Abstract
GABA is the major inhibitory transmitter at CNS synapses. Changes in subunit composition of the pentameric GABAA receptor, including increased levels of α4 subunit in dentate granule cells and associated functional alterations such as increased zinc blockade of GABA currents, are hypothesized to be critical components of epileptogenesis. Here, we report that the minimal promoter of the human α4 subunit gene (GABRA4p), when used to drive reporter gene expression from adeno-associated viral vectors, controls condition-specific up-regulation in response to status epilepticus, defining a transcriptional mechanism for seizure-induced changes in levels of α4 subunit containing GABAA receptors. Transfection studies in primary hippocampal neurons show that inducible early growth response factor 3 (Egr3) up-regulates GABRA4p activity as well as the levels of endogenous α4 subunits. Given that Egr3 knockout mice display ≈50% less GABRA4 mRNAs in the hippocampus and that increases in α4 and Egr3 mRNAs in response to pilocarpine-induced status epilepticus are accompanied by increased binding of Egr3 to GABRA4 in dentate granule cells, our findings support a role for Egr3 as a major regulator of GABRA4 in developing neurons and in epilepsy.
Keywords: epilepsy, dentate gyrus, adeno-associated virus, immediate early genes
GABA is the major inhibitory neurotransmitter in the brain that mediates fast synaptic inhibition, but little is known about the mechanisms underlying the transcriptional regulation of its target, the GABAA receptor (GABAAR). The GABAAR is a heterooligomeric complex composed of five membrane-spanning subunits differentially expressed in various regions of the brain and spinal cord. cDNAs for many GABAAR subunits and subunit subtypes (α1-6, β1-4, γ1-3, δ, ε, π, τ, and ρ1-3) have been isolated. Subunits are encoded by different genes that display regional- and developmental-specific expression (for review, see ref. 1) (2-5). Understanding the factors that control levels of GABAAR subunits is particularly important because different subunits assemble into region-specific isoforms with characteristic responses to GABA and allosteric modulators of the GABA response. In addition, results of several studies have implicated changes in GABAAR function as the basis for a disease phenotype (6-11).
A GABAAR that contains an α4 subunit is insensitive to benzodiazepines (12) and highly sensitive to inhibition by zinc (14, 15). Pharmacological changes in GABAAR function in dentate granule cells (DGCs), including decreased augmentation by type 1 benzodiazepine agonists (such as zolpidem and increased inhibition by zinc), are seen in temporal lobe epilepsy (TLE) and are likely to result from the presence of increased α4-containing receptors (9). Expression of GABAARs containing α4 and δ subunits are associated in normal animals with production of extrasynaptic GABAARs (16, 17). Such receptors are believed to mediate tonic rather than phasic inhibition, and increased tonic inhibition has been associated with a decrease in inhibitory synaptic activity and increased seizure susceptibility (17, 18).
A variety of evidence suggests that GABRs vary their rates of transcription in response to extracellular signals (19-22). In this study, we used stimulation of PKC in primary cultured hippocampal neurons (HNs) to increase levels of GABRA4 transcripts, mirroring changes shown in several epilepsy models (6-8). We relate these observations to those from studies in early growth response factor 3 (Egr3) knockout mice and pilocarpine (PILO)-treated rats that have been injected with adeno-associated viral (AAV) vectors containing GABRA4 promoter fragments.
Materials and Methods
Cell Culture and Transfection. Primary hippocampal, neocortical, and fibroblast cells were derived from E18 rat embryos and grown in defined media as described in refs. 22 and 23. Primary neurons were transfected as described in ref. 22. Expression constructs for Egr3 were generously provided by J. Milbrandt (Washington University, St. Louis), and ZnEgr was provided by J. Baraban (The Johns Hopkins University School of Medicine, Baltimore).
Injection of AAV into the Dentate Gyrus. Adult male Sprague-Dawley rats (Charles River Breeding Laboratories) were used for all in vivo studies. Animals were anesthetized initially with 100 mg/kg ketamine, placed in a stereotaxic frame, and put under continuous halothane (Sigma) for the duration of the procedure. Three burr holes were drilled into one side of the skull at three different locations over the anterior/posterior (A/P) extent of the hippocampus. For the studies of regional expression, sites were injected in the thalamus, the upper and lower blades of the dentate gyrus, and the cortex, and one additional hole was drilled over the cerebellum. For seizure studies, injections were performed only into dentate gyrus of the hippocampus. By using a 30-gauge syringe (Hamilton) under control of a microinjection pump, 2 μl of virus was stereotaxically injected at each site at a rate of 0.2 μl/min. The following coordinates were used to target four different regions: thalamus [anterior site: A/P, -3.0 mm; mediolateral (M/L), 1.4 mm; dorsoventral (D/V), 5.0 mm; middle site: A/P, -4.15 mm; M/L, 2.0 mm; D/V, 5.6 mm; posterior site: A/P, -5.3 mm; M/L, 3.6 mm; D/V, 5.6 mm]; dentate gyrus (anterior site: A/P, -3.14 mm; M/L, 1.4 mm; D/V, 4.1 mm, 3.6 mm; middle site: A/P, -4.15 mm; M/L, 2.0 mm; D/V, 3.8 mm, 3.3 mm; posterior site: A/P, -5.3 mm; M/L, 3.6 mm; D/V, 4.2 mm, 3.7 mm); cortex (anterior site: A/P, -3.0 mm; M/L, 1.4 mm; D/V, 1.5 mm; middle site: A/P, -4.15 mm; M/L, 2.0 mm; D/V, 1.5 mm; posterior site: A/P, -5.3 mm; M/L, 3.6 mm; D/V, 1.5 mm); and cerebellum (A/P, 10.5 mm; M/L, 3.0 mm; D/V, 4.0 mm). Three rats were studied in each group at each time point (2-4 weeks after virus injection) to examine for virus incorporation into DGCs and level of GABRA4p activity. Virus incorporation into DGCs was assessed by using in situ hybridization to detect transcripts of enhanced yellow fluorescent protein (eYFP).
Induction of Status Epilepticus (SE). At 2 weeks after virus injections, a subset of rats was exposed to PILO-induced SE according to a standard protocol (6, 24). PILO injection triggered long-duration (>30 min) seizures within 10-30 min after injection. Rats that did not exhibit behavioral seizures of class 3 or higher on the Racine (25) scale within 1 h of PILO injection were injected i.p. with a second dose of PILO (192.5 mg/kg), which is standard to the use of this model because equivalence is measured in seizures and not in doses of PILO. Diazepam (6 mg/kg; Sigma) was administered i.p. at 1 h after the onset of SE to stop seizure activity, and it was administered again every 2 h until rats completely stopped seizing. Control rats were treated identically to PILO-injected rats, except that a subconvulsive dose of PILO (38.5 mg/kg) was administered.
Detection of eYFP in Vivo. Experiments were performed by using antisense and sense RNA probes for eYFP synthesized by in vitro transcription using a cDNA template containing 300 bp of eYFP coding sequence (Promega). Transcripts were labeled with digoxigenin-11-UTP (Roche Molecular Biochemicals). We fixed 10-μm-thick slide-mounted sections in 4% paraformaldehyde/PBS and permeabilized in 0.02% Triton X-100/PBS. Hybridization was performed by using standard procedures.
Immunocytochemistry. We washed 20-μm-thick slide-mounted sections three times in TBST (50 mM Tris·HCl/150 mM NaCl/0.1% Triton X-100, pH 7.4) for 5 min each and incubated for blocking in 5% horse serum (Vectastain, Vector Laboratories) for 1 h at room temperature. Sections were incubated with Egr3 Ab (Santa Cruz Biotechnology) overnight at 4°C at a 1:300 dilution. Sections were treated by using standard protocols. Slides from all groups were reacted in parallel.
Chromatin Immunoprecipitation (ChIP). ChIP was performed as described in ref. 26. We used 10 million cells for each assay, and they were split into three aliquots for immunoprecipitation in the presence and absence of specific Abs, such as those for Egr1 and Egr3 (Santa Cruz Biotechnology). Quantitative PCR was performed to determine the linearity of amplification before choosing to further dilute the sample 10-fold and use 6.2 μl of the dilution for each experiment. Immunoprecipitated genomic DNAs (gDNAs) were used as templates for PCR to amplify a fragment of GABRA4 surrounding the Egr3 site. PCR was also performed on gDNAs precipitated with unrelated Abs as additional negative controls. The 35S-labeled PCR products were separated on 5% polyacrylamide gels and exposed to x-ray film. For in vivo ChIP, animals were rapidly perfused with 4% paraformaldehyde at 1 or 24 h after the onset of PILO-induced SE. Brains were removed, and slices of dentate were stored in paraformaldehyde until use. Tissue was homogenized and lysed according to standard protocols (Upstate Biotechnology, Lake Placid, NY).
Real-Time PCR. PCR primers were designed by using primer express software (Applied Biosystems). Control probes for relative abundance of rRNA or cyclophilin were used in multiplex assays (Applied Biosystems) with the ABI7900HT (Applied Biosystems). RNA from Egr3 knockout mice hippocampal tissue [generously supplied by J. Milbrandt (Washington University, St. Louis) and W. Tourtellotte (Northwestern University, Chicago)] was extracted by using standard procedures.
Optical Density (OD). The intensity of eYFP mRNA in situ hybridization signal and Egr3 immunolabeling were quantified by using imagepro plus software (Media Cybernectics, Silver Spring, MD). Photomicrographs (×5 magnification) of three sections through the dentate gyrus at equivalent locations were taken from each animal by using identical light settings (control, n = 6; SE group for eYFP in situ hybridization, n = 5; and Egr3 immunohistochemistry, n = 3 per group). OD measurement was performed by an investigator blinded to group identity. DGC layer was marked by using a line tool and intensity of eYFP mRNA signal along that line was measured in OD units. The OD value for each animal was calculated as the average of the OD values of the three sections. The OD value for each animal was normalized to the mean OD for all animals in the control group (percentage of control).
Confocal Microscopy. Primary HNs were transfected with pDsRed2-Monomer vector (BD Biosciences) to identify transfected neurons. Both Egr3 or vector and pDsRed2-Monomer were used at a 1:1 ratio (4 μg per 35-mm dish). At 48 h after transfection, cells were washed and fixed by using a standard protocol. Immunohistochemistry was performed by using an α4-specific Ab (NB 300-194; Novus Biologicals, Littleton, CO) at a 1:200 dilution in 1% BSA for 12 h at 4°C and a secondary Ab conjugated to FITC at a dilution of 1:50 (sc-2012; Santa Cruz Biotechnology). After mounting, cells were viewed by using an Axioscop 100M laser-scanning confocal microscope (Zeiss), equipped with Argon and He/Ne lasers, by using a C Apochromat ×40/1.2 water objective. Photomultiplier gain and pinhole aperture were kept constant. Fluorescence was quantified by using lsm image browser (Zeiss) software and normalized to the total area of a selected cell. Changes in subunit expression were monitored by using a double-blind cell-selection procedure with 38 transfected neurons per condition (n indicates cultures from five different animals).
RACE. PCR products were generated by using a series of adaptor and GABRA4 subunit gene-specific primers with RNA ligase-mediated (RLM) RACE or human RACE-Ready cDNA kits (Ambion, Austin, TX). The following primers were used: rat GABRA4, ATGGTTTCTGTCCAGAAGGTACCT (sense), TTTCCGGGCACAATTTCTCGTCCT (antisense 1), and ATCCAGGACGCAGTCTGTTGTCAT (antisense 1); and human GABRA4, GTTCACGTTTCCAGGCTC (sense), CTTCTTGGCAGAAACCATCTTTGC (antisense 1), and AGCTAGCCATTTCTCCACTTTCCG (antisense 2).
Small Interfering RNA (siRNA) Screening in HEK293 Cells. HEK293 cells were cotransfected with expression vectors (1 μg) for rat GABRA4 cDNA (or an empty vector) and presynthesized siRNAs (2 nM; Ambion) (or scrambled siRNA control). Whole-cell extracts were produced after 48 h. Lysates were run on 10% Tris·glycine gels, and Western blotting was performed by using anti-GABRA4 AB (1:1,000, NB 300-194, Novus Biologicals). Visualization was performed by using secondary Ab conjugated to horseradish peroxidase (sc-2004, Santa Cruz Biotechnology).
Results
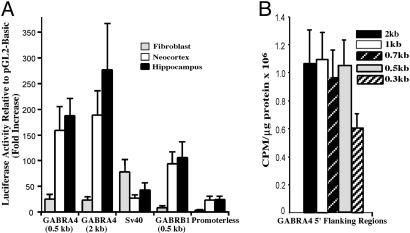
Activity of GABRA4 5′ Flanking Sequence. PCR products containing various sizes of fragments of GABRA4 flanking sequence were cloned into reporter vectors [gene light plasmids (pGL); Promega] and transfected into primary cultured cells. Transcriptional activity of GABRA4 is greater in neurons when compared with fibroblasts with equal amounts in cells derived from embryonic hippocampal formation and neocortex. (Note that neuronal cultures were grown under conditions that do not support growth of nonneuronal cells; Fig. 1A.) Activity driven by a 500-bp fragment of GABRA4 is equal to that of larger 5′ flanking regions and more than those containing 300 bp, suggesting that the minimal promoter (GABRA4p) is contained within the first 500 bp of the 5′ flanking region (Fig. 1B).
Fig. 1.
Transcriptional activity of the GABRA4 5′ flanking region. (A) Primary hippocampal, neocortical, and fibroblast cultures were transfected with pGL2-derived promoter constructs. Specific activity driven by simian virus 40 or GABRB1 promoters, as well as a promoterless basic vector, are shown for comparison. (B) Identification of the minimal promoter by using transfected primary neocortical neurons. Maximal-promoter activity was detected in 5′ flanking regions ≥500 bp. Size of GABRA4 5′ flanking region is as indicated.
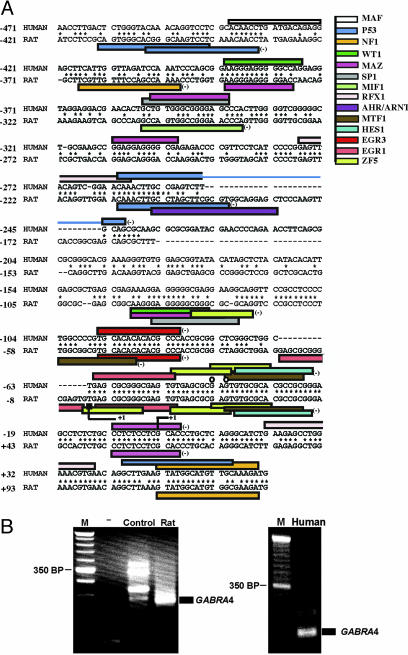
Bioinformatic analysis (Genomatix, Munich) was used to identify potential binding sites in human GABRA4p, and conservation of regulatory elements between human and rat was used to identify sites with highest probability for functional significance (Fig. 2A). Consensus sites associated with cellular signaling, including Egr sites, were identified. Note that Egr consensus sites are found in promoters of other GABRs in which transcripts are increased in the PILO model of TLE (5).
Fig. 2.
Conservation of DNA sequence in human and rat GABRA4 promoters. (A) Alignment and identification of potential regulatory elements was performed by using GENOMATIX software. Only sites from matrix families shared by both the human and rat promoter, irrespective of position, are included. Core similarity of 0.75 and matrix similarity of 1.01 were used for detection. Colored bars indicate consensus sites specific to the factors abbreviated as follows on the right: AHR/ARNT, Aryl hydrocarbon receptor; EGR3, Egr gene 3 product; EGR1, Egr-1/Krox-24/NGFI-A/zif268 immediate-early gene product; WT1, Wilms tumor suppressor; HES1, Drosophila hairy and enhancer of split homologue 1; MAZ, Myc-associated zinc-finger protein; MTF, metal transcription factor 1; NF1, nuclear factor 1; P53, tumor suppressor p53; SP1, stimulating protein 1; RFX1, X-box binding protein R; MIF1, MIBP-1-RFX1 complex; ZF5, zinc finger/poxvirus zinc finger (POZ) domain transcription factor. Detection of major transcription initiation site for human and rat GABRA4 was performed by using RACE with either rat RNA from cultured HNs (most-5′ site indicated by line corner and plus) or human brain RNA (down-stream site indicated by line corner and plus). The 5′ end of the adult mouse hippocampal EST obtained from a search of the National Center for Biotechnology Information database (accession no. BY712705) is indicated by a black rectangle, and circles indicate the 5′ ends of adult human hippocampal ESTs from the National Center for Biotechnology Information database (accession nos. BI545886 and BG699585). Conserved sequence is indicated by stars, and shared sequences between elements are indicated by overlapping bars. The numbering of sequence is in reference to the first nucleotide (+1) of the human GABRA4 gene as determined by RACE. (B) Start-site identification using cDNAs generated from either rat primary hippocampal cultures or human brain total RNA. (Left) Lanes are defined as follows: M, 50 bp DNA ladder marker; -,no template; Control, control mouse thymus cDNAs as template; and Rat, rat hippocampal cDNAs as template. (Right) Lanes are defined as follows: M, 50 bp marker; and Human, human brain cDNAs as template.
cDNA molecules generated from either HNs or from adult human brain were used in high-stringency RACE with polymerases optimized for GC-rich conditions to identify endogenous start sites of transcription (Fig. 2B). PCR products were sequenced for identity. Together with 5′ ends of ESTs (Fig. 2A), there are a number of start sites in GABRA4p, which is indicative of a promoter that does not use TATA for start-site selection. Relative conservation of start sites in rat and human suggests that human GABRA4p uses a similar mechanism for basal transcription.
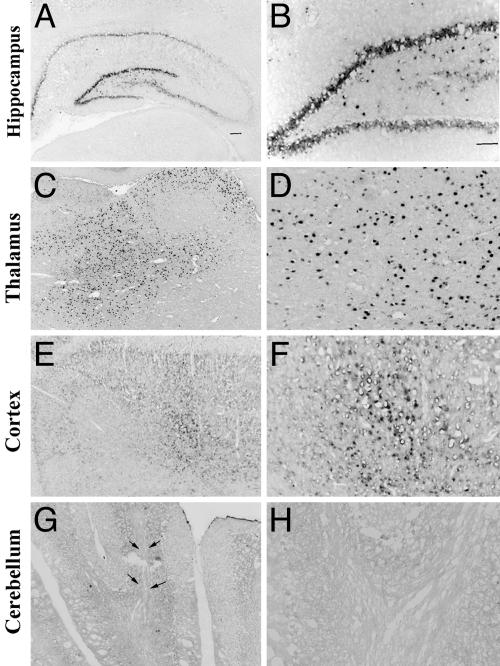
Activity of GABRA4p in AAV Responds to PILO-Induced SE. To test whether minimal GABRA4p is functional in vivo, it was cloned into an AAV serotype 2 (AAV2) vector upstream of the transgene for eYFP. At 3-4 weeks after stereotactic injection of AAV2-A4-eYFP into multiple brain regions, eYFP mRNAs were found in the hippocampal formation, thalamus, and cortex, but not cerebellum (Fig. 3). This region-specific pattern is also seen for endogenous GABRA4 mRNAs (27-30). Studies using a constitutive promoter have shown robust activity in cerebellum, demonstrating that AAV2 vectors can transduce cerebellar neurons (31).
Fig. 3.
The GABRA4 promoter in an AAV2/eYFP viral construct produces region-specific transcription in rat brain. The AAV2-A4-eYFP vector was packaged into an AAV5 serotype capsid, and 2 μl of the generated virus (titer, 5.3 × 1012 genomic particles per μl) was injected into the thalamus, the upper and lower blades of the dentate gyrus in the hippocampus, the cortex, and the cerebellum. At 3-4 weeks after injection, eYFP transcripts driven by GABRA4p were detected by in situ hybridization using an antisense eYFP riboprobe. Note the high levels of expression throughout the hippocampus (A and B) and thalamus (C and D), and modest levels in the cortex (E and F). Transcripts are not present in cerebellum (G and H), as expected based on absence of endogenous GABRA4 expression. Arrows indicate the needle track. Images were taken at ×2.5 (A, C, E, and G) and ×10 (B, D, F, and H) magnification.
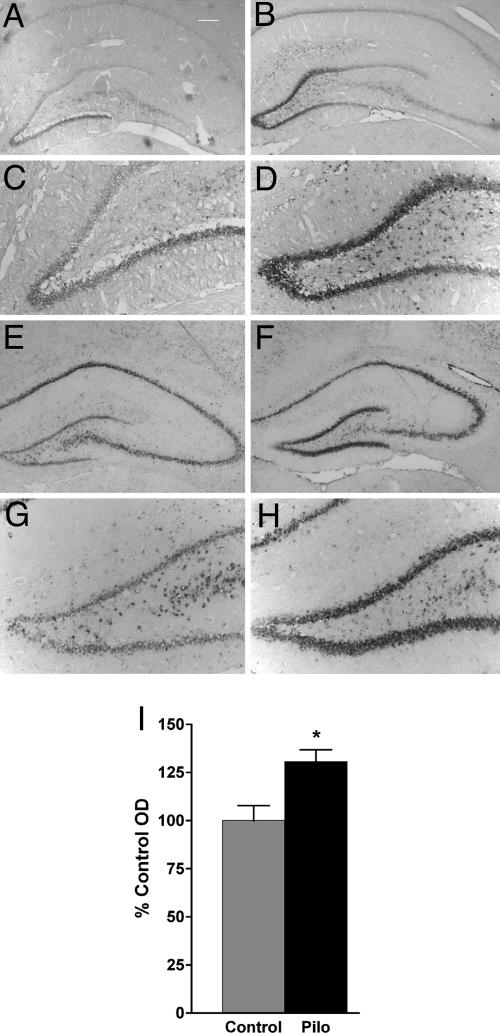
To examine regulation of GABRA4p in epilepsy, AAV2-A4-eYFP was injected into DG of adult rats and SE was induced with PILO 2 weeks later. Latency from PILO injection to seizure onset, severity of SE (on the 1-5 Racine scale), and length of SE were measured. In all cases, there was no difference between AAV-injected and uninjected animals (data not shown). However, there were increases in eYFP mRNAs at 1, 2, and 4 weeks after SE (Fig. 4), indicating that minimal GABRA4p is sufficient to mediate observed up-regulation of α4 mRNAs (6).
Fig. 4.
Activity of GABRA4p in rat hippocampus is up-regulated after PILO-induced SE. The AAV2-A4-eYFP vector was packaged into an AAV5 serotype capsid, and 2 μl of the generated virus (titer, 5.2 × 1012 genomic particles per μl) was injected into the upper and lower blades of the dentate gyrus in the hippocampus. At 2 weeks after injection, SE was induced with PILO. At 1 and 2 weeks after SE (2-4 weeks after virus injection), eYFP transcripts driven by GABRA4p were detected by in situ hybridization using an antisense eYFP riboprobe. Transcripts driven by the GABRA4p in the DGC layer were higher at 1 (B and D) and 2 (F and H) weeks after SE, compared with age-matched AAV2-A4-eYFP-injected animals that were not subjected to SE (controls; A, C, E, and G). Some eYFP transcripts were also detected in CA1 pyramidal layers, likely because of the spread of virus from injection track that passed through CA1. Images were taken at ×2.5 (A, B, E, and F) and ×10 (C, D, G, and H) magnification. (I) Histogram representing levels of eYFP transcripts in DGC layer 4 weeks after AAV injection measured as OD normalized to controls (% Control). *, P = 0.016 (unpaired t test; control, n = 6; SE, n = 5).
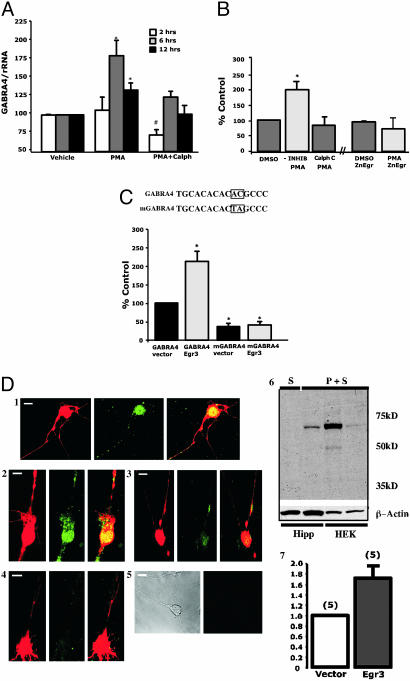
Stimulation of PKC Increases GABRA4 mRNA Levels in Primary Hippocampal Cultures. Distinct isoforms of PKC alter their expression patterns in response to both PILO-(32) and kainic acid-induced seizures (33). Cultured primary HNs were exposed to phorbol myristate acetate (PMA), which activates PKC, as a model to examine mechanisms that may be responsible for up-regulated GABRA4 gene expression after seizures. We found that 6- and 12-h treatments increase GABRA4 mRNA levels, an effect blocked by the specific PKC inhibitor calphostin C (Fig. 5A). Whereas a 2-h stimulation has no effect, PKC inhibition produces a marked decrease, suggesting that PKC may already drive a substantial amount of GABRA4 transcription in neurons.
Fig. 5.
Studies of GABRA4 regulation in primary neurons. (A) Exposure of primary HNs to PMA (1 μM) for 6-12 h increases GABRA4 transcript levels. Effect of PMA is reduced with PKC-specific inhibitor calphostin C (1 μM). * and #, Significant difference from control 95% confidence interval. Data are presented as mean ± SEM from four to eight independent experiments. (B) Primary hippocampal cultures transfected with GABRA4p/reporter were assayed at 24 h after transfection following a 12-h treatment with 1 μM PMA (with and without calphostin C). Data are taken from three to five independent experiments. In a separate set of three independent experiments (as indicated by broken x axis), cells were cotransfected with GABRA4p/reporter and Egr dominant-negative ZnEgr. Results are expressed as the percentage of change with respect to normalized GABRA4 luciferase levels in control dishes (defined as 100%). (C) Vectors containing minimal GABRA4p with (mGABRA4) and without (wtGABRA4) a 2-bp mutation in the Egr3 site (see Fig. 2) (Upper) were transfected into hippocampal cells in the presence and absence of CMV/Egr3 expression construct (Lower). Sister dishes cotransfected with CMV construct minus Egr3 were used as control and indicated as “vector.” Data are displayed as described above, with the exception that it is relative to GABRA4p and cotransfected “vector” control set at 100%. (D)(1-4) Presence of endogenous α4 subunits is up-regulated by Egr3 in DsRed-containing hippocampal cells as detected by immunohistochemistry using anα4-specific Ab (Egr3 construct, 1 and 2; empty vector, 3 and 4). Conditions were as follows (as shown from left to right): DsRed, α4 FITC, Yellow overlay of DsRed, and FITC in representative samples). (5) Secondary Ab alone (white bars, 10 μM). (6) Western blot analysis showing specificity of α4 stain. P, primary Ab; S, secondary Ab; Hipp, hippocampal culture (10μg per lane); and HEK,α4 cDNA transfected HEK293 cells (20 μg per lane) cotransfected with either control (Left)or α4(Right) siRNA. (7) Histogram showing that Egr3-mediated increases in endogenous α4 are significantly different from control 95% confidence interval. y axis displays fold change in α4 immunofluorescence.
GABRA4p/Reporter Activity Is Increased by PKC and Overexpression of Egr3. In parallel to PMA-induced increases in α4 mRNA levels, GABRA4p/reporter activity increases in response to activated PKC (Fig. 5B). Based on the presence of Egr consensus sites (Fig. 2 A), HNs were also cotransfected with the Egr dominant-negative expression construct ZnEgr (34). ZnEgr blocks the effect of PMA on GABRA4p/reporter activity (Fig. 5B). In contrast, overexpression of Egr3 markedly up-regulates promoter activity when the promoter contains an intact Egr3 site (Fig. 5C). Similar results are observed by using a 2-kb fragment of GABRA4p (data not shown).
To establish a direct link between Egr3 and endogenous GABRA4 transcription, HNs were transfected with the CMV/Egr3 expression construct and the level of endogenous α4 subunits was monitored by confocal microscopy (Fig. 5D). A cotransfected CMV/dsRed monomer protein marker (BD Biosciences) was used to identify transfected neurons for analysis. Controls contained transfected vector without the Egr3 transgene. Transfection of an α4 cDNA expression construct into HEK cells and removal of the putative protein by specific α4 siRNA demonstrates that the α4 Ab recognizes a single form of α4 subunit in primary HNs (Figs. 5D and 6).
Fig. 6.
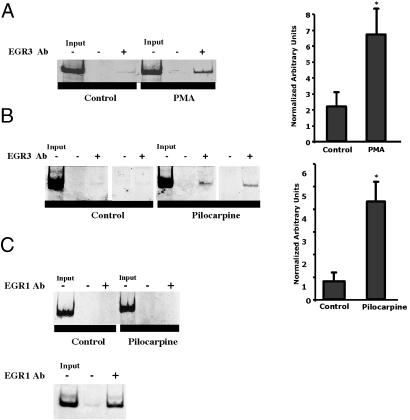
Increased binding of Egr3 to GABRA4 in DGCs of adult rats after PILO parallels changes in PMA-stimulated HNs. (A) ChIP assays were performed by using gDNA extracted from formaldehyde-treated neuronal cultures (with and without 2-h PMA treatment, as indicated). A representative sample from an independent experiment is shown for each precipitation condition. Template (Input) is gDNA before precipitation and gDNA after precipitation with Egr3-specific Abs (Egr3 Ab). The presence and absence of Ab are indicated by + or -, respectively. (Right) Histogram showing quantification of Egr3-specific precipitation. *, P = 0.011 (Student's t test, n = 6 animals per condition). (B) ChIP was performed by using slices of dentate gyrus from animals 24 h after PILO-induced SE and controls. (Right) Egr3-specific precipitation. *, P = 0.025 (Student's t test, n = 4 animals per condition). (C) ChIP was performed with an Egr1 Ab. (Upper) In vivo ChIP, as described in B. (Lower) Binding of Egr1 to the NMDA receptor 1 gene (NMDAR1) in cortical cultures detected by using NMDAR1-specific primers.
Egr3 Is Recruited to Endogenous GABRA4 After PMA and in Vivo After PILO-Induced SE. As a further confirmation that PMA-increased transcription of GABRA4 is mediated through binding of Egr3 to GABRA4p, ChIP was used to show that PMA increases association of Egr3 with GABRA4 in primary neuronal cultures (Fig. 6A). Similarly, adult rats treated with PILO show a robust increase in Egr3 at GABRA4p 24 h after SE (Fig. 6B). In contrast to Egr3, 24 h after SE, there is no increase in the presence of Egr1 (Fig. 6C Upper). This lack of signal is unlikely to be due to poor Ab performance because the Egr1 Ab recognizes Egr1 at the NMDA receptor 1 gene in HNs (Fig. 6C Lower).
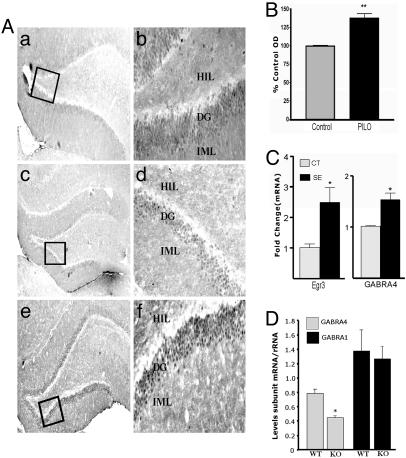
Increased binding of Egr3 to GABRA4 after SE most likely occurs as a result of increased Egr3 synthesis because levels of Egr3 mRNAs and protein increase together (Fig. 7 A-C). Such increases are accompanied by significant changes in levels of GABRA4 mRNAs (Fig. 7C). In contrast, levels of Egr3 are not increased 1 h after SE, which is consistent with our finding that neither Egr3 nor Egr1 binds to GABRA4 1 h after SE and there is no change in levels of GABRA4 mRNAs (data not shown).
Fig. 7.
In vivo changes in levels of Egr3. (A) Levels of Egr3 increase in dentate gyrus 24 h after SE but not 1 h. Photomicrographs show Egr3 immunostaining in hippocampus from a representative control rat (a and b), a rat 1 h after declared SE (c and d), and a rat 24 h after declared SE (e and f). Photographs were taken at magnifications of ×5(a, c, and e) and ×20 (b, d, and f). HIL, hilus; DG, DGC layer; and IML, inner molecular layer. (B) Histogram representing intensity of Egr3 immunostaining in DGCs of control and PILO-treated animals measured as OD normalized to controls (% Control OD). **, P < 0.005 (unpaired t test, n = 3 for each group). (C) Increased Egr3 and GABRA4 mRNA levels accompany increased Egr3 protein (real-time PCR). Egr3, n = 4; Ct, n = 6 (24 h after SE), P = 0.038 Mann-Whitney; GABRA4, P = 0.0025. (D) Egr3 knockout mice express lower levels of GABRA4 mRNAs, with no change in GABRA1. RNA was extracted from hippocampus of three littermate pairs. PCR was performed three times by using the same RNA samples to yield similar results. Statistical significance was determined by Student's t test (P = 0.035).
Knockout of Egr3 Causes Marked Reduction in GABRA4 mRNA Levels. Mice devoid of Egr3 do not have muscle spindles or display sensory ataxia, resting tremor, and scoliosis (35). They also have ≈50% less GABRA4 mRNAs (Fig. 7D), with no change in transcripts for the most abundant α subunit (α1), which is suggestive of a major and specific role of Egr3 at GABRA4 in vivo.
Discussion
Alteration in GABAAR subunit gene expression may have an important role in the etiology of TLE (for review, see ref. 36). Evidence that TLE produces or is the product of alterations in GABAAR function has come from the study of DGCs in adult TLE patients (9, 37) and rodent models of TLE (6, 24, 38). DGCs in chronically epileptic rats display increased density of GABAARs with a unique pharmacology. These changes in receptor activity, including heightened blockade of receptor function by zinc and a marked decrease in type 1 benzodiazepine modulation, are associated with a marked increase in α4 and decrease in α1 subunit gene expression (6). Here, we report that this alteration in α4 mRNAs and protein levels is most likely transcriptionally mediated, and we identify Egr3 as a potential specific regulator.
Egr3 is a member of the Egr family of factors induced in response to immediate early gene activation and multiple isoforms are detected in rat brain (39). Results of our studies strongly suggest that Egr3 controls altered expression of α4 subunits after SE; however, we cannot rule out the possibility that other upstream elements a distance from the start site may also be important in the context of the endogenous GABRA4. The facts that mutating the Egr3 site in GABRA4p markedly reduces activity in developing HNs (Fig. 5C), overexpressing Egr3 protein alters the expression of endogenous α4 subunits (Fig. 5D), and mice devoid of Egr3 have significantly lower levels of α4 mRNAs (Fig. 7D), strongly suggest that Egr3 may be a critical regulator of endogenous GABRA4 during development. The relationship between plastic processes that underlie normal brain development and processes that underlie neuropathology has already been suggested (13).
Given the fact that all GABRs whose mRNA levels increase in response to PILO (6) contain Egr sites (5), a coordinated increase in certain GABR family members may be responsible for observed alterations in GABAergic neurotransmission specific to TLE. The results of these studies have the promise of providing information regarding dynamics of gene expression in the impaired nervous system as well as generating genetic therapies for the treatment, prevention, or cure of some forms of acquired epilepsy.
Acknowledgments
We thank J. Baraban, J. Milbrandt, A. Gallitano-Mendel (Washington University, St. Louis), and W. Tourtellotte for their generous gifts of Egr expression constructs and tissues from Egr3 knockout mice. We especially thank S. C. Martin for her guidance with confocal microscopy studies. We dedicate this article to David H. Farb for his invaluable insight and contributions to the establishment of this area of study. This work was supported by National Institutes of Health Training Grants 2T32 GM00854 (to D.S.R.) and T32 GM07229 (to I.V.L.) and National Institutes of Health Grants NS4236301 (to S.J.R. and A.R.B.-K.) and NS38690 (to J.H.W.).
Author contributions: J.H.W., A.R.B.-K., and S.J.R. designed research; D.S.R., Y.S.R., S.B., E.B., I.V.L., and M.J.P. performed research; J.H.W. contributed new reagents/analytic tools; D.S.R., Y.S.R., A.R.B.-K., and S.J.R. analyzed data; and A.R.B.-K. and S.J.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; GABAAR, GABAA receptor; SE, status epilepticus; DGC, dentate granule cell; Egr, early growth response factor; TLE, temporal lobe epilepsy; PILO, pilocarpine; HN, hippocampal neuron; gDNA, genomic DNA; eYFP, enhanced yellow fluorescent protein; AAV, adeno-associated viral; AAV2, AAV serotype 2; PMA, phorbol myristate acetate; siRNA, small interfering RNA.
References
- 1.Rabow, L. E., Russek, S. J. & Farb, D. H. (1995) Synapse 21, 189-274. [DOI] [PubMed] [Google Scholar]
- 2.Davies, P. A., Hanna, M. C., Hales, T. G. & Kirkness, E. F. (1997) Nature 385, 820-823. [DOI] [PubMed] [Google Scholar]
- 3.Bonnert, T. P., McKernan, R. M., Farrar, S., le Bourdelles, B., Heavens, R. P., Smith, D. W., Hewson, L., Rigby, M. R., Sirinathsinghji, D. J., Brown, N., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 9891-9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russek, S. J. (2003) in Nature Encyclopedia of the Human Genome, ed. Cooper, D. N. (Macmillan, Basingstoke, England).
- 5.Steiger, J. L. & Russek, S. J. (2004) Pharmacol. Ther. 101, 259-281. [DOI] [PubMed] [Google Scholar]
- 6.Brooks-Kayal, A. R., Shumate, M. D., Jin, H., Rikhter, T. Y. & Coulter, D. A. (1998) Nat. Med. 10, 1166-1172. [DOI] [PubMed] [Google Scholar]
- 7.Clark, M. (1998) Neurosci. Lett. 250, 17-20. [DOI] [PubMed] [Google Scholar]
- 8.Smith, S. S., Gong, Q. H., Hsu, F. C., Markowitz, R. S., Ffrench-Mullen, J. M. & Li, X. (1998) Nature 392, 926-930. [DOI] [PubMed] [Google Scholar]
- 9.Brooks-Kayal, A. R., Shumate, M. D., Jin, H., Lin, D. D., Rikhter, T. Y., Holloway, K. L. & Coulter, D. A. (1999) J. Neurosci. 19, 8312-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsian, A. & Zhang, Z. H. (1999) Am. J. Med. Genet. 88, 533-538. [DOI] [PubMed] [Google Scholar]
- 11.Kapur, J. (2000) Epilepsia 41, Suppl. 6, S86-S89. [DOI] [PubMed] [Google Scholar]
- 12.Benke, D., Michel, C. & Mohler, H. (1997) J. Neurochem. 69, 806-814. [DOI] [PubMed] [Google Scholar]
- 13.Schwartzkroin, P. A. (2001) Int. Rev. Neurobiol. 45, 1-15. [DOI] [PubMed] [Google Scholar]
- 14.Knoflach, F., Benke, D., Wang, Y., Scheurer, L., Luddens, H., Hamilton, B. J., Carter, D. B., Mohler, H. & Benson, J. A. (1996) Mol. Pharmacol. 50, 1253-1261. [PubMed] [Google Scholar]
- 15.Fisher, J. L. & Macdonald, R. L. (1998) J. Neurosci. 18, 2944-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng, Z., Hauer, B., Mihalek, R. M., Homanics, G. E., Sieghart, W., Olsen, R. W. & Houser, C. R. (2002) J. Comp. Neurol. 446, 179-197. [DOI] [PubMed] [Google Scholar]
- 17.Wisden, W., Cope, D., Klausberger, T., Hauer, B., Sinkkonen, S. T., Tretter, V., Lujan, R., Jones, A., Korpi, E. R., Mody, I., et al. (2002) Neuropharmacology 43, 530-549. [DOI] [PubMed] [Google Scholar]
- 18.Sinkkonen, S., Vekovischeva, O. Y., Möykkynen, T., Ogris, W., Sieghart, W., Wisden, W. & Korpi, E. R. (2004) Eur. J. Neurosci. 20, 2168-2178. [DOI] [PubMed] [Google Scholar]
- 19.Tseng, Y. T., Wellman, S. E. & Ho, I. K. (1994) J. Neurochem. 63, 301-309. [DOI] [PubMed] [Google Scholar]
- 20.Harris, B., Costa, E. & Grayson, D. (1995) Mol. Brain Res. 28, 338-342. [DOI] [PubMed] [Google Scholar]
- 21.Poulter, M. O., Ohannesian, L., Larmet, Y. & Feltz, P. (1997) J. Neurochem. 68, 631-639. [DOI] [PubMed] [Google Scholar]
- 22.Russek, S. J., Bandyopadhyay, S. & Farb, D. H. (2000) Proc. Natl. Acad. Sci. USA 97, 8600-8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia, Z., Dudek, H., Miranti, C. K. & Greenberg, M. E. (1996) J. Neurosci. 16, 5425-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs, J. W., III, Shumate, M. D. & Coulter, D. A. (1997) J. Neurophysiol. 77, 1924-1938. [DOI] [PubMed] [Google Scholar]
- 25.Racine, R. (1972) Electroencephalogr. Clin. Neurophysiol. 32, 281-294. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, M. H. & Allis, C. D. (1999) Methods 19, 425-433. [DOI] [PubMed] [Google Scholar]
- 27.Wisden, W., Laurie, D. J., Monyer, H. & Seeburg, P. H. (1992) J. Neurosci. 12, 1040-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntsman, M. M., Munoz, A. & Jones, E. G. (1999) Neuroscience 91, 1223-1245. [DOI] [PubMed] [Google Scholar]
- 29.Arnot, M. I., Davies, M., Martin, I. L. & Bateson, A. N. (2001) J. Neurosci. Res. 64, 617-625. [DOI] [PubMed] [Google Scholar]
- 30.Merali, Z., Du, L., Hrdina, P., Palkovits, M., Faludi, G., Poulter, M. O. & Anisman, H. (2004) J. Neurosci. 24, 1478-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passini, M. A. & Wolfe, J. H. (2001) J. Virol. 75, 12382-12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, F. R., Lee, W. L., Gao, H., Chen, Y., Loh, Y. T. & Chia, S. C. (2004) Hippocampus 14, 87-98. [DOI] [PubMed] [Google Scholar]
- 33.Guglielmetti, F., Rattray, M., Baldessari, S., Butelli, E., Samanin, R. & Bendotti, C. (1997) Brain Res. Mol. Brain Res. 49, 188-196. [DOI] [PubMed] [Google Scholar]
- 34.Rolli-Derkinderen, M., Machavoine, F., Baraban, J. M., Grolleau, A., Beretta, L. & Dy, M. (2003) J. Biol. Chem. 278, 18859-18867. [DOI] [PubMed] [Google Scholar]
- 35.Tourtellotte, W. G. & Milbrandt, J. (1998) Nat. Genet. 20, 87-91. [DOI] [PubMed] [Google Scholar]
- 36.Coulter, D. A. (2000) Epilepsia 41, Suppl. 6, S96-S99. [DOI] [PubMed] [Google Scholar]
- 37.Shumate, M. D., Lin, D. D., Gibbs, J. W., III, Holloway, K. L. & Coulter, D. A. (1998) Epilepsy Res. 32, 114-128. [DOI] [PubMed] [Google Scholar]
- 38.Buhl, E. H., Otis, T. S. & Mody, I. (1996) Science 271, 369-373. [DOI] [PubMed] [Google Scholar]
- 39.O'Donovan, K. J. & Baraban, J. M. (1999) Mol. Cell. Biol. 19, 4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]