Abstract
In humans, type I interferon (IFN) is a family of 17 cytokines, among which the α subtypes and the β subtype are differentially expressed. It has been suggested that IFN-β activates a specific signaling cascade in addition to those activated by all type I IFNs. Nevertheless, no true biological relevance for a differential activity of α and β IFN subtypes has been identified so far. Because type I IFNs are critical for the regulation of osteoclastogenesis in mice, we have compared the effect of IFN-α2 and IFN-β on the differentiation of human monocytes into osteoclasts. Primary monocytes undergoing osteoclastic differentiation are highly and equally sensitive to both α2 and β IFNs as determined by measuring the induction levels of several IFN-stimulated genes. However, IFN-β was 100-fold more potent than the α2 subtype at inhibiting osteoclastogenesis. Expression profiling of the genes differentially regulated by IFN-α2 and IFN-β in this cellular system revealed the chemokine CXCL11 as the only IFN-induced gene differentially up-regulated by IFN-β. We show that recombinant CXCL11 by itself inhibits osteoclastic differentiation. These results indicate that autocrine-acting CXCL11 mediates, at least in part, the regulations of osteoclastogenesis by type I IFNs.
Keywords: cytokine, osteoclast
The type I interferons (IFNs) belong to the large family of cytokines that sense pathogens and orchestrate an integrated immune response. They are synthesized by almost all cells on viral infection or by specialized cells on stimulation of several Toll-like receptors. Originally described for their direct antiviral activities, the type I IFNs are now recognized as major elements of the immune response, mainly for their profound effect on differentiation of the myeloid and lymphoid tissues (1, 2).
In all eutherian mammals, the type I IFN family shows a high level of complexity. In humans, there are 13 α, 1 β, 1 ω, 1 κ, and 1 τ subtypes (3). All type I IFNs act through a single housekeeping cell surface receptor composed of the Ifnar1 and Ifnar2 subunits and two associated cytoplasmic tyrosine kinases, Tyk2 and Jak1 (4). The activation of the receptor is followed by the phosphorylation of Stat1 and Stat2, which will associate with the IFN regulatory factor 9 to form the IFN-stimulated transcriptional factor 3 (ISGF3). ISGF3 binds to a large number of promoters of IFN-stimulated genes (ISGs) to launch a first transcription wave. Depending of the responding cell type, several other signaling effectors can be activated, including Stat3, Stat4, Stat5, and Stat6 (5).
The nature of the selective pressure for the existence of type I IFN as a multigene family during evolution is largely unknown. It may be a simple means to increase the concentration of circulating IFN or, perhaps, it may provide the necessary flexibility to control biological activities as diverse as nonspecific antiviral effect or subtle regulations of cellular differentiation. Interestingly, all mammalian orders possess at least one IFN-α and one IFN-β gene (6, 7). The IFN-β forms a distinct complex with the IFN receptor as compared with the IFN-αs, suggesting a differential recruitment of downstream signaling components (8–10). Furthermore, a fundamental hallmark distinguishing the IFN-β and -α is their different promoter structure, giving rise to differential expression patterns, a prerequisite for the biological relevance of potential differential activities (11, 12). In human monocyte-derived dendritic cells, IFN-β is the only type I IFN subtype induced on stimulation of Toll-like receptor 3 or 4 with dsRNA and LPS, respectively, whereas influenza A or Sendai virus infection induces all type I IFN subtypes (13).
Another physiological context where the IFN-β is specifically expressed is during the regulation of osteoclastogenesis in mice. Bone mass regulation depends on the balance between bone formation and bone resorption. Bone resorption is primarily the activity of osteoclasts, which are multinucleated giant cells derived from the monocyte/macrophage cell lineage in response to receptor activator of NF-κB ligand (RANKL) (14). Takayanagi and colleagues (15, 16) have shown that the expression of IFN-β in osteoclast is induced by c-Fos, the central effector of osteoclast differentiation. IFN-β in turn inhibits osteoclastogenesis by inhibiting the activity of c-Fos itself. The biological consequence of this negative feedback loop is important because mice lacking Ifnar1, which are resistant to type I IFNs, develop osteopenia due to an enhanced osteoclastogenesis (16). Such a mechanism constitutes a unique IFN-β activity exerted in the absence of pathogen aggression.
During the course of studies aimed at characterizing functional differences between the IFNs α and β, we have compared the effect of human IFNs α2 and β on the in vitro differentiation of CD14-selected monocytes into osteoclasts. IFN-α2 and the IFN-β were chosen because they exhibit comparable antiviral specific activities in conventional cell line-based IFN assay systems. Moreover, these two type I IFNs are currently used in clinic for the treatment of several diseases, including viral hepatitis (IFN-α2) or multiple sclerosis (IFN-β) (17, 18). We show here that the IFN-β is 100-fold more potent than the α2 subtype at inhibiting the differentiation of monocytes into osteoclasts. We performed a microarray analysis to compare the profile of genes differentially regulated by IFN-α2 and IFN-β in this primary cell system. The analysis revealed that the chemokine CXCL11, also called I-TAC, is the only ISG differentially up-regulated by IFN-β as compared with IFN-α2. We further showed that recombinant CXCL11 is sufficient to inhibit osteoclast differentiation. Based on these results, we propose that the potent inhibition of osteoclastogenesis by IFN-β is mediated, at least in part, through the chemokine CXCL11.
Methods
Cytokines and Chemokines. Human IFN-α2c was from Gunter Adolf (Ernst Boehringer Institute, Vienna). Human IFN-β was from Laura Runkel (Biogen). Both were purified to specific activities >108 international units/mg of protein. Human soluble RANKL (sRANKL) and macrophage colony-stimulating factor (M-CSF) were from PeproTech EC (London), and human CXCL9, CXCL10, and CXCL11 were from R & D Systems.
Cell Culture and Osteoclastic Differentiation. Peripheral blood samples were obtained from healthy volunteers through the Etablissement Français du Sang (Montpellier, France). Monocytes were purified by positive sorting using anti-CD14-conjugated microbeads (Miltenyi Biotec, Paris). Osteoclasts were generated by culturing monocytes at 100,000 cells per cm2 in α-MEM medium (Invitrogen) supplemented with 10% FCS, 100 μg/ml penicillin, 100 μg/ml streptomycin, 25 ng/ml sRANKL, and 25 ng/ml M-CSF, as described by Karsdal et al. (19). Medium was replaced every 2 days, until we observed a clear formation of giant cells (6–9 days, depending on the blood donor). Cells were then fixed in 3.7% paraformaldehyde, permeabilized in ethanol/acetone, and stained for tartrate-resistant acid phosphatase (TRAP) as described in ref. 19, and nuclei with Hoechst stain solution (Sigma). The number of TRAP+ multinucleated cells (MNCs) with at least three nuclei was counted under a Zeiss Axiovert 200M fluorescence microscope with A-plan ×10 lens.
Gene Array Study. RNAs were extracted by using the High Pure RNA Isolation kit (Roche Diagnostics). RNA integrity was assessed by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). cRNAs were prepared according to One-Cycle Target Labeling protocol (Affymetrix, Santa Clara, CA) starting from 5 μg of total RNA and hybridized to HG-U133 plus 2.0 GeneChip oligonucleotide arrays (Affymetrix). HG-U133 plus 2.0 array contains 54,675 sets of oligonucleotide probes that correspond to ≈39,000 unique human genes. GeneChip Operating Software, Version 1.1 (Affymetrix), was used for the primary image analysis of the arrays, for the normalization (global scaling method, target value of 100), and for the comparison between IFN-α2- and IFN-β-treated samples.
Reverse Transcription and PCR. DNase I-treated total RNAs were extracted by using the High Pure RNA Isolation kit (Roche Diagnostics). Reverse transcriptions were primed with 10-mer random primer and performed by using SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR were performed with a Light Cycler (Roche Diagnostics) using the Platinum Taq DNA polymerase (Invitrogen) and SYBR Green I (BioWittaker) as described in ref. 13. Sequences of the primer pairs used for the quantification of GAPDH and the 6-16, MxA, 69-kDa 2′–5′ oligoadenylate synthetase (25A69) were published in ref. 20. The other primer pairs used were as follows: PKR forward, 5′-TCTACGCTTTGGGGCTAA-3′; PKR reverse, 5′-GCCATCCCGTAGGTCTGT-3′; ISG20 forward, 5′-GAGCAGTGGCAGCAGAGAGG-3′; ISG20 reverse, 5′-GGCCGGATGAACTTGTCGTA-3′; GBP5 forward, 5′-GGTTGGCGGCGATTCAAAG-3′; GBP5 reverse, 5′-ACAGTCCTCTGGGCGTGCTG-3′; CXCL11 forward, 5′-CGATGCCTAAATCCC-3′; CXCL11 reverse, 5′-CACAAAACCATAGAAAAGTC-3′; CD69 forward, 5′-TTCTCAATGCCATCAGACAG-3′; CD69 reverse, 5′-CCTCTCTACCTGCGTATCGT-3′; MGC22805 forward, 5′-GCCTGTGAAATGAAAAACCA-3′; MGC22805 reverse, 5′-CCGTGCAATATCCAGTGAG-3′; KIAA0040 forward, 5′-CCAGCCCCAGCCCTTTATTC-3′; KIAA0040 reverse, 5′-TGTCCCCCGTGAACTTACCC-3′; SOCS1 forward, 5′-AACTGCTTTTTCGCCCTTA-3′; SOCS1 reverse, 5′-GCCACGTAGTGCTCCA-3′; PCD1L1 forward, 5′-ATGTGGCATCCAAGATACAA-3′; PCD1L1 reverse, 5′-GCCAGGTTCCATTTTCAGT-3′; GBP4 forward, 5′-CCCCAGACCTGATGAAGC-3′; GBP4 reverse, 5′-GTAGGCCGGTCAAAGACAAA-3′; CCR1 forward, 5′-GATGACTGGGTTTTTGGTGA-3′; CCR1 reverse, 5′-AATGATGATGCTGGTGATGA-3′; CCR5 forward, 5′-TTCTCTTCTGGGCTCCCTAC-3′; CCR5 reverse, 5′-CCCGACAAAGGCATAGATG-3′; CCRL2 forward, 5′-AGCTGGTGCCATCACTCTG-3′; CCRL2 reverse, 5′-ACTGTACAGGCCCACGAAGT-3′; CCL3 forward, 5′-CACCTCCCGGCAGATTCC-3′; CCL3 reverse, 5′-CCTCACTGGGGTCAGCACAG-3′; and CXCR3 forward, 5′-TTGACCGCTACCTGAACATA-3′; CXCR3 reverse, 5′-GGGAAGTTGTATTGGCAGTG-3′. The specificities of the primer pairs were validated by DNA sequencing of the PCR products. All data are expressed as a ratio to the GAPDH level. The standard errors of the ratios were calculated using Student's t test. The 95% confidence limits are always <0.2 log10. Statistical significances of comparisons of expression levels of a given gene among cells from different blood donors were calculated by using a two-tailed nonparametric Mann–Whitney test performed with instat 3.0 (GraphPad, San Diego). The identification of monocytes carrying the Δ32 mutation in the CCR5 gene was done with the PCR condition and primer pair described by Eri et al. (21).
Results
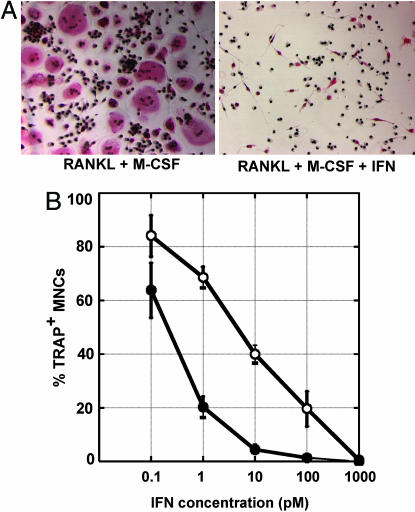
IFN-α2 and -β Differentially Inhibit Osteoclast Differentiation of CD14+ Monocytes. Monocytes from human peripheral blood were isolated by CD14+ magnetic cell sorting. Osteoclast differentiation was induced by cultivating monocytes in the presence of sRANKL and M-CSF. After 6–9 days, monocytes fused into giant MNCs expressing TRAP, a characteristic marker of osteoclast differentiation (19). The positive selection method of CD14+ monocytes transiently affected the expression of several cell surface markers, notably class I and II MHC markers, which returned to their initial levels after 2 days of culture in the presence of M-CSF and sRANKL (data not shown). To compare the capacity of the IFN-α2 and -β subtypes to inhibit osteoclastic differentiation, CD14+ cells were cultured in the presence of sRANKL and M-CSF for 2 days, and then the IFNs were added at different concentrations. After 4–6 additional days in culture, cells were fixed and stained for TRAP and nuclei, and the number of large multinucleated TRAP+ cells was counted. As reported by several groups studying osteoclast precursors isolated from murine bone marrow (16, 22, 23), type I IFNs strongly inhibited the osteoclastic differentiation process of human monocytes (Fig. 1A). However, IFN-β was 100-fold more potent than the α2 subtype at inhibiting the differentiation of monocytes into osteoclasts (Fig. 1B).
Fig. 1.
IFN-α2 and IFN-β exhibit a 100-fold difference in their specific activities toward the inhibition of the differentiation of monocytes in osteoclasts. Freshly purified monocytes from human blood donors were cultured in the presence of sRANKL and M-CSF for 2 days, and with different concentrations of IFN-α2 or IFN-β for an additional 4–6 days. Cells were then fixed and stained for TRAP and nuclei. (A) Photomicrographs of IFN-free culture (Left) or culture with 1 nM IFN (Right). (B) Quantification of the number of TRAP+ MNCs in IFN-α2-treated culture (○) or IFN-β treated culture (•). Each point represents the mean osteoclast number relative to IFN-free cultures ± SEM from 5 to 29 blood donors. The osteoclastic differentiation is inhibited by 30% with 1 pM IFN-α2, 80% with 1 pM IFN-β, 60% with 10 pM IFN-α2, and 95% with 10 pM IFN-β.
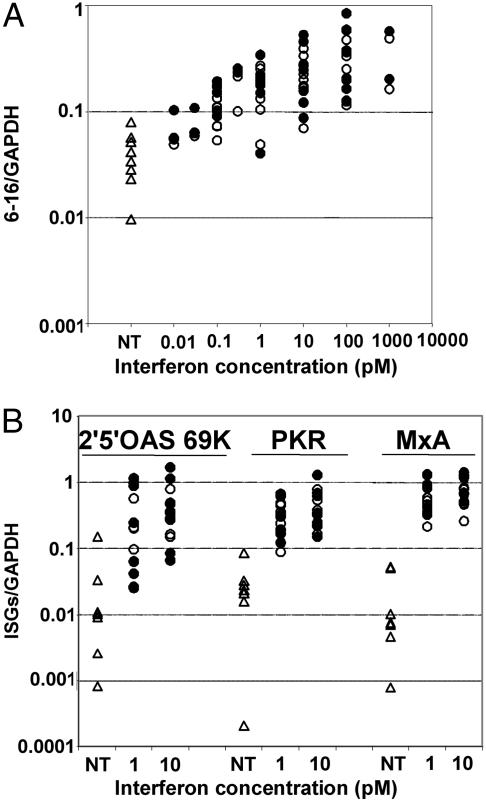
To determine whether the differential effect of α2 and β IFNs was specific for the inhibition of osteoclastogenesis or a general characteristic of sRANKL/M-CSF-treated monocytes, the activities of IFN-α2 and -β were compared for the induction of several well known ISGs. Monocytes cultured in the presence of sRANKL and M-CSF for 2 days were treated with IFN-α2 or -β for 4 h, and the expression of some classical ISGs was quantified by quantitative RT-PCR. These cells responded to both α2 and β IFNs in the 10 fM to 1 pM range, and IFN-α2 and -β did not show any differential response for the induction of the 6-16 (Fig. 2A), 2′–5′ oligoadenylate synthetase, PKR, or MxA genes (Fig. 2B).
Fig. 2.
IFN-α2 and IFN-β exhibit the same specific activities for the induction of the expression of IFN-stimulated genes. Freshly purified monocytes from human blood donors were cultured in the presence of sRANKL and M-CSF for 2 days, and then left untreated (▵) or treated for 4 h with IFN-α2 (○) or IFN β (•). RNAs were then extracted and the levels of 6-16 (A) or 2′–5′ oligoadenylate synthetase, PKR, and MxA (B) transcripts were measured by quantitative RT-PCR. Each point in the vertical represents a different blood donor.
Taken together, these results establish that IFN-α2 and -β exhibit a substantial difference in their ability to inhibit the differentiation of monocytes into osteoclasts. This differential activity is not a consequence of an overall differential in the early cellular response to these two IFN subtypes.
CXCL11 Is the only Early ISG Differentially Up-Regulated by IFN-α2 and -β. We analyzed the differential expression of 30,000 genes on Affymetrix microarrays to determine whether the differential effect of α2 and β IFNs on the inhibition of osteoclastogenesis could be assigned to an early differential at the transcriptional level.
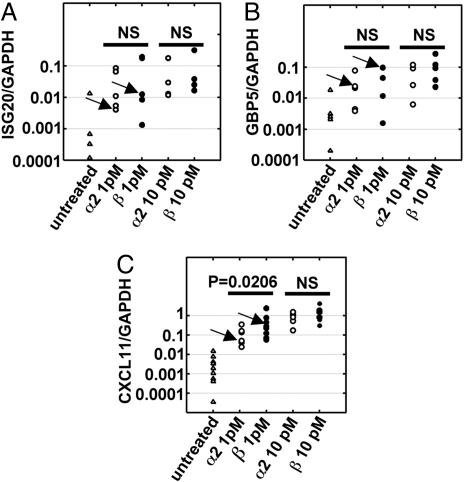
The osteoclastic differentiation was initiated for 2 days in the presence of sRANKL and M-CSF, and cells were then treated for 4 h with 1 pM of either α2 or β IFNs. At this concentration, IFN-β inhibited almost completely the differentiation, whereas IFN-α2 had a marginal effect (Fig. 1B). The gene expression profiles of IFN-α2- and -β-treated osteoclasts were compared. As expected from quantitative RT-PCR data (Fig. 2), most of the classical ISGs were equally expressed in IFN-α2 and -β treated cells, indicating that the two IFN subtypes were equally potent in initiating the ISGF3-driven response. Several genes appeared up-regulated by IFN-β by a factor of at least 3, as compared with IFN-α2 (Table 1). This differential up-regulation was confirmed by quantitative RT-PCR performed on RNA samples isolated from the blood donor used for the microarray hybridization (Table 1). The differential effect of IFN-α2 and -β on these genes was then analyzed by quantitative RT-PCR on RNA samples from several blood donors. For most genes, the differential became nonsignificant when a population of independent blood donors was analyzed (Table 1). As an example, Fig. 3 A and B show that the induction levels of ISG20 and GBP5 by IFN-α2 and -β were not significantly different among cells isolated from five different blood donors, even if the differential found on the microarray was confirmed by quantitative RT-PCR for cells isolated from the particular blood donor used in the microarray study.
Table 1. Differential transcriptional induction by IFN-α2 and -β in sRANKL/M-CSF-treated monocytes.
| Gene symbol | Name | Ratio β/α2 in Affymetrix data* | Ratio β/α2 in Q-RT-PCR data† | Significance among blood donors in Q-RT-PCR data‡ | IFN induction in Q-RT-PCR data§ |
|---|---|---|---|---|---|
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | 7.73 | 4.35 | P = 0.0206, n = 9 | Yes |
| CD69 | CD69 antigen (p60, early T cell activation antigen) | 7.06 | 4.43 | NS P = 0.4127, n = 5 | No |
| MGC22805 | Hypothetical protein MGC22805 | 5.53 | 4.14 | NS P = 0.6286, n = 4 | No |
| KIAA0040 | KIAA0040 gene product | 3.65 | 1.99 | NS P = 0.1508, n = 5 | Yes |
| ISG20 | IFN-stimulated gene of 20 kDa | 3.63 | 3.01 | NS P = 0.6905, n = 5 | Yes |
| SOCS1 | Suppressor of cytokine signaling 1 | 3.63 | 3.69 | NS P > 0.9999, n = 5 | Yes |
| GBP5 | Guanylate binding protein 5 | 3.60 | 4.09 | NS P = 0.9048, n = 5 | Yes |
| PDCD1L1 | Programmed cell death 1 ligand 1 | 3.43 | 4.24 | NS P = 0.5476, n = 5 | Yes |
| GBP4 | Guanylate binding protein 4 | 3.10 | 1.94 | NS P = 0.6905, n = 5 | Yes |
Data from hybridization of Affymetrix HG-U133 plus 2.0 GeneChip oligonucleotide arrays with cRNA from cells treated with 1 pM IFN-α2 or -β
Confirmation of the differential induction by quantitative RT-PCR (Q-RT-PCR) analysis on the RNA used for the GeneChip study
Statistical significance of the differential induction among several blood donors using cells treated with 1 pM IFN-α2 or -β. NS, nonsignificant; P, probability of the null hypothesis (Mann–Whitney test)
IFN inductibility determined by comparing gene expression levels in IFN-treated cells to untreated cells
Fig. 3.
CXCL11 is the only ISG differentially up-regulated by IFN-β compared with IFN-α2 in monocytes purified from several blood donors. Candidate genes identified as preferentially up-regulated by 1 pM IFN-β in the gene array study (Table 1) were analyzed by quantitative RT-PCR. Freshly purified monocytes from human blood donors were cultured in the presence of sRANKL and M-CSF for 2 days, and then left untreated (▵) or treated for 4 h with IFN-α2 (○) or IFN-β (•). RNAs were then extracted and the levels of ISG20 (A), GBP5 (B), and CXCL11 transcripts (C) were quantified by quantitative RT-PCR. Each point in the vertical represents a different blood donor. Samples used for the gene array study are indicated by arrows. Statistical significance of the difference of the expression levels induced by IFN-α2 or IFN-β have been analyzed by the Mann–Whitney test. NS, nonsignificant. The median of the ratio of CXCL11 induced by IFN-β/CXCL11 induced by IFN-α2 is 4.2 for IFNs at 1 pM and 1.5 for IFNs at 10 pM.
CXCL11 was the only gene significantly up-regulated in IFN-β-treated preosteoclasts. Quantitative RT-PCR in cells isolated from nine different blood donors revealed a significant differential induction of CXCL11 by 1 pM IFN-α2 and -β, with IFN-β being the most potent subtype (Fig. 3C). In cells treated with 10 pM IFN, activities of IFN-α2 and -β were no longer significantly different. Because the differential activity between IFN-α2 and -β on the inhibition of osteoclastogenesis was mostly apparent at low IFN concentration (Fig. 1B), this result is consistent with a correlation between the expression level of CXCL11 and the inhibition of osteoclastic differentiation by type I IFNs.
CXCL11 Inhibits Osteoclast Differentiation of CD14+ Monocytes. We have established that IFN-α2 and -β display differential potency to inhibit sRANKL/M-CSF-induced monocyte differentiation into osteoclasts, and that this phenomenon does not reflect a general differential specific activity of the two IFN subtypes. Because CXCL11 was the only gene differentially induced by IFN-α2 and -β, we sought to investigate whether CXCL11 by itself could effect osteoclastic differentiation. CXCL11 (I-TAC) is a chemokine originally characterized as an inducer of chemotactic responses in Th1 cells. It acts through the chemokine receptor CXCR3, which is also the receptor for CXCL9 (MIG) and CXCL10 (IP-10) (24).
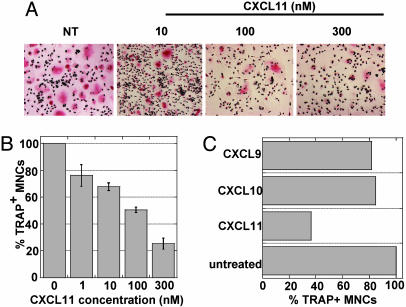
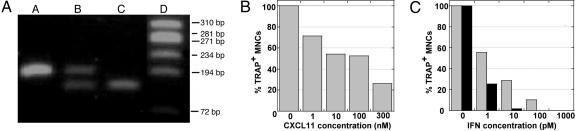
As for the IFNs, CD14+ monocytes were cultured for 2 days in the presence of sRANKL and M-CSF, and then recombinant CXCL11 was added at different concentrations. After 4–6 days, cells were fixed and stained for TRAP and nuclei, and TRAP+ MNCs were counted. CXCL11 inhibited osteoclastic differentiation in a dose-dependent manner (Fig. 4 A and B). The morphology of CXCL11-treated monocytes resembled that of IFN-treated cells (compare Fig. 1 A and Fig. 4A). With 100 nM CXCL11, 50% inhibition of osteoclastic differentiation was obtained (Fig. 4B). Interestingly, the two other CXCR3 agonists, CXCL9 and CXCL10, had no effect on the osteoclastic differentiation of monocytes (Fig. 4C).
Fig. 4.
CXCL11 but not CXCL9 or CXCL10 inhibits osteoclastic differentiation. Freshly purified monocytes from human blood donors were cultured in the presence of sRANKL and M-CSF for 2 days, and with different concentrations of the chemokine for an additional 4–6 days. Cells were then fixed and stained for TRAP and nuclei. (A) Photomicrographs of CXCL11-treated cultures. (B) Quantification of the number of TRAP+ MNCs in CXCL11-treated cultures; mean osteoclast number relative to untreated cultures ± SEM from eight blood donors. (C) Comparison of the effect of 100 nM CXCL11, CXCL9, and CXCL10 on the inhibition of osteoclastic differentiation.
CXCL11 Inhibits Osteoclast Differentiation in the Absence of CXCR3. CXCR3 is the only functional receptor of CXCL11 known so far (24). Nevertheless, according to the Affymetrix chip data, CXCR3 was not expressed in sRANKL/M-CSF-cultured CD14+ monocytes, in the presence of IFN-α2 or -β. Moreover, quantitative RT-PCR experiments using an oligonucleotide couple detecting all described alternative splice variants of CXCR3 (25), and performed on RNA samples from several blood donors, did not amplify any product above the 30th cycle (PCR efficiency close to 1) at any stages of the osteoclastic differentiation process (data not shown). This result suggests that CXCL11 effects are mediated through a receptor other than CXCR3 to inhibit osteoclastogenesis.
Extensive expression profiles of chemokines and chemokine receptors were established from the hybridization of the Affymetrix chip with cRNAs from IFN-α2 or -β treated preosteoclasts (Table 2). CCR1, CCR5, and CCRL2 were the only chemokine receptors expressed in this cellular system. The expression of these three receptors was confirmed by quantitative RT-PCR (data not shown). CCRL2 is still an orphan chemokine receptor whose function is unknown. In this cellular system, four potential autocrine-acting ligands were described for CCR1 and CCR5: CCL3 (MIP-1α), CCL4 (MIP-1β), CCL7 (MCP-3), and CCL8 (MCP-2).
Table 2. Unambiguously expressed chemokines and chemokine receptors in IFN-treated preosteclasts.
| Expressed chemokine | Expressed chemokine receptor |
|---|---|
| CCL2 (MCP-1) | CCR1 |
| CCL3*(MIP-1 α) | CCR5 |
| CCL4* (MIP-1 β) | CCRL2 |
| CCL7* (MCP-3) | |
| CCL8* (MCP-2) | |
| CXCL1 (GRO-α) | |
| CXCL2 (GRO-β) | |
| CXCL3 (GRO-γ) | |
| CXCL5 (ENA-78) | |
| CXCL6 (GCP-2) | |
| CXCL7 (NAP-2) | |
| CXCL8 (IL-8) | |
| CXCL9 (MIG) | |
| CXCL10 (IP-10) | |
| CXCL11 (I-TAC) | |
| CXCL16 | |
| CKLF | |
| CKLFSF6 | |
| CKLFSF7 |
Data are from Affymetrix HG-U133 plus 2.0 hybridization. Boldface type, expression confirmed by quantitative PCR analysis.
Chemokine known to interact with CCR1 or CCR5
Interestingly, CXCL11 has been shown to be a natural antagonist for CCR5 in CD14+ human monocytes (26). We thus tested the hypothesis that CXCL11 could inhibit osteoclast differentiation through an antagonistic effect on autocrine-acting CCR5 ligands. The Δ32 mutation in the CCR5 gene is a relatively frequent loss-of-function mutation in the human population. In homozygous individuals, the truncated CCR5 is not processed to the cell surface and accumulates in internal membrane components (27). A blood donor homozygous for the Δ32 CCR5 mutation was identified (Fig. 5A). The differentiation of CD14+ monocytes homozygous for the Δ32 CCR5 mutation was inhibited by CXCL11 in the same dose range as wild-type cells (compare Fig. 5B and Fig. 4B). Moreover, the differential effect of IFN-α2 and -β on osteoclast differentiation was still observed (compare Fig. 5C and Fig. 1B). We have checked by quantitative RT-PCR analysis that neither CXCR3 nor CCRL2 mRNAs were overexpressed to compensate CCR5 mutation (data not shown). Therefore, the absence of functional CCR5 did not affect the inhibition of osteoclast differentiation by CXCL11.
Fig. 5.
The inhibitory effect of CXCL11 on osteoclastogenesis is independent of CCR5. (A) Identification of monocytes heterozygous (lane B) or homozygous (lane C) for the CCR5 Δ32 mutation. Lanes: A, wild type; D, molecular weight marker in base pairs. (B and C) Inhibitory activity of CXCL11 (B), IFN-α2(C, gray bars) and IFN-β (C, black bars) on osteoclastic differentiation of monocytes homozygous for the CCR5 Δ32 mutation. Cellular differentiation assay and analysis were performed as for Figs. 1 and 4.
Discussion
This study describes a physiological differentiation process where two type I IFN subtypes display a 2-log10 difference in potency. The differential inhibition of primary monocyte differentiation into multinucleated osteoclasts by IFN-α2 and -β was consistently observed in cells isolated from >25 blood donors. Microarray technology and RNA quantification identified the chemokine CXCL11 as the only transcript differentially up-regulated by IFN-β.
Global gene expression analyses aimed to identify genes differentially regulated by IFN-α2 and -β have been performed in the HT1080 cell line (28), melanoma cell lines (29), and primary human umbilical vein endothelial cells (HUVEC) (30). These studies have identified a large number of genes preferentially induced by IFN-β. In contrast to HUVEC, which were found to exhibit a 2- to 3-log10 overall greater sensitivity to IFN-β (30), the differential effect described here is uniquely toward osteoclastogenesis inhibition. It does not represent a general loss in IFN-α2 sensitivity, because most of the ISGs were equally activated by IFN-α2 and -β in monocytes cultured in the presence of sRANKL and M-CSF. Using the microarray technology, we have identified several candidate genes in differentiating monocytes isolated from a single blood sample, and in general, real time RT-PCR quantifications of these transcripts have confirmed their differential expression. However, when the quantifications were performed with cells isolated from five to nine independent blood donors, only the chemokine CXCL11 was consistently up-regulated by IFN-β.
CXCL11 was originally identified as an IFN-γ-induced chemokine that stimulates a chemotactic response in activated T cells (24). It is also up-regulated preferentially by IFN-β in the HT1080 cell line (31). Besides the common type I IFN-induced ISGF3 pathway, IFN-β-specific transcriptional induction of CXCL11 also relies on the activation of NF-κB transcription factor through a phosphatidylinositol-3-kinase and Akt-dependent pathway (32).
CXCL11 per se inhibits the differentiation of monocytes into osteoclasts, suggesting that the particularly high activity of IFN-β is, in fact, mediated by an autocrine action of CXCL11. In mice, the IFN-mediated inhibition of osteoclastogenesis was shown to be dependent on ISGF3, because the differentiation of osteoclast precursors isolated from Stat1-/- or IFN regulatory factor 9-/- mice is resistant to the IFN-β effect (16). However, the links between activation of ISGs expression and inhibition of c-Fos activity that blocks differentiation are not known. It is tempting to speculate from our data that the IFN-induced CXCL11 plays a role.
The concentration range at which exogenous CXCL11 inhibits osteoclastic differentiation is high (ED50, 100 nM). Assuming that CXCL11 acts through an autocrine loop, this concentration range loses its significance because it is unlikely that chemokines act in soluble state. Rather, they are expected to act as aggregates on the glycoaminoglycan component of the extracellular matrix or the cell surface, significantly increasing the local concentration of chemokines presented to chemokine receptors (33). The receptor through which CXCL11 inhibits osteoclastogenesis is unknown. CXCL11 is a potent agonist of CXCR3 (24), which is not expressed in human monocytes undergoing osteoclastic differentiation. Interestingly, the two other CXCR3 agonists, CXCL9 and CXCL10, do not inhibit osteoclastic differentiation. It rules out the possibility that the potential CXCL11 receptor in this cellular system is shared with CXCL9 and CXCL10. CXCL11 was precisely described as a specific antagonist of CCR5 (26). The hypothesis that CXCL11 acts through an antagonist effect on CCR5 was appealing because the CCR5 agonists CCL3 and CCL4 are expressed by monocytes in our differentiation system (Table 2) and have been implicated in the severe bone destruction due to inappropriate osteoclastogenesis occurring in multiple myeloma (34). However, this hypothesis can be ruled out because cells homozygous for the CCR5 Δ32 mutation are as sensitive as wild-type cells to CXCL11 and its inducer IFN-β. Apart from CCR5, the other well characterized chemokine receptor expressed in human monocytes undergoing osteoclastic differentiation is CCR1, which was shown not to interact with CXCL11 (26). Finally, the orphan chemokine receptor CCRL2 (Table 2), which has considerable homology to CCR2, remains a candidate to mediate the CXCL11 inhibitory effect. The expression of CCRL2 is up-regulated in leukocytes that infiltrate the joints of patients with rheumatoid arthritis, a disease characterized by the destruction of cartilage and bone, in which osteoclasts have a major role (35). Furthermore, CCL2, the agonist ligand of CCR2, plays an essential function in the in vitro fusion of adherent peripheral blood mononuclear cells, a necessary step in osteoclast formation (36). CCRL2/CCR2 activities are thus likely to play a role in the osteoclastic differentiation, and a reasonable hypothesis would be that in the cellular system described here, the orphan CXCL11 antagonizes the orphan CCRL2. Unfortunately, there are no available molecular tools to investigate the CXCL11–CCRL2 relationship.
Like CXCL11 in osteoclasts, other orphan chemokines have been described. For instance, CXCL10 is active in human umbilical vein endothelial cells in the absence of CXCR3 expression (37). It is remarkable that CXCL10 is precisely differentially induced by IFN-α1 and IFN-α2 or -α21 in human T cells and dendritic cells (38), and that IFN-α2 and -α8 were shown to differentially affect T cell motility (39). These results suggest that differential activities between IFN subtypes acquire their biological relevance in the context of a cross-talk with the chemokine network to ultimately regulate “chemokine-to-cytokine-to-chemokine” cascades necessary for immune and developmental processes (40).
The differential activity between IFN-α2 and -β on the inhibition of osteoclastic differentiation is the largest differential activity described so far for primary cells that do not present an overall difference at the level of the early transcriptional response. The behavior of the other type I IFN-α subtypes tested (α8 and ω) resembles that of IFN-α2 (data not shown). Thus, in accordance with their different structure, receptor complex formation, expression regulation, and evolutionary history, IFN-α and -β form two families that can be functionally distinguished for their distinct effect on the regulation of bone mass formation.
Targeting osteoclast formation is a prioritized therapeutic approach for the treatment of malignant osteolytic pathologies such as multiple myeloma or breast carcinoma (41). Based on this work, the use of IFN-β could be clinically relevant. Given its extremely high activity in the inhibition of osteoclastogenesis, it could be used at a low dose, sufficient to exert a beneficial effect on osteolysis, but not causing the side effects generally associated with IFN therapies.
Acknowledgments
We thank Drs. John de Vos, Pierre Corbeau, and Christelle Turriere for their help and advice; Drs. Gideon Schreiber and Jacob Piehler for discussion; Dr. Sandra Pellegrini and José Van der Heyden for critical reading of the manuscript; and Danièle Monneron for her help. L.F.L.C. and G.M.d.F.A. are Ph.D. students from Federal University of Minas Gerais. This work was supported by Human Frontier Science Program Grant RGP60/2002.
Author contributions: G.U. designed research; L.F.L.C. and G.M.d.F.A. performed research; F.J.D.M., A.B., and G.U. contributed new reagents/analytic tools; L.F.L.C. and G.U. analyzed data; and G.U. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ISGF3, IFN-stimulated transcriptional factor 3; ISG, IFN-stimulated genes; RANKL, receptor activator of NF-κB ligand; sRANKL, soluble RANKL; TRAP, tartrate-resistant acid phosphatase; MNC, multinucleated cell; M-CSF, macrophage colony-stimulating factor.
References
- 1.Gresser, I. & Belardelli, F. (2002) Cytokine Growth Factor Rev. 13, 111-118. [DOI] [PubMed] [Google Scholar]
- 2.Santini, S. M., Di Pucchio, T., Lapenta, C., Parlato, S., Logozzi, M. & Belardelli, F. (2002) J. Interferon Cytokine Res. 22, 1071-1080. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J., Baig, E. & Fish, E. N. (2004) J. Interferon Cytokine Res. 24, 687-698. [DOI] [PubMed] [Google Scholar]
- 4.Langer, J. A., Cutrone, E. C. & Kotenko, S. (2004) Cytokine Growth Factor Rev. 15, 33-48. [DOI] [PubMed] [Google Scholar]
- 5.Platanias, L. C. & Fish, E. N. (1999) Exp. Hematol. (Charlottesville, Va) 27, 1583-1592. [DOI] [PubMed] [Google Scholar]
- 6.Harrison, G. A., McNicol, K. A. & Deane, E. M. (2004) Immunol. Cell Biol. 82, 112-118. [DOI] [PubMed] [Google Scholar]
- 7.Hughes, A. L. & Roberts, R. M. (2000) J. Interferon Cytokine Res. 20, 737-739. [DOI] [PubMed] [Google Scholar]
- 8.Domanski, P., Nadeau, O. W., Platanias, L. C., Fish, E., Kellum, M., Pitha, P. & Colamonici, O. R. (1998) J. Biol. Chem. 273, 3144-3147. [DOI] [PubMed] [Google Scholar]
- 9.Lewerenz, M., Mogensen, K. E. & Uze, G. (1998) J. Mol. Biol. 282, 585-599. [DOI] [PubMed] [Google Scholar]
- 10.Platanias, L. C., Uddin, S., Domanski, P. & Colamonici, O. R. (1996) J. Biol. Chem. 271, 23630-23633. [DOI] [PubMed] [Google Scholar]
- 11.Levy, D. E., Marie, I. & Prakash, A. (2003) Curr. Opin. Immunol. 15, 52-58. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald, N. J., Kuhl, D., Maguire, D., Naf, D., Gallant, P., Goswamy, A., Hug, H., Bueler, H., Chaturvedi, M., de la Fuente, J., et al. (1990) Cell 60, 767-779. [DOI] [PubMed] [Google Scholar]
- 13.Coccia, E. M., Severa, M., Giacomini, E., Monneron, D., Remoli, M. E., Julkunen, I., Cella, M., Lande, R. & Uze, G. (2004) Eur. J. Immunol. 34, 796-805. [DOI] [PubMed] [Google Scholar]
- 14.Teitelbaum, S. L. & Ross, F. P. (2003) Nat. Rev. Genet. 4, 638-649. [DOI] [PubMed] [Google Scholar]
- 15.Takayanagi, H., Kim, S., Koga, T. & Taniguchi, T. (2005) J. Cell. Biochem. 94, 232-240. [DOI] [PubMed] [Google Scholar]
- 16.Takayanagi, H., Kim, S., Matsuo, K., Suzuki, H., Suzuki, T., Sato, K., Yokochi, T., Oda, H., Nakamura, K., Ida, N., et al. (2002) Nature 416, 744-749. [DOI] [PubMed] [Google Scholar]
- 17.Billiau, A., Kieseier, B. C. & Hartung, H. P. (2004) J. Neurol. 251, Suppl. 2, 10-14. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer, L. M., Dinarello, C. A., Herberman, R. B., Williams, B. R., Borden, E. C., Bordens, R., Walter, M. R., Nagabhushan, T. L., Trotta, P. P. & Pestka, S. (1998) Cancer Res. 58, 2489-2499. [PubMed] [Google Scholar]
- 19.Karsdal, M. A., Hjorth, P., Henriksen, K., Kirkegaard, T., Nielsen, K. L., Lou, H., Delaisse, J. M. & Foged, N. T. (2003) J. Biol. Chem. 278, 44975-44987. [DOI] [PubMed] [Google Scholar]
- 20.Dondi, E., Rogge, L., Lutfalla, G., Uze, G. & Pellegrini, S. (2003) J. Immunol. 170, 749-756. [DOI] [PubMed] [Google Scholar]
- 21.Eri, R., Jonsson, J. R., Pandeya, N., Purdie, D. M., Clouston, A. D., Martin, N., Duffy, D., Powell, E. E., Fawcett, J., Florin, T. H., et al. (2004) Genes Immun. 5, 444-450. [DOI] [PubMed] [Google Scholar]
- 22.Fox, S. W., Haque, S. J., Lovibond, A. C. & Chambers, T. J. (2003) J. Immunol. 170, 3679-3687. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, T., Kaneda, T., Toyama, Y., Kumegawa, M. & Hakeda, Y. (2002) J. Biol. Chem. 277, 27880-27886. [DOI] [PubMed] [Google Scholar]
- 24.Cole, K. E., Strick, C. A., Paradis, T. J., Ogborne, K. T., Loetscher, M., Gladue, R. P., Lin, W., Boyd, J. G., Moser, B., Wood, D. E., et al. (1998) J. Exp. Med. 187, 2009-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehlert, J. E., Addison, C. A., Burdick, M. D., Kunkel, S. L. & Strieter, R. M. (2004) J. Immunol. 173, 6234-6240. [DOI] [PubMed] [Google Scholar]
- 26.Petkovic, V., Moghini, C., Paoletti, S., Uguccioni, M. & Gerber, B. (2004) J. Leukocyte Biol. 76, 701-708. [DOI] [PubMed] [Google Scholar]
- 27.Samson, M., Libert, F., Doranz, B. J., Rucker, J., Liesnard, C., Farber, C. M., Saragosti, S., Lapoumeroulie, C., Cognaux, J., Forceille, C., et al. (1996) Nature 382, 722-725. [DOI] [PubMed] [Google Scholar]
- 28.Der, S. D., Zhou, A., Williams, B. R. & Silverman, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leaman, D. W., Chawla-Sarkar, M., Jacobs, B., Vyas, K., Sun, Y., Ozdemir, A., Yi, T., Williams, B. R. & Borden, E. C. (2003) J. Interferon Cytokine Res. 23, 745-756. [DOI] [PubMed] [Google Scholar]
- 30.da Silva, A. J., Brickelmaier, M., Majeau, G. R., Lukashin, A. V., Peyman, J., Whitty, A. & Hochman, P. S. (2002) J. Interferon Cytokine Res. 22, 173-188. [DOI] [PubMed] [Google Scholar]
- 31.Rani, M. R., Foster, G. R., Leung, S., Leaman, D., Stark, G. R. & Ransohoff, R. M. (1996) J. Biol. Chem. 271, 22878-22884. [DOI] [PubMed] [Google Scholar]
- 32.Rani, M. R., Hibbert, L., Sizemore, N., Stark, G. R. & Ransohoff, R. M. (2002) J. Biol. Chem. 277, 38456-38461. [DOI] [PubMed] [Google Scholar]
- 33.Ali, S., Palmer, A. C., Banerjee, B., Fritchley, S. J. & Kirby, J. A. (2000) J. Biol. Chem. 275, 11721-11727. [DOI] [PubMed] [Google Scholar]
- 34.Abe, M., Hiura, K., Wilde, J., Moriyama, K., Hashimoto, T., Ozaki, S., Wakatsuki, S., Kosaka, M., Kido, S., Inoue, D., et al. (2002) Blood 100, 2195-2202. [PubMed] [Google Scholar]
- 35.Galligan, C. L., Matsuyama, W., Matsukawa, A., Mizuta, H., Hodge, D. R., Howard, O. M. & Yoshimura, T. (2004) Arthritis Rheum. 50, 1806-1814. [DOI] [PubMed] [Google Scholar]
- 36.Kyriakides, T. R., Foster, M. J., Keeney, G. E., Tsai, A., Giachelli, C. M., Clark-Lewis, I., Rollins, B. J. & Bornstein, P. (2004) Am. J. Pathol. 165, 2157-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soejima, K. & Rollins, B. J. (2001) J. Immunol. 167, 6576-6582. [DOI] [PubMed] [Google Scholar]
- 38.Hilkens, C. M., Schlaak, J. F. & Kerr, I. M. (2003) J. Immunol. 171, 5255-5263. [DOI] [PubMed] [Google Scholar]
- 39.Foster, G. R., Masri, S. H., David, R., Jones, M., Datta, A., Lombardi, G., Runkell, L., de Dios, C., Sizing, I., James, M. J., et al. (2004) J. Immunol. 173, 1663-1670. [DOI] [PubMed] [Google Scholar]
- 40.Salazar-Mather, T. P., Hamilton, T. A. & Biron, C. A. (2000) J. Clin. Invest. 105, 985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittrant, Y., Theoleyre, S., Chipoy, C., Padrines, M., Blanchard, F., Heymann, D. & Redini, F. (2004) Biochim. Biophys. Acta 1704, 49-57. [DOI] [PubMed] [Google Scholar]