Abstract
At the onset of M phase, the activity of somatic Wee1 (Wee1A), the inhibitory kinase for cyclin-dependent kinase (CDK), is down-regulated primarily through proteasome-dependent degradation after ubiquitination by the E3 ubiquitin ligase SCFβ-TrCP. The F-box protein β-TrCP (β-transducin repeat-containing protein), the substrate recognition component of the ubiquitin ligase, binds to its substrates through a conserved binding motif (phosphodegron) containing two phosphoserines, DpSGXXpS. Although Wee1A lacks this motif, phosphorylation of serines 53 and 123 (S53 and S123) of Wee1A by polo-like kinase 1 (Plk1) and CDK, respectively, are required for binding to β-TrCP. The sequence surrounding phosphorylated S53 (DpSAFQE) is similar to the conserved β-TrCP-binding motif; however, the role of S123 phosphorylation (EEGFGSSpSPVK) in β-TrCP binding was not elucidated. In the present study, we show that phosphorylation of S123 (pS123) by CDK promoted the binding of Wee1A to β-TrCP through three independent mechanisms. The pS123 not only directly interacted with basic residues in the WD40 repeat domain of β-TrCP but also primed phosphorylation by two independent protein kinases, Plk1 and CK2 (formerly casein kinase 2), to create two phosphodegrons on Wee1A. In the case of Plk1, S123 phosphorylation created a polo box domain-binding motif (SpSP) on Wee1A to accelerate phosphorylation of S53 by Plk1. CK2 could phosphorylate S121, but only if S123 was phosphorylated first, thereby generating the second β-TrCP-binding site (EEGFGpS121). Using a specific inhibitor of CK2, we showed that the phosphorylation-dependent degradation of Wee1A is important for the proper onset of mitosis.
Keywords: polo-like kinase 1, ubiquitin, cell cycle, CK2, β-TrCP
Wee1 family protein kinases that inhibit Cdc2 during the G2 phase of the cell cycle must be down-regulated at the onset of mitosis. At that time, somatic Wee1 (Wee1A) is degraded by the proteasome after ubiquitination by E3 ubiquitin ligase SCFβ-TrCP (1) (in which β-TrCP is β-transducin repeat-containing protein). Although Wee1A lacks the conserved β-TrCP-binding motif (DpSGXXpS), phosphorylation of serines S53 and S123 of Wee1A by polo-like kinase 1 (Plk1) and cyclin-dependent kinase (CDK), respectively, is required for binding to β-TrCP and Wee1A turnover (1, 2). The sequence surrounding S53 of Wee1A is similar to the consensus binding motif for β-TrCP when S53 is phosphorylated (DpSAFQE) and acts as a phosphodegron (PD53), with E57 serving as a phosphomimetic in place of the downstream phosphoserine (Fig. 1A). But in the second Wee1A phosphodegron surrounding S123 [PD123 (EEGFGSSpSPVK)], although the S123 phosphorylation must be important for the recognition because even the S123A single mutation markedly stabilizes Wee1A (see Fig. 1B), the precise mechanism of PD123 recognition and the role of pS123 was not clear.
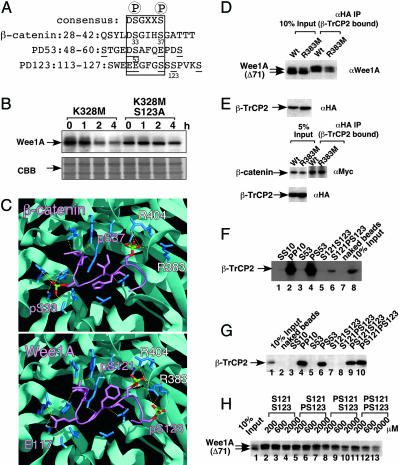
Fig. 1.
Phosphorylated S123 in Wee1A interacts with the WD40 domain of β-TrCP. (A) Two phosphodegrons (PD53 and PD123) in the Wee1A sequence and a phosphodegron in β-catenin are aligned with the consensus binding sequence (boxed) for β-TrCP. The important amino acids in Wee1A for binding to β-TrCP are underlined (1). (B) HeLa cells were transfected with kinase-negative Wee1A (K328M) or its S123A mutant. Two days after transfection, cells were treated with nocodazole (0.1 μg/ml) for 21 h. Cycloheximide (10 μg/ml) was then added, and the stability of Wee1A was examined by immunoblotting at the times indicated. (C) Structures of the phosphodegron peptide of β-catenin (Upper) and PD123 phosphopeptide of Wee1A (Lower) bound to the β-TrCP WD40 domain. The β-TrCP WD40 domain is shown in light blue, with its side chains in dark blue, and the phosphodegron peptides of β-catenin or Wee1A are in purple [phosphate groups on serines are shown in green (P) and red (O)]. Hydrogen bonds are represented by yellow dotted lines. The crystal structure of the interface between the WD40 domain and phosphodegron peptide of β-catenin is as published in ref. 23, although the numberings of the WD40 domain are for β-TrCP2, whereas the interface between the WD40 domain and PD123 is a representative model structure calculated by an energy minimization using sybyl 6.91 with the Kollman amber force field (30). The calculated intermolecular interaction energy values between β-TrCP and each of the Wee1A peptides PS121PS123, PS121S123, S121PS123, and S121S123 were –2.33 ± 0.33, –1.96 ± 0.13, –1.79 ± 0.37, and –1.38 ± 0.26 × 103 kcal/mol, respectively; those between R383M and R404M mutated β-TrCP and PS121PS123 peptides were –1.98 ± 0.21 and –2.09 ± 0.29 × 103 kcal/mol, respectively. (D) Wee1A lacking 71 amino acids from the N terminus (Δ71Wee1A) was coexpressed with HA-tagged β-TrCP2 or its R383M mutant in HtTA-1 cells. Cell lysates (10% of total input) and αHA immunoprecipitates of the lysates were analyzed by immunoblotting. HA-β-TrCP2 is hard to detect in αHA immunoprecipitates, because it migrates at a similar position to that of Ig heavy chain. (E) HA-taggedβ-TrCP2 or its R383M mutant was coexpressed with Myc-tagged β-catenin and axin in HtTA-1 cells and immunoprecipitated as in D.β-Catenin bound to HA-β-TrCP2 was detected by using αMyc antibody. (F and G) Peptides as indicated were coupled to Sepharose beads. The beads were mixed with HtTA-1 cell lysates expressing HA-tagged β-TrCP2 and washed. Cell lysates (10% of total input) and bound proteins were examined by immunoblotting with αHA antibody. (H) Δ71Wee1A was coexpressed with HA-tagged β-TrCP2 in HtTA-1 cells. The ability of peptides to abolish the association of Wee1A and β-TrCP2 was examined by the addition of peptides at the indicated concentration to the cell lysates before the addition of αHA antibody in the coimmunoprecipitation assay as in D.
Plk1 is a protein kinase that is active at late G2 phase and in M phase of the cell cycle and has several roles in the initiation, progression, and termination of mitosis (reviewed in ref. 3). Its activity is regulated at both the protein level and by phosphorylation. In addition, Plk family kinases, including Plk1, have a unique domain, PBD (polo box domain), in their C-terminal noncatalytic region, which is important not only for regulation of their intrinsic kinase activity and cellular localization but also for substrate recognition (4–7). PBD functions as a phospho-Ser/Thr-binding motif (6). Based on in vitro phosphopeptide-binding studies, the optimal phosphopeptide-binding motif for the human Plk1 PBD showed strong selectivity for a serine at the –1 position of the phosphorylation site and some specificity for proline at the +1 position, suggesting that PBD recognizes a substrate that is prephosphorylated by proline-directed kinases, such as CDKs and mitogen-activated protein kinases. Indeed, Plk1 has been shown to bind Cdc25C by means of a PBD-binding motif in a CDK phosphorylation-dependent manner, and the phosphorylation of Cdc25C by Plk1 is important for the initiation of mitosis (6, 8). A PBD-binding motif-dependent phosphorylation by Cdc2 of Myt1 is also important for the initiation of mitosis (9). Plk1 also binds to and phosphorylates proteins that are important for the termination of mitosis or cytokinesis, such as Mklp2, Emi1, Nir2, GRASP65, and MEI-S332, after prephosphorylation by Cdc2 during mitosis (10–14). However, these proteins seem to use more than one suboptimal PBD-binding motif, rather than optimal motifs, for binding to Plk1. Thus, the cumulative evidence suggests that whether a Plk1 substrate has optimal PBD-binding motifs or suboptimal ones may relate to the timing of its phosphorylation by Plk1 during mitosis. The Plx1 PBD binds Claspin by means of a site phosphorylated by a checkpoint-activated kinase (not proline-directed), promoting Plx1-mediated phosphorylation of Claspin, which is required for the adaptation of the replication checkpoint in the Xenopus system (15).
CK2 (formerly casein kinase 2) is a highly conserved serine/threonine protein kinase that is ubiquitously expressed in both the cytoplasm and nucleus of eukaryotic cells (reviewed in refs. 16 and 17). It is constitutively active and independent of either second messenger or phosphorylation events. The preferential CK2 phosphorylation site is Ser/Thr with an acidic amino acid at the n + 1, 2, or 3 position (16, 17). Because phosphorylated Ser or Thr serves as an acidic residue in this connection, priming phosphorylation by other protein kinases can create CK2 sites. CK2 is probably the most pleiotropic protein kinase, with >300 putative polypeptide substrates; CK2 is involved in a wide variety of cellular processes, including cell cycle, transcriptional regulation, apoptosis, and signal transduction (16, 17). However, for CK2 substrates, the molecular mechanisms through which CK2 phosphorylation regulates their physiological functions have not been elucidated yet.
In the present study, we have examined how CDK phosphorylation of S123 destabilizes Wee1A. We show that this phosphorylation promotes binding of Wee1A to β-TrCP through three independent mechanisms: namely, direct interaction with the basic residues of β-TrCP, binding to PBD by means of an optimal binding motif to accelerate Plk1 phosphorylation, and priming of CK2 phosphorylation.
Materials and Methods
Plasmids. Plasmids encoding GST-PBD (amino acids 345–603 of human Plk1) and its pincer sequence mutant (H538A/K540M) (7) were from Mike Yaffe (Massachusetts Institute of Technology, Cambridge, MA). Myc-tagged β-catenin and axin were from Keiichi Nakayama (Kyushu University, Fukuoka, Japan) (18). For construction of CK2α expression vectors, PCR-amplified CK2α (WT or V66/I174A) ORFs (provided by Enzo Pinna, University of Padova, Padova, Italy) (19) were inserted into pcDNA3.1/myc-His A (Invitrogen). Other plasmids are described in ref. 1.
Peptides. Peptides corresponding in sequences to PD53 (GSGHSTGEDSAFQEPDSP; S53 is set in italics) and PD123 (EEEGFGSSSPVKSPAAP; S121, S122, and S123 are set in italics) are described in ref. 1. All derivatives (with phosphorylation or serine-to-alanine mutation) of these two peptides had an additional cysteine at their N terminus.
Cell Culture and Immunological Techniques. Cell culture and immunological techniques were as described in refs. 1 and 20. Plk1 and CK2 antibodies were from Oncogene Research Products (San Diego) and Calbiochem, respectively. To inhibit CK2 activity, 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB) (from David Shugar, Polish Academy of Sciences, Warsaw) (21) and 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (from Enzo Pinna) (22) were used at concentrations described. Alternatively, CK2α and α′ were inactivated by small interfering RNA (siRNA) (silencer-validated siRNAs no. 1337 and no. 183, Ambion).
For αpS53 antibody production, PS53 peptides (PD53 phosphopeptides with pS53) were used both as antigen and for affinity purification. The αpS121 antibody was affinity-purified from antiserum of rabbits immunized with PS121PS123 on the PS121 phosphopeptide; αpS121/123 antibodies were affinity-purified from the same antiserum on the PS121PS123 phosphopeptide. Specificities of the phosphospecific antibodies were analyzed by ELISA, using each peptide bound to Reacti-Bind maleic anhydride-activated polystyrene plates (Pierce), horseradish peroxidase (HRP)-labeled second antibody, and 2, 2′-azinobis(3-ethylbenzoline-6-sulfonic acid) diammonium salt for substrates of HRP. For the detection of endogenous phosphorylated Wee1A, immunoreaction enhancer solution (Can Get Signal, Toyobo, Japan) was used.
In Vitro β-TrCP-Binding Assay. Peptides were covalently coupled to N-hydroxysuccinimide (NHS)-activated Sepharose 4 Fast Flow Matrix (Amersham Pharmacia). Beads were added to HtTA-1 cell lysates expressing hemagglutinin (HA)-tagged β-TrCP2 and washed with wash buffer (20 mM Hepes, pH 7.4/150 mM NaCl/0.1% Triton X-100/10% glycerol/1 mM DTT). Bound proteins were analyzed by immunoblotting with 12CA5 αHA monoclonal antibody.
Expression of PBD Proteins and Association Assay. GST-PBD and its mutant were expressed in Escherichia coli BL21 (DE3). The bacteria were lysed by sonication in sonication buffer (20 mM Tris, pH 7.5/125 mM NaCl/0.5% Nonidet P-40/1 mM EDTA/1 mM DTT/200 μM Na3VO4/50 mM NaF) supplemented with complete protease inhibitor mixture (Roche Applied Science, Indianapolis). The bacterial lysate was mixed with lysates of HeLa cells expressing Wee1A, and then glutathione beads were added to the mixture and washed with the sonication buffer. Proteins bound to the beads were examined by immunoblotting with αWee1A antiserum. For the analysis of the association between GST-PBD and phosphorylated PD123 peptides, agarose beads coated with the (phospho) peptides were mixed with bacterial lysates expressing GST-PBD and washed with sonication buffer. Bound proteins were analyzed by immunoblotting using αGST antibody (Amersham Pharmacia).
Phosphorylation of Synthetic Peptides. Synthesized peptides were phosphorylated by GST-cyclin B1/Cdc2 purified from insect cells or CK2α (human CK2α, no. 14-445, Upstate Charlottesville, VA) and analyzed by cellulose thin-layer plate electrophoresis as described in ref. 1.
Results
pS123 Directly Interacts with the WD40 Domain of β-TrCP in a Mode That Is Unique to Wee1A. Based on the finding (1) that E116 and E117 are essential for binding of Δ71 Wee1A (lacking the N-terminal 71 residues) to β-TrCP and alignment of PD123 with the consensus motif (Fig. 1 A), one would predict that phosphorylation of S121 should promote binding to β-TrCP. Consistent with this, the S121A mutation reduced binding approximately to the same extent as S123A (1). Moreover, when we superimposed the aligned PD123 peptide on the recently determined structure of β-TrCP bound to a β-catenin phosphopeptide (the structure in ref. 23 was redrawn in Fig. 1C Upper), which has a consensus β-TrCP-binding motif, we found that PD123 fit the binding pocket reasonably well (Fig. 1C Lower). However, in an energy-minimized structure of the complex between PD123 phosphopeptide and the WD40 domain, we noticed that pS121 and pS123 both interacted with R383 of β-TrCP2 in addition to the interaction between pS121 and R404, which is analogous to that with pS37 of β-catenin (23) [Fig. 1C; our numbering is for β-TrCP2 (24), which is shorter than β-TrCP1 (variant 2) by 27 aa]. The existence of an additional acidic residue, pS123, two residues downstream of S121 that is unique to Wee1A (i.e., the +2 position of pS37 is alanine in β-catenin) could account for this difference and may explain why phosphorylation of S123 is important for the binding.
To examine whether R383 actually contributes to β-TrCP2 binding to Wee1A but not to β-catenin, R383 was mutated to methionine in β-TrCP2, and the binding to Wee1A or β-catenin was examined. As predicted from our model, binding of β-TrCP2 to Δ71Wee1A was significantly weakened in the R383M mutant (Fig. 1D). In contrast, the binding of β-TrCP2 to β-catenin was not decreased by the R383M mutation (Fig. 1E). These results strongly support the significance of our model for the interface between Wee1A and β-TrCP, in which R383M interacts with both pS121 and pS123.
pS121, Rather than pS123, Is Mainly Responsible for the Binding to β-TrCP. Next, we examined the strength of the interaction between pS123 of Wee1A and β-TrCP by using a set of synthetic peptides covalently attached to agarose beads. As reported in ref. 25, a doubly phosphorylated peptide with the consensus binding motif [PP10, IκBα-derived sequence (25)], but not its unphosphorylated form (SS10), bound to β-TrCP (Fig. 1F). Similarly, a PD53 phosphopeptide with pS53 (PS53), but not its unphosphorylated form (S53), bound to β-TrCP. In contrast, however, a PD123 phosphopeptide with pS123 (S121PS123) bound to β-TrCP very weakly, although significantly more strongly than an unphosphorylated peptide with the same sequence (S121S123). Rather, a doubly phosphorylated PD123 phosphopeptide with pS121 and pS123 (PS121PS123) bound to β-TrCP strongly and slightly better than PS121S123 (Fig. 1G). When the affinity of these phosphopeptides for β-TrCP was examined by the ability to abolish the binding between β-TrCP and Δ71 Wee1A, PS121S123 and PS121PS123, but not S121PS123, reduced the binding, with PS121PS123 being more effective (Fig. 1H). These results indicate that phosphorylation at S121 is mainly responsible for the interaction between PD123 and β-TrCP, with phosphorylation at S123 having only an accessory role in binding per se.
Phosphorylation of S53, S121, and S123 During the Cell Cycle. If the direct contribution of pS123 to β-TrCP binding is small, why does mutation of S123 to alanine have such a large effect on Wee1A binding and stability (ref. 1 and Fig. 1B)? Does phosphorylation of S123 affect phosphorylation of other residues that promote β-TrCP binding? To examine this possibility, we set out to raise antisera against PS53, PS121S123, S121PS123, and PS121PS123 peptides. Assessment of the specificities of antibodies affinity-purified on appropriate peptides by ELISA (Fig. 2A) indicated that we were successful in generating antibodies that recognize pS53 and pS121, respectively, but not antibodies that specifically recognize pS123. Using PS121PS123 as an antigen, however, we obtained antibodies that recognize Wee1A only when both pS121 and pS123 were phosphorylated (αpS121/123) (Fig. 2 A).
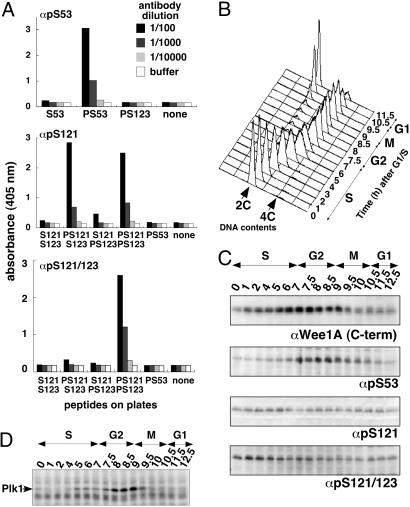
Fig. 2.
Phosphorylation of S53, S121, and S123 in synchronized HeLa cells. (A) Serially diluted affinity-purified antibodies as indicated were added to wells of ELISA plates, which are coated with each phosphopeptide as shown in the horizontal axis. Bound antibodies were quantified by the peroxidase activity of horseradish peroxidase-conjugated secondary antibody. (B) HeLa cells were synchronized at the G1/S boundary and released. Cell cycle progression was monitored by DNA content determined by FACS analysis. (C) Levels of Wee1A or phosphorylation at S53, S121, and S121 and S123 was examined by immunoblotting of total cell lysates [equivalent to the same number of cells at the G1/S border (time 0); the levels of total proteins were confirmed by Coomassie blue staining of the filter (not shown)] from each time point. (D) Levels of Plk1 were examined by immunoblotting of total cell lysates from each time point as in C.
Using these antibodies, phosphorylation of the serines in PD53 and PD123 was examined in synchronized HeLa cells (Fig. 2B). As reported in ref. 20, the level of Wee1A increased during the S and G2 phases and decreased at M phase (Fig. 2C, uppermost blot). Phosphorylation of S53 rose at the S/G2 boundary, consistent with the expression and activation of Plk1 at this time (ref. 26 and Fig. 2 C and D). The signal decreased at M phase, when Wee1A was degraded. In contrast, phosphorylation of S121, which was detected by using αpS121 antibodies, rose in early S phase and gradually decreased toward M phase (Fig. 2C). An almost identical pattern was obtained by using antibodies that recognize pS121 and pS123, indicating that phosphorylation of S121 and S123 occurs with similar timing (Fig. 2C).
pS123 Creates a PBD-Binding Motif and Enhances S53 Phosphorylation by Plk1. The sequence surrounding pS123 (S122-pS123-P124) fits the recently identified PBD-binding motif consensus (6), S-pS/pT-(P). Thus, pS123 might induce binding of Plk1 to Wee1A and thereby accelerate phosphorylation of S53 by Plk1. To examine this possibility, WT and H538A/K540M mutant (7) GST-PBD fusion proteins were expressed in bacteria, and their binding to Δ71Wee1A was tested. WT Δ71Wee1A effectively bound to WT, but not mutant, Plk1 PBD (Fig. 3A). Moreover, S122A and S123A mutant Δ71Wee1As failed to bind GST-PBD, whereas S121A did bind, indicating that this binding is depends exclusively on the PBD-binding motif surrounding S123, even though several other potential PBD-binding motifs exist downstream in Δ71Wee1A.
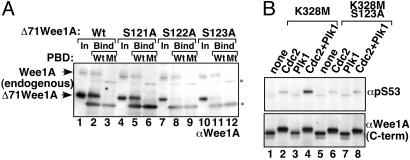
Fig. 3.
S123 phosphorylation creates a PBD-binding motif and accelerates S53 phosphorylation by Plk1. (A) WT GST-PBD (WT) and its pincer sequence mutant form (Mt, H538A/K540M) were expressed in bacteria. The bacterial lysates were mixed with HeLa cell lysates expressing WT or mutant forms of Δ71Wee1A. Cell lysates (In, 50% of total input is shown) and GST-pulled-down proteins (Bind) were analyzed by immunoblotting with αWee1A antiserum. Endogenous HeLa cell Wee1A also bound detectably to WT PBD. Asterisks indicate nonspecific signals derived from bacterial lysates. (B) Kinase-negative mutant Wee1A (K328M) or its S123A derivative was expressed in HeLa cells, immunopurified, and phosphorylated by purified cyclin B1/Cdc2 and/or Plk1 as indicated. Phosphorylation of S53 and the level of Wee1A were analyzed on the same filter by sequentially immunoprobing with αpS53 antibody and α-C-terminal antiserum, respectively.
As shown above, S121 and S123 are phosphorylated with similar timing during the cell cycle. Although a specific amino acid at the –2 position (corresponding to S121) from the phosphorylation site (S123) is not strongly required for PBD binding (6, 7), we examined the possibility that pS121 perturbs the PBD binding. As expected, neither the S121A mutation nor the S121 phosphorylation affected the binding of PBD (Fig. 3A; see also Fig. 6, which is published as supporting information on the PNAS web site).
We also examined the extent to which PBD-dependent binding of Plk1 to Wee1A contributed to S53 phosphorylation. There was a low level of S53 phosphorylation, as determined by immunoblotting with αpS53 antibodies, when Wee1A (K328M, kinase negative) expressed and immunopurified from HeLa cells was phosphorylated by Plk1 in vitro (Fig. 3B, lanes 1 and 3). S53 phosphorylation was greatly enhanced when Cdc2 was also present in the reaction, although Cdc2 alone did not increase phosphorylation at S53 (Fig. 3B, lanes 2 and 4). In contrast, Cdc2 did not significantly enhance Plk1 phosphorylation of S53 in S123A mutant Wee1A under the same conditions (Fig. 3B, lanes 5–8). These results indicate that phosphorylation at S123 facilitates recruitment of Plk1 and accelerates the phosphorylation of S53 of Wee1A. However, because there was weak phosphorylation of S53 in S123A Wee1A expressed in HeLa cells (data not shown) and because the combined S53A/S123A mutation reduces β-TrCP binding to Wee1A more than the S123A single mutation (1), it appears that phosphorylation at S123 enhances but is not absolutely essential for the phosphorylation of S53.
CK2 Phosphorylates S121 only When S123 Is Phosphorylated. How does phosphorylation of S121 occur? During the search for a protein kinase that phosphorylates S121, we noticed that CK2 phosphorylated the PD123 peptide only when S123 is prephosphorylated (SSpS), whereas Cdc2 phosphorylated S123 in the unphosphorylated peptide (SSS) (Fig. 4A, lanes 4 and 6). Thus, pS123 seems to have a priming effect for the CK2 phosphorylation. Because CK2 preferentially phosphorylates Ser/Thr with an acidic amino acid at the n + 1, 2, or 3 position (reviewed in ref. 17), S121 and/or S122 is predicted to be phosphorylated by CK2. However, when S121 was changed to alanine (ASpS), CK2 was unable to phosphorylate the pS123 containing phosphopeptide, despite the presence of other serines, including S122, in the peptide, indicating that CK2 specifically phosphorylates S121 when S123 is phosphorylated in vitro (Fig. 4B, lane 4). The result that S121, but not S122, was phosphorylated by CK2 in pS123 containing phosphopeptide was rather unexpected, because a Ser/Thr with acidic residue at the n + 1 position is usually a better substrate for CK2 than a Ser/Thr with an acidic residue at the n + 2 position (17). Possibly, the proline at 124 (the n + 3 position for S121) affects CK2 substrate sequence specificity.
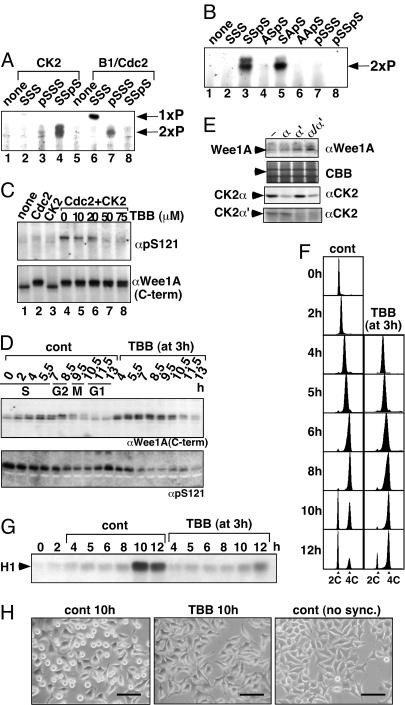
Fig. 4.
Phosphorylation at S123 promotes S121 phosphorylation by CK2, which is essential for the proper onset of Wee1A degradation and M phase. (A) Synthetic PD123 peptide [(C)EEEGFGSSSPVKSPAAP] and its phosphorylated versions (status of the three italicized serines is indicated) were phosphorylated by CK2 and cyclin B1/Cdc2 and analyzed by electrophoresis on a thin-layer cellulose plate at pH 3.5 toward the anode (bottom). The positions of singly (1xP) and doubly (2xP) phosphorylated peptides are shown. Doubly phosphorylated peptides migrated as doublets for unknown reasons. (B) PD123 peptide or its derivatives (phosphoserine or alanine substitution of three serines italicized in A are indicated) were phosphorylated by CK2 and analyzed as in A. (C) Kinase-negative mutant Wee1A (K328M) was expressed in HeLa cells, immunopurified, and phosphorylated by cyclin B1/Cdc2 and/or CK2α in the presence of 100 μM ATP. In some reactions, TBB was added at a final concentration as indicated. After the reaction and Western transfer, phosphorylation of S121 and level of Wee1A were analyzed on the same filter by sequentially immunoprobing with αpS121 antibody and α-C-terminal antiserum, respectively. (D) HeLa cells were synchronized as in Fig. 2 and cultured in the absence or presence of TBB (75 μM, added at 3 h after the release). The level of Wee1A and phosphorylation of S121 were analyzed by immunoblotting as in Fig. 2C. (E) HeLa cells were transfected with siRNA for CK2α and/or α′. Two days after transfection, the levels of Wee1A, CK2α, and CK2α′ were analyzed by immunoblotting. Coomassie blue (CBB) staining of the filter is also shown (arrowhead indicates the position of Wee1A). (F) Effect of TBB (75μM, added at 3 h after the release) on the cell cycle progression was monitored by DNA content determined by FACS analysis. DNA contents of G1 (2C) and G2/M (4C) cells are indicated by triangles. (G) Effect of TBB (75 μM, added at 3 h after the release) on the onset of M phase was monitored by H1 histone kinase activity of cyclin B1 immunoprecipitates. (H) TBB (75 μM, added at 3 h after the release)-treated cells (Center) appear “angular” at 10 h after the release, when most of the control cells (Left) were rounded up. TBB-treated cells started to round up and divide at ≈12 h (not shown). Nonsynchronized control cells are also shown (Right). (Scale bars, 100 μm.)
S123 phosphorylation-dependent phosphorylation of S121 by CK2 was also confirmed in full-length Wee1A by using αpS121 antibodies. Phosphorylation of S121 was not significantly detected in Wee1A (K328M, kinase negative) overexpressed and immunoprecipitated from HeLa cells. As expected, however, S121 phosphorylation was significantly increased after incubation with both CK2 and Cdc2, but not with either alone (Fig. 4C Upper, lanes 1–4). TBB, one of the most specific CK2 inhibitors available (21), inhibited S121 phosphorylation in a dose-dependent manner, although the characteristic shift of Wee1A upon Cdc2 phosphorylation was not significantly affected by TBB (Fig. 4C, lanes 4–8).
Inhibition of CK2 Suppresses S121 Phosphorylation and Retards Wee1A Degradation and the Onset of Mitosis in Vivo. Phosphorylation of S121 was examined in HeLa cells synchronized at the G1/S boundary and released in the absence or presence of TBB (Fig. 4D). As predicted, the S121 phosphorylation at G2 phase was suppressed when TBB was added at mid-S phase (3 h after G1/S), indicating that S121 is phosphorylated by CK2 in vivo.
In cells treated with TBB, the degradation of Wee1A at M phase (≈10 h after release in control cells) was significantly retarded, consistent with CK2-dependent phosphorylation of Wee1A being important for Wee1A degradation at M phase (Fig. 4D Upper). In addition, the level of Wee1A was significantly increased by TBB treatment even in the S and G2 phases, possibly because phosphorylation at S121 by CK2 is responsible for Wee1A turnover during interphase [Wee1A is unstable even during interphase (20), and S121/123A mutation stabilizes Wee1A during interphase (data not shown)]. A recently developed, more specific and potent CK2 inhibitor, 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (19, 22), also increased the Wee1A level, and this increase was reverted by overexpression of CK2α (Fig. 7, which is published as supporting information on the PNAS web site). Moreover, the increase in Wee1A level was detected by CK2α and/or CK2α′ knockdown by siRNA (Fig. 4E). From these results, we concluded that CK2 is responsible for the stability of Wee1A through S121 phosphorylation in vivo.
Although the progression through S phase was not significantly affected in TBB-treated cells (Fig. 4F), the onset of M phase was retarded >2 h as evidenced by FACS analysis, the delayed increase in cyclin B1-dependent histone H1 kinase activity, and cell morphology (Fig. 4 F–H). During normal M phase, HeLa cells first become angular and refractile and then round up and divide (Fig. 4H; for angular refractile cells, compare non-rounded-up cells in Left and nonsynchronized cells in Right). When most of the control cells were rounding up and dividing (10 h after the release), almost all of the TBB-treated cells became angular and refractile but did not round up, suggesting that TBB-treated cells arrest just before M phase. CK2 is known to be a pleiotropic protein kinase, and its inhibition may have effects on several CK2 substrates that are important for the G2/M progression. Nevertheless, the effect on the Wee1A degradation in vivo, together with the in vitro analysis of the role of CK2 on Wee1A phosphorylation, suggests that the CK2-mediated degradation of Wee1A is an important event for the proper onset of M phase as well.
Discussion
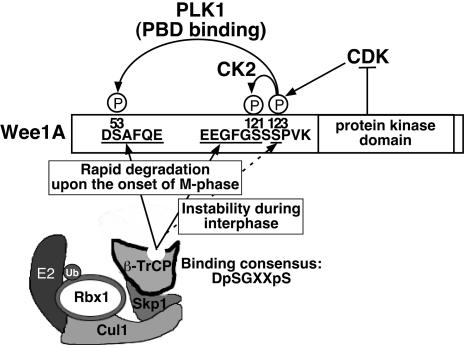
The ubiquitination of Wee1A is regulated by the binding of Wee1A to β-TrCP in a phosphorylation-dependent manner. Of the three identified protein kinases involved, phosphorylation by a CDK appears to be the key step, having three roles in promoting β-TrCP binding. CDK not only phosphorylates a residue on Wee1A that enhances interaction with β-TrCP directly, but it triggers a “phosphorylation cascade” by two other protein kinases, Plk1 and CK2, to create two phosphodegrons for β-TrCP recognition (Fig. 5).
Fig. 5.
Phosphorylation at S123 by CDK promotes the binding of β-TrCP to Wee1A via three independent pathways. During the S and G2 phases, S123 is phosphorylated by a CDK (possibly CDK2). pS123 primes CK2 to phosphorylate S121, resulting in creation of a β-TrCP phosphodegron (EEGFGpS121) that is responsible for the instability of Wee1A during interphase. PD123 binding involves interaction of pS123 with basic residues in the WD40 repeat domain of β-TrCP in a manner unique to Wee1A (dotted line). At the onset of M phase, when activated Plk1 accumulates, Plk1 binds to Wee1A at the PBD-binding motif surrounding pS123 (SpSP) by means of its PBD and phosphorylates S53, resulting in generation of the second phosphodegron (DpSAFQE). These two phosphodegrons act cooperatively to promote β-TrCP binding and the turnover of Wee1A, which is important for the proper onset of M phase.
Phosphorylation at S123 was observed during the S and G2 phases, as well as during mitosis. Phosphorylation of S123 during interphase seems likely to be mediated by CDK2. Once S123 is phosphorylated during interphase, S121 will be rapidly phosphorylated by CK2, which is constitutively active throughout the cell cycle (Fig. 2C). Phosphorylation at S121 in conjunction with pS123 promotes binding to β-TrCP by means of PD123, which may be responsible for the basal rate of Wee1A turnover during the S and G2 phases (Figs. 4 D and E and 7). At the onset of M phase, when Plk1 accumulates and is activated, Plk1 binds to Wee1A by means of the pS123 PBD-binding motif and phosphorylates S53, thus creating the second phosphodegron, PD53 (Figs. 2 C and D and 3). When both phosphodegrons are activated simultaneously, they can act cooperatively to promote strong binding of Wee1A to β-TrCP and induce efficient ubiquitination and degradation of Wee1A (Fig. 5). At this time, Cdc2 becomes the CDK responsible for promoting Wee1A degradation, and the decrease in Wee1A levels, in turn, enhances Cdc2 activation by Cdc25; this Cdc2-mediated negative feedback on Wee1A is an important facet of the switch-like activation of Cdc2 at mitosis. In addition, Wee1A catalytic activity can be directly inhibited by phosphorylation at M phase, and, although Cdc2 phosphorylation alone cannot inhibit Wee1A, Cdc2 phosphorylation at sites other than S123 may also contribute to Wee1A inactivation in cooperation with phosphorylation by other M-phase kinases (20).
Because CK2 is also known to be important for G2/M progression in budding yeast (27), the phosphorylation cascade promoting the degradation of Wee1 family protein kinases may be conserved. In this regard, Cdc5p (Plk1 homolog) activity has been shown to be important for the ubiquitination-dependent degradation of Swe1p (Wee1 homolog) (28). In addition, noncatalytic regions of these two kinases are known to interact with each other (29), suggesting that the PBD-dependent interaction of these two kinases is very likely to be conserved as well. Because binding of substrates to the PBD stimulates the activity of Plk1 (7), pS123-dependent binding of Plk1 is important not only for Wee1A degradation but also potentially for activation of Plk1 at the onset of M phase.
In summary, we have identified three roles of S123 phosphorylation for the ubiquitination-dependent degradation of Wee1A. pS123 not only directly interacts with WD40 domains of β-TrCP in a mode unique to Wee1A, but it also triggers a phosphorylation cascade to create phosphodegrons for β-TrCP recognition. To our knowledge, this study provides the first example of a single phosphorylated residue that primes multiple protein kinases for the phosphorylation of the molecule itself.
Supplementary Material
Acknowledgments
We thank D. Shugar, L. A. Pinna, M. Yaffe, and K. I. Nakayama for reagents; members of the RIKEN Antibiotics Laboratory for discussions; Y. Ichikawa and R. Nakazawa (Bioarchitect Research Group, RIKEN) for DNA sequencing; N. Hirotani (Brain Science Institute, Research Resources Center. RIKEN) for peptide synthesis; M. Kumai (Brain Science Institute, Research Resources Center, RIKEN) for antibody production; and M. Watanabe and Y. Ikawa for encouragement. T.H. is a Frank and Else Schilling American Cancer Society Research Professor.
Author contributions: N.W., M.S., K.O., and T.H. designed research; N.W., H.A., J.-i.I., and M.S. performed research; N.W., M.S., and K.O. analyzed data; and N.W., T.H., and H.O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CDK, cyclin-dependent kinase; Wee1A, somatic Wee1; β-TrCP, β-transducin repeat-containing protein; Plk1, polo-like kinase 1; PBD, polo box domain; PDn, phosphodegron n; TBB, 4,5,6,7-tetrabromo-2-azabenzimidazole; siRNA, small interfering RNA.
Note Added in Proof. While this manuscript was under review, Asano et al. (31) reported that phosphorylation of Swe1p (Wee1 homolog) by Cdc5p (Plk homolog) at the onset of M phase was primed by Clb2-Cdc28 (mitotic CDK) phosporylation in a manner dependent on Cdc5p's PBD, and that this was important for Swe1p degradation in budding yeast.
References
- 1.Watanabe, N., Arai, H., Nishihara, Y., Taniguchi, M., Watanabe, N., Hunter, T. & Osada, H. (2004) Proc. Natl. Acad. Sci. USA 101, 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Vugt, M. A., Bras, A. & Medema, R. H. (2004) Mol. Cell 15, 799–811. [DOI] [PubMed] [Google Scholar]
- 3.Barr, F. A., Sillje, H. H. & Nigg, E. A. (2004) Nat. Rev. Mol. Cell. Biol. 5, 429–440. [DOI] [PubMed] [Google Scholar]
- 4.Lane, H. A. & Nigg, E. A. (1997) Trends Cell Biol. 7, 63–68. [DOI] [PubMed] [Google Scholar]
- 5.Lee, K. S., Grenfell, T. Z., Yarm, F. R. & Erikson, R. L. (1998) Proc. Natl. Acad. Sci. USA 95, 9301–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elia, A. E. H., Cantley, L. C. & Yaffe, M. B. (2003) Science 299, 1228–1231. [DOI] [PubMed] [Google Scholar]
- 7.Elia, A. E. H., Rellos, P., Haire, L. F., Chao, J. W., Ivins, F. J., Hoepker, K., Mohammad, D., Cantley, L. C., Smerdon, S. J. & Yaffe, M. B. (2003) Cell 115, 83–95. [DOI] [PubMed] [Google Scholar]
- 8.Kumagai, A. & Dunphy, W. G. (1996) Science 273, 1377–1380. [DOI] [PubMed] [Google Scholar]
- 9.Inoue, D. & Sagata, N. (2005) EMBO J. 24, 1057–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neef, R., Preisinger, C., Sutcliffe, J., Kopajtich, R., Nigg, E. A., Mayer, T. U. & Barr, F. A. (2003) J. Cell Biol. 162, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen, D. V., Loktev, A. V., Ban, K. H. & Jackson, P. K. (2004) Mol. Biol. Cell 15, 5623–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litvak, V., Argov, R., Dahan, N., Ramachandran, S., Amarilio, R., Shainskaya, A. & Lev, S. (2004) Mol. Cell 14, 319–330. [DOI] [PubMed] [Google Scholar]
- 13.Preisinger, C., Korner, R., Wind, M., Lehmann, W. D., Kopajtich, R. & Barr, F. A. (2005) EMBO J. 24, 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke, A. S., Tang, T. T., Ooi, D. L. & Orr-Weaver, T. L. (2005) Dev. Cell 8, 53–64. [DOI] [PubMed] [Google Scholar]
- 15.Yoo, H. Y., Kumagai, A., Shevchenko, A. & Dunphy, W. G. (2004) Cell 117, 575–588. [DOI] [PubMed] [Google Scholar]
- 16.Litchfield, D. W. (2003) Biochem. J. 369, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meggio, F. & Pinna, L. A. (2003) FASEB J. 17, 349–368. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa, M., Hatakeyama, S., Shirane, M., Matsumoto, M., Ishida, N., Hattori, K., Nakamichi, I., Kikuchi, A., Nakayama, K. & Nakayama, K. (1999) EMBO J. 18, 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagano, M. A., Andrzejewska, M., Ruzzene, M., Sarno, S., Cesaro, L., Bain, J., Elliott, M., Meggio, F., Kazimierczuk, Z. & Pinna, L. A. (2004) J. Med. Chem. 47, 6239–6247. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe, N., Broome, M. & Hunter, T. (1995) EMBO J. 14, 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szyszka, R., Grankowski, N., Felczak, K. & Shugar, D. (1995) Biochem. Biophys. Res. Commun. 208, 418–424. [DOI] [PubMed] [Google Scholar]
- 22.Pagano, M. A., Meggio, F., Ruzzene, M., Andrzejewska, M., Kazimierczuk, Z. & Pinna, L. A. (2004) Biochem. Biophys. Res. Commun. 321, 1040–1044. [DOI] [PubMed] [Google Scholar]
- 23.Wu, G., Xu, G., Schulman, B. A., Jeffrey, P. D., Harper, J. W. & Pavletich, N. P. (2003) Mol. Cell 11, 1445–1456. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki, H., Chiba, T., Kobayashi, M., Takeuchi, M., Suzuki, T., Ichiyama, A., Ikenoue, T., Omata, M., Furuichi, K. & Tanaka, K. (1999) Biochem. Biophys. Res. Commun. 256, 127–132. [DOI] [PubMed] [Google Scholar]
- 25.Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., Andersen, J. S., Mann, M., Mercurio, F. & Ben-Neriah, Y. (1998) Nature 396, 590–594. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K. S., Yuan, Y. L., Kuriyama, R. & Erikson, R. L. (1995) Mol. Cell. Biol. 15, 7143–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanna, D. E., Rethinaswamy, A. & Glover, C. V. (1995) J. Biol. Chem. 270, 25905–25914. [DOI] [PubMed] [Google Scholar]
- 28.Sakchaisri, K., Asano, S., Yu, L. R., Shulewitz, M. J., Park, C. J., Park, J. E., Cho, Y. W., Veenstra, T. D., Thorner, J. & Lee, K. S. (2004) Proc. Natl. Acad. Sci. USA 101, 4124–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartholomew, C. R., Woo, S. H., Chung, Y. S., Jones, C. & Hardy, C. F. (2001) Mol. Cell. Biol. 21, 4949–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiner, S. J., Kollman, P., Case, D. A., Singh, U. C., Ghio, C., Alagona, G., Profeta, S. & Weiner, P. (1984) J. Am. Chem. Soc. 106, 765–784. [Google Scholar]
- 31.Asano, S., Park, J.-E., Sakchaisri, K., Yu, L.-R., Song, S., Supavilai, P., Veenstra, T. D. & Lee, K. S. (2005) EMBO J. 24, 2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.