Abstract
AlaX is the homologue of the class II alanyl-tRNA synthetase editing domain and has been shown to exhibit autonomous editing activity against mischarged tRNAAla. Here, we present the structures of AlaX from the archaeon Pyrococcus horikoshii in apo form, complexed with zinc, and with noncognate amino acid l-serine and zinc. Together with mutational analysis, we demonstrated that the conserved Thr-30 hydroxyl group located near the β-methylene of the bound serine is responsible for the discrimination of noncognate serine from cognate alanine, based on their chemical natures. Furthermore, we confirmed that the conserved Gln-584 in alanyl-tRNA synthetase, which corresponds to Thr-30 of AlaX, is also critical for discrimination. These observations strongly suggested conservation of the chemical discrimination among trans- and cis-editing of tRNAAla.
Keywords: alanyl-tRNA synthetase, class II tRNA synthetase, crystal structure, trans-editing
Aminoacyl-tRNA synthetases (aaRSs) establish the genetic code through aminoacylation of cognate tRNA (1). However, in some aaRSs, the affinity difference of the active site is not large enough to distinguish among similar amino acids with sufficient accuracy. Therefore, during evolution, an additional editing domain that specifically hydrolyzes mischarged tRNAs has assembled with the catalytic domain to comprise contemporary aaRSs (2). In accordance with this model, the genes that autonomously encode an editing domain were distributed in many organisms (3–5), and, indeed, some of them are shown to be responsible for the trans-editing activity of mischarged tRNAs (4, 5). AlaX is the one such protein that shows homology to the class II alanyl-tRNA synthetase (AlaRS) editing domain (Fig. 1) and is widely scattered among all three kingdoms of life (3). The specific activities of the archaeal AlaXs from Methanosarcina barkeri and Sulfolobus solfataricus have recently been shown to specifically hydrolyze mischarged Ser- and Gly-tRNAAla in vitro (4).
Fig. 1.
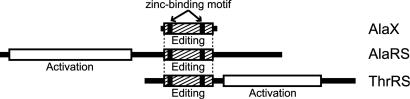
Domain organizations of AlaX, AlaRS, and ThrRS. The boxes with hatched lines indicate the homologous editing domains. The two vertical black stripes in the boxes show the conserved HXXXH and CXXXH motifs.
In contrast to the well established class I aaRSs, information regarding editing of the evolutionarily distinct class II aaRSs (to which AlaRS belongs) has only recently begun to emerge. The editing mechanism of the threonyl system has been investigated by structural analyses of the bacterial threonyl-tRNA synthetase (ThrRS) editing domain (hereafter called ThrRS-N2) complexed with the serine product or with the substrate analogue seryl-3′-aminoadenosine (SerA76) (6). These analyses showed that (i) cognate threonine and noncognate serine are discriminated by steric exclusion of the additional γ-methyl group of threonine by the conserved His-77, Tyr-104, and Asp-180, and (ii) the HXXXH and CXXXH motifs characteristic of ThrRS-N2 should not bind a zinc ion for catalysis, despite the capability to bind zinc (7). The AlaX/AlaRS editing domains are evolutionarily related to ThrRS-N2 and share the characteristic HXXXH and CXXXH motifs (Fig. 1) (4, 8), which were also shown to be important for the deacylation activity of AlaRS (9). The inclusion of noncognate serine and glycine in the AlaRS activation site was reported in ref. 10 and was recently confirmed structurally (11). The structures of AlaRS complexed with amino acids suggested that serine hydroxyl is accommodated by an induced fit of Asn-194 that allows expansion of the activation pocket and that the smaller glycine is also included, based simply on a canonical size-sieving model. Therefore, the editing site in the alanyl system should recognize both serine and glycine but not alanine (9). However, the discrimination cannot be explained by simple size exclusion, because the side chain of serine is larger than that of alanine. The mechanism of editing in the alanyl system still remains unclear, because no structure of the editing site has yet been determined.
To understand the structural basis of amino acid discrimination in an alanyl system, we determined the structure of AlaX from the archaeon Pyrococcus horikoshii (PhoAlaX), the minimum region of the class II-specific editing domain, in complex with the noncognate amino acid l-serine. Our results demonstrated that the discrimination of serine and alanine depends critically on Thr-30, which is located in the vicinity of the β-methylene of the bound serine. Therefore, the chemical repulsion of an alanine side chain was suggested. The proposed model could also be extended to the cis-editing in AlaRS, because mutation of the residue corresponding to Thr-30 had a similar effect.
Materials and Methods
Preparation of Proteins. Wild-type and C-terminal His6-tagged PhoAlaX genes were expressed in Escherichia coli BL21-Codonplus-(DE3)-RIL-X (Stratagene) at 37°C. His6-tagged PhoAlaX was captured by Ni-NTA (Qiagen) after a heat treatment (70°C for 30 min) of the cell extract and further purified by using a Superdex 200 gel filtration column (Amersham Pharmacia Biosciences) at pH 6.0. Dimer and monomer were separated during the chromatography. Each fraction was dialyzed against the final buffer (20 mM sodium phosphate, pH 7.0, for the monomer form; 20 mM sodium phosphate, pH 7.0/0.2 M NaCl for the dimer form) and concentrated to 8 mg/ml. The wild-type PhoAlaX was purified by heat treatment (70°C for 30 min) by using the SP-XL cation-exchange column, Superdex 200 gel filtration column, and RESOURCE PHE column (all from Amersham Pharmacia Biosciences) at pH 6.0. Before the cation-exchange chromatography, removal of nucleic acids by adding 0.5% polyethyleneimine (pH 6.0) was necessarily for effective binding of PhoAlaX to the column. Dimer and monomer were also separated during the gel filtration as well as the His6-tagged protein. Each fraction was dialyzed against 10 mM Tris·HCl, pH 7.5 (monomer) or 10 mM Tris·HCl, pH 7.5/0.2 M NaCl (dimer) and concentrated to 8 mg/ml.
The plasmid encoding C-terminal-truncated P. horikoshii AlaRS (PhoAlaRS) was a kind gift from O. Nureki (Tokyo Institute of Technology, Tokyo). For enhancement of the solubility of PhoAlaRS during expression, the region after the editing domain (754–915), which encodes the oligomerization domain that has been believed to be not significant in both the aminoacylation and the editing activities, was eliminated during the expression. The purified PhoAlaRS was confirmed to be efficient in both the aminoacylation and the editing reactions (data not shown), indicating that the C-terminal truncation did not greatly affect the functions of AlaRS. PhoAlaRS was expressed in E. coli BL21-Codonplus-(DE3)-RIL-X and purified by heat treatment (70°C for 30 min) by using the Q-XL anion-exchange column, Superdex 200 gel filtration column, and RESOURCE PHE column at pH 7.5.
The mutations were introduced by site-directed mutagenesis. Each mutant was expressed and purified essentially the same as were the wild-type enzymes.
Deacylation Assay. The plasmid for the template of P. horikoshii tRNAAla transcription was also a kind gift from O. Nureki. tRNAAla was prepared by a standard in vitro transcription, essentially as described in ref. 9, and purified by using a RESOURCE Q column (Amersham Pharmacia Biosciences). 3H-labeled aminoacyl-tRNAs were prepared by using PhoAlaRS-C717A mischarging mutant, essentially as described in ref. 9. tRNAAla (10 μM) was incubated with PhoAlaRS-C717A (2 μM) and ATP (2 mM) in the presence of 10.5 μM [3H]serine or 9.3 μM [3H]alanine (Amersham Pharmacia Biosciences) in a charging buffer (50 mM NaHepes, pH 7.5/20 mM KCl/10 mM MgCl2/5 mM 2-mercapthoethanol/0.1 mg/ml BSA) at 60°C for 20 min. The reaction mixture was phenol/chloroform extracted, ethanol precipitated, and stored at –80°C.
The deacylation assay was performed, essentially as described in ref. 9. The 3H-labeled aminoacyl-tRNA (5 μM) was incubated with 5 μM PhoAlaX or 2 μM PhoAlaRS in the charging buffer at 55°C. The reaction was quenched by spotting a 10-μl aliquot of the mixture to a Whatman 3MM filter disk saturated with 5% trichloroacetic acid (TCA). The filters were washed with 5% TCA and ethanol and dried. Radioactivity was quantified by liquid scintillation counting.
Crystallization and Data Collection. The crystals were initially obtained for His6-tagged PhoAlaX dimer (PhoAlaXD) by the hanging-drop method within a week at 20°C with a reservoir containing 0.1 M Tris·HCl, pH 8.6, 24% (wt/vol) polyethylene glycol (PEG) 4000, and 3% (vol/vol) 2-methyl-2,4-pentanediol (MDP). The crystal was soaked in cryoprotectant containing an additional 15% MPD in the reservoir for 1 min and flash-cooled under a liquid nitrogen stream. For the ligand-complexed structure, the crystal was presoaked in the mother liquor containing 5 mM ZnSO4 and 5 mM l-serine for 3 h before soaking in the cryoprotectant. The diffraction experiments were performed at 100 K by using synchrotron radiation sources at BL38B1 (SPring-8, Harima, Japan) or BL6A (Photon Factory, Tsukuba, Japan). The diffraction images were analyzed and processed by using the program hkl2000 (12). The crystals belonged to a space group of P212121, with cell dimensions of a = 34.2 Å, b = 88.7 Å, and c = 110.1 Å, indicating two molecules per asymmetric unit with a VM value of 2.17 (Å3/Da). The crystals of wild-type PhoAlaX monomer (PhoAlaXM) were obtained by the hanging-drop method at 20°C with a reservoir containing 0.1 M NaMes, pH 6.4, and 14% (wt/vol) PEG 8000. The data were collected by using BL41XU (SPring-8) by essentially the same method as His-tagged PhoAlaX crystal, except that the buffer containing 25% (vol/vol) glycerol was used for the cryoprotectant. The data were analyzed and processed with hkl2000, and the results indicated that the crystal belongs to a space group of P21 with the unit cell parameters of a = 42.5 Å, b = 98.0 Å, c = 60.6 Å, and β = 95.8° and containing three molecules in an asymmetric unit. Data-collection statistics are summarized in Table 1.
Table 1. Statistics of the structure analysis.
| Data set
|
|||||
|---|---|---|---|---|---|
| apo-PhAlaXD-HIS
|
|||||
| Parameter | Peak | Edge | Remote | PhAlaXD-HIS-Ser-Zn2+ | WT-PhAlaXM-Zn2+ |
| Data collection statistics* | 0.9797 | 0.9800 | 0.9000 | 1.0000 | 0.9793 |
| Wavelength, Å | SPring-8 BL38B1 | PF BL6A | SPring-8 BL41XU | ||
| X-ray source | P212121 | P212121 | P21 | ||
| Unit cell, Å | 34.2, 88.7, 110.1 | 34.0, 88.5, 109.9 | 42.5, 98.0, 60.6, β = 95.8° | ||
| Unique reflections | 10,320 | 10,576 | 9,907 | 8,871 | 39,904 |
| Resolution, Å | 50.0-2.64 | 50.0-2.62 | 50.0-2.68 | 50.0-2.80 | 50.0-1.88 |
| Completeness, % | 99.7 (99.8) | 99.7 (99.6) | 99.7 (99.9) | 100.0 (100.0) | 99.8 (98.8) |
| Redundancy | 6.4 (6.0) | 6.3 (5.8) | 6.4 (5.9) | 8.8 (8.6) | 5.5 (4.7) |
| Rmerge,† % | 9.0 (39.1) | 8.1 (39.4) | 8.4 (39.6) | 9.3 (40.5) | 7.1 (38.2) |
| Refinement statistics | |||||
| Resolution, Å | 15.0-2.62 | 15.0-2.80 | 50.0-1.88 | ||
| Number of atoms (Protein/water/l-serine/zinc) | 2,514/107/0/0 | 2,497/107/14/2 | 3,737/422/0/3 | ||
| R factor‡/Rfree,§ % | 23.5/27.3 | 22.7/26.6 | 19.3/23.2 | ||
| rmsd bond lengths, Å/bond angles, ° | 0.011/1.4 | 0.0084/1.3 | 0.0050/1.3 | ||
| Ramachandran plot, % (favored/allowed/generous/disallowed) | 84.4/15.3/0.4/0 | 90.9/9.1/0/0 | 93.9/6.1/0/0 | ||
| Average B factor, Å2 | 51.8 | 41.0 | 26.7 | ||
rmsd, rms deviation.
Values in parentheses are for the highest resolution shell.
Rmerge = {ΣhΣi |〈I(h)〉 - I(h)i|}/ΣhΣi |I(h)i, where I(h)i is the ith observation of reflection h and 〈I(h)〉 is the mean intensity of all observations of reflection h.
R factor = {Σ∥Fobs|-|Fcalc∥} |Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factor amplitudes.
Rfree was calculated for 6.5%, 7.0%, and 10% randomly selected reflections of free, ser-Zn2+, and Zn2+ complex data sets that were not used in the refinement.
Phasing and Refinement. The structure of PhoAlaXD was determined by Se-MAD (SeMet multiple-wavelength anomalous diffraction). Five of six Se sites were identified by using the program solve (13). The remaining Se site was identified from the difference Fourier map calculated with these five Se sites by using the program sharp (14). The initial phase was recalculated by sharp with all six Se sites and improved by solvent-flattening using the program solomon (15) and noncrystallographic symmetry (NCS) averaging using the program dm (16). The initial model was manually traced and refined semiautomatically by using the program lafire (M.Y., Y. Zhou, and I.T., unpublished work) with the program cns (17). The NCS restraint was applied to the main chain during the refinement. Finally, the structure was refined to 2.62-Å resolution with an R factor of 23.5% and an Rfree factor of 27.3% and was validated by the program procheck (18). The refined model of PhoAlaX contains 155 and 153 residues in two molecules and 107 water molecules. The C-terminal residues (156–157, 154–157, and fused His-tags) could not be built because the electron densities were poor.
The structure of the serine–Zn2+ complex was solved by positioning the ligand-free protein model with rigid-body refinement by using cns. After several cycles of positional and B factor refinement with manual fitting, the electron density of serine and zinc was clearly shown in the Fo–Fc map (the omit map of serine is shown in Fig. 4A). The models of the ligands were manually placed. The model of the serine–Zn2+ complex was refined to an R factor of 22.7% and an Rfree factor of 26.6% at 2.80-Å resolution, with 154 and 152 residues in two molecules, one l-serine and zinc per molecule, and 107 water molecules.
Fig. 4.
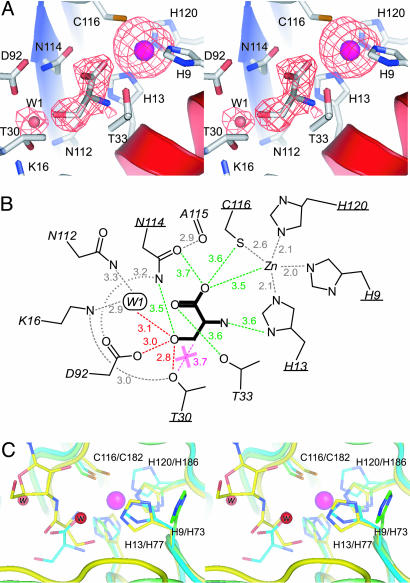
The editing site. (A) Stereo diagram of the editing site of PhoAlaX–serine–Zn2+. The omit map of serine, zinc, and W1 (resolution 2.8 Å, contoured at 2.0σ) is also presented. (B) Schematic representation of the editing site. Distances are indicated (in Å) with dashed lines. The red dashed lines indicate the hydrogen bonds with serine. The gray dashed lines indicate the internal hydrogen bonds or the metal coordination that also exist in the structures without serine. The magenta dashed line with the X mark indicates the chemical repulsion. Note that green dashed lines do not represent hydrogen bonds but indicate only distances with each residue. The conserved residues are underlined. (C) Stereo diagram of the superposition of the zinc-binding motifs of apo-PhAlaX (green), PhoAlaX–serine–Zn2+ (cyan), and ThrRS-N2–SerA76 (yellow) (6). The red spheres indicate the catalytic water molecules in ThrRS-N2-SerA76 (w, the nucleophile that binds to the His-73; w, the proton donor). The magenta ball indicates the zinc ion in PhoAlaX–serine–Zn2+.
The structure of PhoAlaXM was solved by molecular replacement by using the PhoAlaXD structure as a search model. Three solutions were found unambiguously by using the program amore (19). The model was refined to an R factor of 19.3% and an Rfree factor of 23.2% at 1.88-Å resolution by lafire with cns and following manual fitting. The final model contained 152, 154, and 152 residues in three molecules, one zinc per molecule, and 422 water molecules. Refinement statistics are summarized in Table 1.
Results
Deacylation of Aminoacyl-tRNAAla by PhoAlaX. Although two AlaXs from archaea, S. solfataricus (SsoAlaX1) and M. barkeri AlaX1 (MbaAlaX), were identified as trans-editing protein of mischarged tRNAAla in vitro, phylogenetic analysis suggested that AlaXs are widely divergent, and our protein of interest, indeed, belonged to a different subfamily from SsoAlaX1 or MbaAlaX (4). Therefore, to assess whether PhoAlaX could be a trans-editing protein of tRNAAla, we first tested whether PhoAlaX could deacylate Ser-tRNAAla in vitro. Because we could have separated PhoAlaXD and PhoAlaXM by gel filtration and confirmed that the former corresponds to the apo form and the latter corresponds to the metal (possibly zinc)-bound form by the structural analyses (see below), we analyzed the activities of each fraction. Both PhoAlaXD and PhoAlaXM deacylated Ser-tRNAAla at a certain level (Fig. 2A), although spontaneous thermal degradation was high under the current conditions (55°C), because the enzyme was from a hyperthermophile. Interestingly, the activity of the zinc-bound monomer form was much higher than that of the apo dimer form. To clarify the reason for the poor activity of PhoAlaXD, we analyzed the effects of 100 μM ZnSO4/10 mM EDTA on the deacylation activity or on the elution pattern of gel filtration in both forms. The results showed that the activity of AlaX is not largely affected by the absence/presence of zinc and that the quaternary structure is also independent from zinc, suggesting that the poor activity of PhoAlaXD is because of the dimerization rather than the absence of zinc.
Fig. 2.
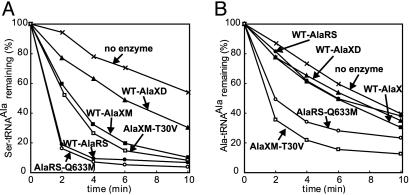
Ser-/Ala-tRNAAla deacylation by AlaX/AlaRS. Shown are the deacylation of Ser-tRNAAla (A) and Ala-tRNAAla (B) by wild-type PhoAlaXM (WT-AlaXM), wild-type PhoAlaXD (WT-AlaXD), PhoAlaXM-T30V mutant (AlaXM-T30V), wild-type PhoAlaRS (WT-AlaRS), and PhoAlaRS-Q633M mutant (AlaRS-Q633M), or without the enzymes (no enzyme) at 55°C.
The deacylation of Gly/Ala-tRNAAla was almost at the background level (the result of only Ala-tRNAAla is shown in Fig. 2B), indicating that PhoAlaX could efficiently discriminate between noncognate serine and cognate alanine but not between noncognate glycine and alanine. The defection of Gly-tRNAAla deacylation implies that PhoAlaX has diverged and has a different role in vivo, as discussed below.
Two Forms of PhoAlaXs. As mentioned above, we purified two forms of PhoAlaX by gel filtration. Each fraction could be crystallized with different space groups, and their structures were solved. The most striking difference between the two structures is the presence of the metal ion tetracoordinated by the conserved HXXXH and CXXXH motifs at the editing site of PhoAlaXM. Neither zinc nor any other heavy metals are included in the buffer used during purification and crystallization of PhoAlaXM, indicating that PhoAlaXM tightly bound the metal from the host cells. In the structure of the serine–Zn2+ complex, the zinc ion, which was systematically introduced by soaking, bound tightly to this motif in the same tetracoordination manner. Therefore, we regarded the metal in the PhoAlaXM as zinc ion. In agreement with this, the structure of PhoAlaX recently deposited in the PDB data bank by another group (PDB ID code 1V4P) also contained zinc ion tetracoordinated by the HXXXH and CXXXH motifs. The crystal lattice and the packing of 1V4P were almost the same as that of PhoAlaXM, suggesting that the crystal of 1V4P was prepared similarly to our PhoAlaXM crystals. The rms displacements among apo, Zn2+, and the serine–Zn2+ complex were within 0.5 Å, showing that the binding of neither zinc nor serine induces a significant conformational change at the main-chain level.
Structure of PhoAlaX. The PhoAlaX monomer consists of two antiparallel β-sheets connected by an α2-helix surrounding a central α1-helix (Fig. 3), showing almost the same topology as the ThrRS-N2 (8), with the rms displacements of 1.8 Å for the 118 Cα atoms compared. The editing site is located at the center cleft, which consists mainly of two parts: the zinc-binding motif, consisting of the conserved His-9, His-13, Cys-116, and His-120 cluster, and another part forming an extremely hydrophilic pocket, consisting of side chains of conserved Thr-30, Asn-114, nonconserved Asp-92, and one water molecule (hereafter called W1). W1 is hydrogen-bonded to the side chains of Lys-16 and Asn-112, and is observed within 1 Å, with an average B factor of ≈24 Å2 in all structures described here, except for the one subunit in apo-PhoAlaXD. The side chains of Thr-30, Asn-114, and Asp-92 are tightly held by internal hydrogen-bond networks, as shown in Fig. 4B, and are almost identical in all of the structures containing W1.
Fig. 3.
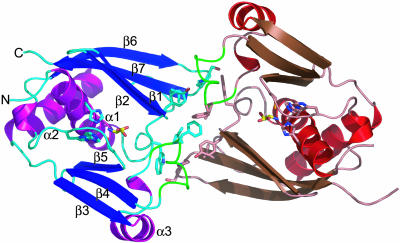
The ribbon diagram of the PhoAlaX dimer–serine–Zn2+ complex. The zinc-binding motif, l-serine, and the residues involved in dimer interactions are shown as stick models. The secondary structures are designated as indicated. The loop between α3 and β4 is colored in green. The noncrystallographic symmetry twofold axis is perpendicular to the paper.
The dimer has a noncrystallographic twofold symmetry and is mainly stabilized by hydrophobic interactions of Tyr-31, Leu-93, Phe-94, and Leu-143 side chains from each subunit and hydrogen bonds between the Tyr-31 and Lys-141 side chains and the Leu-93′ main-chain carbonyl (the prime sign indicates the second subunit) and between the Trp-29 and Ser-140′ side chains (Fig. 3). Most of these residues are located at the loop region and are not conserved.
Discrimination of Serine from Alanine. In the serine–Zn2+ complex structure, three obvious peaks were seen around the editing site in the Fo–Fc map calculated after the refinement of the protein structure (Fig. 4A). Two of them were clearly defined as zinc ion and W1, by comparison with the structures without serine. Consequently, the remaining peak, which appears only when the crystal is soaked in serine with zinc, was defined as the serine molecule. The shape of the side chain and carboxyl group could be recognized unambiguously, and, thus, the serine molecule was modeled as shown in Fig. 4A. The electron density at the amino group was a little poor, even at the final structure, possibly because of the loose recognition of the main-chain moiety by PhoAlaX, as described below.
l-serine binds deeply within the center of the editing site by orienting its main-chain moiety toward the zinc motif, whereas the side-chain hydroxyl is directed to the opposite hydrophilic pocket (Fig. 4 A and B). No obvious conformational change was induced upon serine binding, including zinc coordination. The side-chain hydroxyl of the l-serine is recognized by hydrogen bonds with Thr-30 hydroxyl, Asp-92 carboxyl, and W1, with distances of 2.8, 3.0, and 3.1 Å, respectively. Remarkably, Thr-30 hydroxyl, which is highly conserved among prokaryotic AlaXs (Fig. 5), is located at the “entrance” of the hydrophilic pocket and, thus, is located near the β-methylene of the serine, with a distance of 3.7 Å, approximately an atomic radius. Therefore, the chemical repulsion of the β-carbon by Thr-30 hydroxyl is envisaged. To test this idea, we mutated Thr-30 to Val, which replaces the hydroxyl group by the methyl group with minimal change in size. Consequently, the T30V mutant degraded cognate Ala-tRNAAla with higher efficiency than wild-type PhoAlaX, without affecting its activity against Ser-tRNAAla (Fig. 2 A and B). This strongly supported the notion that serine and alanine are chemically, not sterically, discriminated by the Thr-30 hydroxyl; an aliphatic β-methyl of alanine would not be accommodated with the Thr-30 hydroxyl and would be rejected from the editing site by chemical repulsion, whereas a serine hydroxyl would be held strongly by the dense hydrogen-bond network with the hydrophilic pocket that compensates the unfavorable interaction between the Thr-30 hydroxyl and the β-methylene. The previous observation that yeast AlaX had no deacylation activity (4) can be explained partly by the divergence of the residue corresponding to Thr-30 (Fig. 5).
Fig. 5.
Multiple alignment of AlaXs and AlaRSs. Completely conserved residues among AlaXs are boxed in orange, and >75%-identical residues are in yellow. *, the conserved zinc-binding motif; #, the conserved Thr 30/Gln-584. The secondary structures of PhoAlaX are indicated at the tops of the columns. Each protein was aligned by clustalw (33) and was manually refined. The species aligned are as follows: Pho-, P. horikoshii; Mba-, M. barkeri; Sso-, S. solfataricus; Bce-, Bacillus cereus; Rso-, Ralstonia solanacearum; Hsa-, Homo sapiens; Sce-, Saccharomyces cerevisiae; Eco-, E. coli; Cel-, Caenorhabditis elegans.
Recognition of the Main-Chain Moiety of l-Serine. The main chain moiety of l-serine is not strongly bound to the enzyme but loosely recognized by hydrophilic residues Asn-114, Thr-33, Cys-116, and His-13 in the zinc-binding motif, with distances of ≈3.6 Å (Fig. 4B). Although no obvious hydrogen bond with the carboxyl or the amino group within ≈3.2 Å is observed, these residues would weakly contribute to the recognition of the main-chain moiety by surrounding the serine molecule at near-interacting distances. Such loose recognition of the main-chain moiety would be related to the defection of Gly-tRNAAla deacylation in PhoAlaX.
A clear difference between ThrRS-N2 and PhoAlaX is the coexistence of serine and zinc, which is not consistent with the proposed catalytic mechanism of ThrRS-N2 (6), because His-73 in the structure of ThrRS-N2–SerA76, which corresponds to His-9 in the zinc-binding motif of PhoAlaX, binds the catalytic water molecule but not zinc. However, we have confirmed that (i) the presence of the zinc ion is necessary for the introduction of l-serine into PhoAlaX crystals (no electron density of the serine was observed without zinc), and (ii) the fraction containing the zinc-complexed enzyme (PhoAlaXM) has a substantial deacylation activity (Fig. 2A). In fact, the structure shows that the serine is bound more deeply to the editing site than in the ThrRS-N2 (Fig. 4C) and, thus, is distant from the position of the nucleophilic water in ThrRS-N2. Moreover, in the structure of apo-PhoAlaX, His-9, which corresponds to the catalytic His-73 in ThrRS-N2, is flipped away from the serine carboxyl for ≈8 Å (Fig. 4C). All these observations imply that the zinc-binding motif is a little too far from serine to participate in the catalysis and that the zinc-binding does not interfere with the binding/deacylation of Ser-tRNAAla. Although the role of zinc is not clear, it is reasonable to say that the presence of zinc does (at least) not inhibit the deacylation activity of PhoAlaX.
Discussion
Discrimination in the AlaRS Cis-Editing Site. Because AlaX is a close structural and functional homologue of the AlaRS editing domain, the mechanism of AlaRS cis-editing, which remains unclear, could be deduced from comparison of the two proteins, although PhoAlaX has no obvious activity against glycine. In AlaRS, the residue corresponding to Thr-30 in PhoAlaX is completely conserved as Gln-584 (residue numbering refers to E. coli AlaRS; Fig. 5). It was reported that the Q584H/C666A double mutation enhances the mischarging phenotype of the C666A mutant (C666 corresponds to C116 in the zinc-binding motif of PhoAlaX), but the single Q584H mutation had no marked effect on the editing activity (9). However, the authors did not precisely describe the Q584H mutant, and, thus, we designed the PhoAlaRS-Q633M mutant that corresponds to the mutation in E. coli AlaRS Gln-584. Mutation of a Gln to a Met is paralleled by the T30V mutation in PhoAlaX, which reduces its hydrophilic nature with a minimal change in size. Notwithstanding the change, the deacylation activity of the Q633M mutant against Ser-tRNAAla is comparable to that of the wild-type enzyme (Fig. 2A). However, the Q633M mutant efficiently deacylated cognate Ala-tRNAAla (Fig. 2B). Therefore, the phenotype of the Q633M mutation closely resembles that of the PhoAlaX-T30V mutant. Such “misediting” has never been observed in wild-type AlaRS, even with an increase in the concentration of the enzyme up to 5 μM (data not shown). These results clearly demonstrate that the chemical discrimination model in AlaX can be extended to AlaRS editing; cognate alanine is discriminated from serine by the chemical repulsion of aliphatic β-carbon by the hydrophilic side chain of Gln-584. In this situation, glycine could also be accommodated in the editing site, because it has no side chain that conflicts with Gln-584, although glycine should be tightly held by interactions of main-chain moiety, which are lacking in our AlaX structure. Similar inclusion of smaller amino acids has been shown in a class I valyl-tRNA synthetase editing site (20), showing that smaller noncognate α-aminobutyrate or cysteine could sterically be accommodated within the editing site, whereas a larger cognate, valine, is chemically excluded. Furthermore, we observed that the Q584M mutation slightly impairs the deacylation of Gly-tRNAAla (data not shown), suggesting that the Gln-584 not only prevents the entry of an alanine side chain but also could interact with an α-hydrogen of glycine by so-called weak hydrogen bonding. Similar discrimination of glycine from alanine has been reported in a glycyl-tRNA synthetase activation site (21), showing that the conserved Glu-359 not only occupies the space for any larger side chains to bind but also interacts with an α-hydrogen of cognate glycine.
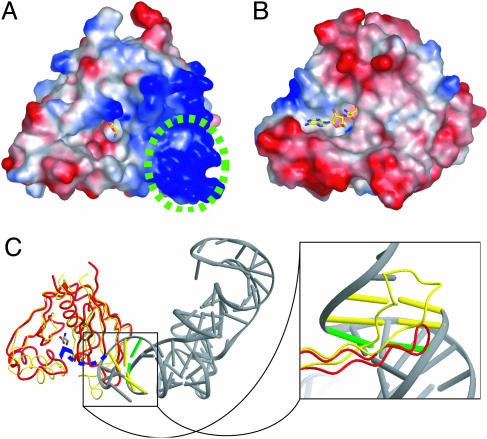
Recognition of RNA Moiety. In the ThrRS–tRNA complex structure (8), the editing domain interacts with the tRNA acceptor stem from the minor-groove side and specifically recognizes the first and second base pairs of the tRNA by a hairpin motif between Thr-201 and Met-214 (corresponding to the region between β6 and β7 in PhoAlaX). It is thought that the tRNA CCA terminus could be translocated to the editing site without breaking any contacts between the acceptor stem and the editing domain (6). We found that (i) the hairpin motif in PhoAlaX is extensively positively charged (Fig. 6A), generally accommodating the negative charge of the RNA backbone, (ii) the basic residues in this positively charged surface, Lys-134, Lys-139, and Lys-141, are highly conserved (Fig. 5), and (iii) a concave region similar to the adenosine-binding surface of ThrRS-N2 was also discernible in the PhoAlaX structure (Fig. 6 A and B). These findings suggested that AlaX would bind a tRNA in a similar fashion to ThrRS-N2 and allowed us to model tRNA to PhoAlaX by superimposition of the ThrRS-tRNA structure (Fig. 6C). A tRNA molecule was compatible with the surface of the PhoAlaX monomer without any severe steric clashes, although, in the dimer form, the loop between α3 and β4 from the adjacent subunit seems to interfere with the coming aminoacyl-CCA end of the substrate (compare Figs. 3 and 6C), partly explaining the poor activity of PhoAlaXD. Because the hairpin motif in AlaX is shorter than that of ThrRS-N2, it was located around the third base pair of tRNA from the minor-groove side. This is quite intriguing, because the major identity determinant of tRNAAla has been shown to be the third wobble base pair G3·U70, in particular the 2′-amino group of G3 that is located at the minor-groove side in ordinary base pairing (22). This model is also consistent with the previous report that the tRNA-specificity of previously identified trans-editing AlaXs was significantly affected by the sequence of the acceptor stem (4).
Fig. 6.
Recognition of tRNA moiety by AlaX. Shown is a surface potential representation of PhoAlaX (A) and ThrRS-N2 (8) (B), where acidic and basic potentials are represented in red and blue, respectively. The serine and the SerA76 are shown as stick models. The region corresponding to the hairpin motif is circled with a green dashed line. (C) Superposition of PhoAlaX (red tube) on ThrRS-N2–tRNA (yellow tube). (Left) The overview of the superposed image. The third base pair is shown as a green bar. (Right) An enlarged view of the hairpin motif interacting with tRNA.
However, the recognition of G3·U70 by an N-terminal active fragment of AlaRS, not by the editing domain, has been well established (23–26). Indeed, the region in AlaRS corresponding to the hairpin motif has a characteristic conserved insertion of ≈20 amino acids harboring invariant Arg-702 and Gly-708 (Fig. 5), suggesting that this region has a different structure. Therefore, the mode of recognition of tRNAAla by AlaX and AlaRS would be divergent.
Biological Implications. The important subject remaining to be studied is the in vivo function of AlaXs. The cis-editing domain of AlaRS is universally conserved, with only one exception observed in Nanoarchaeum equitans, which has a separated editing domain, shown to be necessary for the activity of AlaRS (27), and, thus, the editing activity is apparently duplicated in most organisms that carry the AlaX gene (or genes). It is likely that the AlaRS cis-editing activity is more efficient than that of trans-acting AlaXs. In fact, PhoAlaX and previously characterized trans-editing AlaXs (4) slightly deacylated cognate Ala-tRNAAla when the concentration of the enzyme and temperature were increased (data not shown). Moreover, it has been shown that charged tRNAs are channeled from aaRSs to ribosomes without releasing free charged tRNAs (28, 29). Therefore, the editing should collaborate with aminoacylation, and it is likely that duplication of the editing activities by the free-standing domains is not necessary for the ordinary translation pathway. However, at least for PhoAlaX, it is likely that the gene is expressed differentially under certain conditions, as shown by DNA microarray analysis of Pyrococcus furiosus (30), which showed that the close homologue of PhoAlaX, PF0428 (90% strong similarity), was expressed and regulated under certain conditions. It is possible that the deacylation activities of AlaXs are involved in the regulatory processes of unknown functional RNAs by analogy with the regulation of the phosphorylation/dephosphorylation of proteins, as proposed recently by Geslain and Ribas de Pouplana (31) Unlike the cis-editing of AlaRS, which requires the overall L-shaped structure of the tRNA molecule for efficient deacylation (32), the tRNA-docking model shown in Fig. 6C suggests that an acceptor-stem moiety would be sufficient for AlaX deacylation. This suggests that AlaX deacylates an aminoacylated RNA rather than that a mischarged tRNAAla mimics the structure of the acceptor stem.
Acknowledgments
We thank Dr. O. Nureki (Tokyo Institute of Technology) for advice on the preparation of tRNA, Dr. T. Tamura (National Institute of Advanced Industrial Science and Technology) for advice on the expression of the His-tagged protein, and the staff of SPring-8 BL38B1 and BL44B2 and the Photon Factory BL6A beamlines for help during the x-ray diffraction experiments. This work was supported by a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Human Frontier Science Program.
Author contributions: M.S., M.Y., T.N., and I.T. designed research; M.S., A.O., and M.Y. performed research; M.S., A.O., and M.Y. analyzed data; and M.S. and I.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: aaRS, aminoacyl-tRNA synthetase; AlaRS, alanyl-tRNA synthetase; PhoAlaRS, Pyrococcus horikoshii AlaRS; PhoAlaX, P. horikoshii AlaX; PhoAlaXD, PhoAlaX dimer; PhoAlaXM, PhoAlaX monomer; SerA76, seryl-3′-aminoadenosine; ThrRS, threonyl-tRNA synthetase; ThrRS-N2, threonyl-tRNA synthetase editing domain; W1, water molecule no. 1.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1V7O, 1WXO, and 1WNU).
References
- 1.Carter, C. W., Jr. (1993) Annu. Rev. Biochem. 62, 715–748. [DOI] [PubMed] [Google Scholar]
- 2.Sankaranarayanan, R. & Moras, D. (2001) Acta Biochim. Pol. 48, 323–335. [PubMed] [Google Scholar]
- 3.Schimmel, P. & Ribas de Pouplana, L. (2000) Trends Biochem. Sci. 25, 207–209. [DOI] [PubMed] [Google Scholar]
- 4.Ahel, I., Korencic, D., Ibba, M. & Söll, D. (2003) Proc. Natl. Acad. Sci. USA 100, 15422–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korencic, D., Ahel, I., Schelert, J., Sacher, M., Ruan, B., Stathopoulos, C., Blum, P., Ibba, M. & Söll, D. (2004) Proc. Natl. Acad. Sci. USA 101, 10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dock-Bregeon, A., Rees, B., Torres-Larios, A., Bey, G., Caillet, J. & Moras, D. (2004) Mol. Cell 16, 375–386. [DOI] [PubMed] [Google Scholar]
- 7.Dock-Bregeon, A., Sankaranarayanan, R., Romby, P., Caillet, J., Springer, M., Rees, B., Francklyn, C. S., Ehresmann, C. & Moras, D. (2000) Cell 103, 877–884. [DOI] [PubMed] [Google Scholar]
- 8.Sankaranarayanan, R., Dock-Bregeon, A. C., Romby, P., Caillet, J., Springer, M., Rees, B., Ehresmann, C., Ehresmann, B. & Moras, D. (1999) Cell 97, 371–381. [DOI] [PubMed] [Google Scholar]
- 9.Beebe, K., Ribas de Pouplana, L. & Schimmel, P. (2003) EMBO J. 22, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsui, W. C. & Fersht, A. R. (1981) Nucleic Acids Res. 9, 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swairjo, M. A. & Schimmel, P. (2005) Proc. Natl. Acad. Sci. USA 102, 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowsky, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 13.Terwilliger, T. C. (1994) Acta Crystallogr. D 50, 17–23. [DOI] [PubMed] [Google Scholar]
- 14.De La Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 15.Abrahams, J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52, 30–42. [DOI] [PubMed] [Google Scholar]
- 16.Cowtan, K. D. & Main, P. (1996) Acta Crystallogr. D 52, 43–48. [DOI] [PubMed] [Google Scholar]
- 17.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 19.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 20.Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D. G. & Yokoyama, S. (2000) Cell 103, 793–803. [DOI] [PubMed] [Google Scholar]
- 21.Arnez, J. G., Dock-Bregeon, A. C. & Moras, D. (1999) J. Mol. Biol. 286, 1449–1459. [DOI] [PubMed] [Google Scholar]
- 22.Musier-Forsyth, K., Usman, N., Scaringe, S., Doudna, J., Green, R. & Schimmel, P. (1991) Science 253, 784–786. [DOI] [PubMed] [Google Scholar]
- 23.Regan, L., Bowie, J. & Schimmel, P. (1987) Science 235, 1651–1653. [DOI] [PubMed] [Google Scholar]
- 24.Francklyn, C. & Schimmel, P. (1989) Nature 337, 478–481. [DOI] [PubMed] [Google Scholar]
- 25.Buechter, D. D. & Schimmel, P. (1995) Biochemistry 34, 6014–6019. [DOI] [PubMed] [Google Scholar]
- 26.Lovato, M. A., Swairjo, M. A. & Schimmel, P. (2004) Mol. Cell 13, 843–851. [DOI] [PubMed] [Google Scholar]
- 27.Waters, E., Hohn, M. J., Ahel, I., Graham, D. E., Adams, M. D., Barnstead, M., Beeson, K. Y., Bibbs, L., Bolanos, R., Keller, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 12984–12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negrutskii, B. S. & Deutscher, M. P. (1991) Proc. Natl. Acad. Sci. USA 88, 4991–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrushenko, Z. M., Budkevich, T. V., Shalak, V. F., Negrutskii, B. S. & El'skaya, A. V. (2002) Eur. J. Biochem. 269, 4811–4818. [DOI] [PubMed] [Google Scholar]
- 30.Schut, G. J., Brehm, S. D., Datta, S. & Adams, M. W. (2003) J. Bacteriol. 185, 3935–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geslain, R. & Ribas de Pouplana, L. (2004) Trends Genet. 20, 604–610. [DOI] [PubMed] [Google Scholar]
- 32.Beebe, K., Merriman, E. & Schimmel, P. (2003) J. Biol. Chem. 278, 45056–45061. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]