PubMed] [Google Scholar]
- 44.Marker C., Zemann A., Terhorst T., Kiefmann M., Kastenmayer J.P., Green P., Bachellerie J.P., Brosius J., Huttenhofer A. Experimental RNomics: identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 2002;12:2002–2013. doi: 10.1016/s0960-9822(02)01304-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.