Abstract
The current model of basal ganglia rests on the idea that the striatofugal system is composed of two separate (direct and indirect) pathways originating from distinct cell populations in the striatum. The striatum itself is divided into two major compartments, the striosomes and the matrix, which differ by their neurochemical makeup and input/output connections. Here, neurons located in either striosomes or the extrastriosomal matrix in squirrel monkeys were injected with biotin dextran amine, and their labeled axons were entirely reconstructed with a camera lucida. Twenty-four of 27 reconstructed axons arborized into the three main striatal targets (external pallidum, globus pallidus, and substantia nigra pars reticulata), a finding that is at odds with the concept of a dual striatofugal system. Axons of striosomal neurons formed several columnar terminal fields in the substantia nigra pars reticulata. These data indicate that the substantia nigra pars compacta is neither the only nor the main target of striosomal neurons, a finding that calls for a reevaluation of the organization of the striatonigral projection system.
Keywords: anatomy, basal ganglia, matrix, striatum, striosomes
The striatum is the largest and main integrative component of the basal ganglia. It is often the site of massive cell losses or major transmitter deficits that lead to severe motor disorders, such as the excess of involuntary movements (hyperkinesia) encountered in Huntington's disease or the poverty of movements (hypokinesia) that typifies Parkinson's disease (1, 2). The striatal projection system is currently viewed as arising from two distinct but intermingled neuronal populations. A first set of striatal projection neurons targets the internal segment of the globus pallidus (GPi) or the substantia nigra pars reticulata (SNr), and the second set is directed at the external pallidal segment (GPe). These projections have been referred to as the direct and indirect striatofugal pathways (3). The direct pathway is thought to originate from GABAergic neurons that coexpress substance P and/or dynorphin and project monosynaptically to the GPi/SNr. The indirect pathway is believed to arise from GABAergic striatal neurons that coexpress enkephalin and project to the GPi/SNr complex via relays in the GPe and the subthalamic nucleus.
The striatum can also be divided into two compartments: the striosomes and the surrounding extrastriosomal matrix, which are known to have distinct chemical compositions and connections (4–6). The function of this compartmentalization is still poorly understood, but anatomical studies in rats have shown differential relations of these two compartments with the substantia nigra (SN). The matrix is believed to receive inputs from sensory and motor cortical areas and to project to the SNr, whereas striosomes are thought to receive inputs from limbic cortical areas and to project to the SN pars compacta (SNc) (4, 7, 8). However, these concepts derive mainly from studies undertaken in rodents by means of bulk injections of anterograde and/or retrograde axonal tracers. Hence, these views need to be validated in other species with the help of more refined neuroanatomical procedures, such as single-cell labeling techniques that allow the reconstruction of the entire axonal arborization of singly labeled neurons. Previous investigations in rats using this single-cell labeling approach have challenged the concept of a dual striatofugal system by showing that the majority of striatal neurons project to the three main striatal target structures (GPe, GPi, and SN) (9, 10). These two sets of contradictory findings prompted us to investigate the anatomical organization of the striatal projection system in primates with a single-axon tracing approach. During our study, we paid particular attention to the axonal arborization of striatofugal neurons located in striosomes, for which no information is available.
Methods
Animals and Stereotaxic Injections. Seven adult squirrel monkeys (Saimiri sciureus) of both sexes, with body weights ranging from 0.8 to 1.5 kg, were used in the present study. Animal work was performed in accordance with the Canadian Guide for the Care and Use of Laboratory Animals, and the Laval University Institutional Animal Care Committee approved all surgical and animal care procedures. The animals were first anesthetized with ketamine (75 mg/kg) plus xylazine (5 mg/kg) and underwent a ventriculography to localize the baseline formed by anterior and posterior commissures in each animal. One week after ventriculography, the animals were anesthetized as above and then maintained under propofol anesthesia (10 mg/ml, i.v.). One to two microiontophoretic injections of biotin dextran amine (BDA) (Molecular Probes) were made bilaterally in different portions of the putamen and caudate nucleus. We used the stereotaxic coordinates of the atlas of Emmers and Akert (11) as modified by the data collected from ventriculography. Microiontophoretic labeling was carried out with glass micropipettes (tip diameter, 2–3 μm) filled with a solution of potassium acetate (0.5 M) plus 2% BDA (12). After surgery, the monkeys were isolated and kept under close surveillance by the animal keepers until they fully recovered from anesthesia. They remained isolated and were fed ad libitum until killing.
Tracer Visualization and Immunohistochemistry. After a survival period of 7 days, the animals were deeply anesthetized with a mixture of ketamine (150 mg/kg, i.m.) and xylazine (10 mg/kg, i.m.) and perfused transcardially with a saline solution followed by a fixative solution containing 4% paraformaldehyde in phosphate buffer (PB) and a 10% sucrose solution in PB. The brains were dissected out and placed in a cryoprotective solution (30% solution in PB) for 24 h at 4°C. The brains were then cut along the sagittal plane at 70 μm with a freezing microtome. The sections were collected serially in PBS and processed for the visualization of BDA according to the avidin–biotin–peroxidase method (ABC, Vector Laboratories) with nickel-intensified 3-,3′-diaminobenzidine tetrachloride as the chromogen. To determine the precise location of labeled neurons in either striosome or matrix compartment of the striatum, sections with injection sites were treated to reveal calbindin-D28k (Sigma; 1:2,500) according to standard immunohistochemical procedures with nonintensified 3-,3′-diaminobenzidine tetrachloride as the chromogen (brown precipitate). Sections containing labeled axon in SN were also treated to visualize SNc neurons expressing tyrosine hydroxylase, which is a faithful marker of dopaminergic neurons. Tyrosine hydroxylase immunostaining (Incstar; 1:2,000) served as a means to delineate the border between SNc and SNr. Labeled axons were drawn with a light microscope equipped with a camera lucida by using a ×40 objective. Five axons emerging from neurons located in a striosome of the caudate nucleus were entirely reconstructed with a computerized image-analysis system (Neurolucida, MicroBrightField) to delineate their branching pattern and 3D organization.
Results
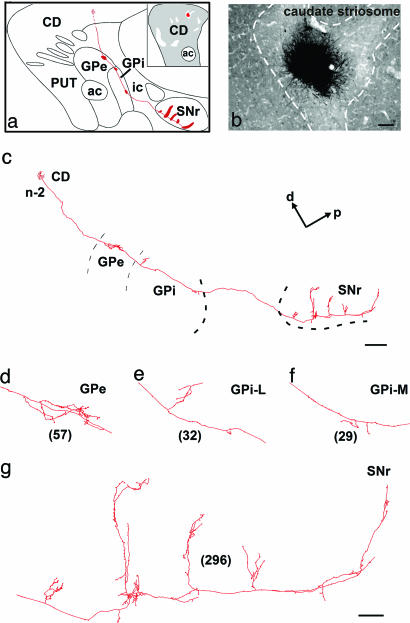
General Labeling Feature. Twelve BDA injection sites located bilaterally in the precommissural and postcommissural sectors of the striatum of the seven injected animals have been retained for analysis in the present study. These injection sites were small and consisted of a dense core of BDA precipitate (100–300 μm) surrounded by a few (1–10) medium-sized spiny neurons labeled in a Golgi-like manner. The intensely labeled axons that emerged from these sites could easily be followed individually throughout each section and were traced entirely along the sagittal plane with a camera lucida. A total of 27 axons were totally reconstructed from their parent cell body in the striatum to their most distant terminal fields outside the striatum. Particular attention was paid to the striatal compartments (striosome or matrix) in which the cell body of each of these neurons was located. The number of terminal boutons of individually labeled axon in each striatal target was counted to assess the relative strength of the various axonal branches. Some striatofugal axons yielded as many terminals in GPe as in GPi and SNr together. Most frequently, however, axons of caudate nucleus neurons yielded a much larger number of terminals at the nigral level than at the pallidal level, whereas the inverse was true for axons of putamen neurons. Many striatal projection neurons yielded multiple distinct axonal arborizations at the pallidal level, principally in the anterior and posterior sectors of the GPe and in medial and lateral subregions of the GPi (Figs. 1 and 2). Furthermore, a large proportion of striatal projection neurons exhibited a local network of collaterals in the striatum. Most of these highly branched varicose collaterals appeared to remain within their striatal compartment of origin (Fig. 3b).
Fig. 1.
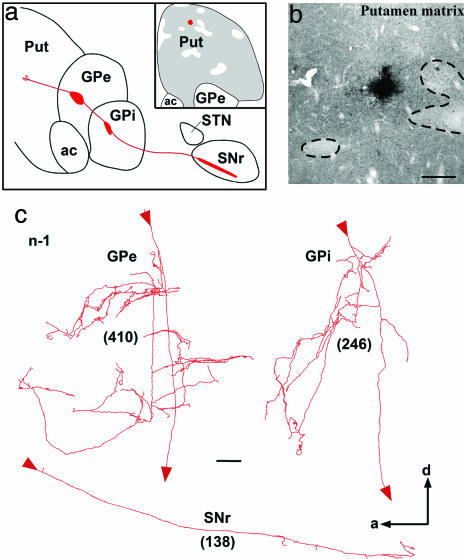
Camera lucida reconstruction from serial sagittal sections of a single axon (n-2) that projects to GPe, GPi, and SNr and whose cell body is located in the striosome compartment of the caudate nucleus. The main axon crosses the internal capsule and penetrates the GPe and GPi, where it leaves collaterals in the dorsomedial border of the nuclei. The main axon continues its route through the SNr, where it leaves a large number of varicosities and five collaterals oriented along the dorsal–ventral axis (c). More detailed views of the axonal arborization in GPe (d), GPi-L (e), GPi-M (f), and SNr (g) are provided. The total number of terminal boutons in each target structure is indicated in parentheses. The neuron drawn here was located at the border of an injection site that was confined to a single caudate striosome, as shown in the a Inset and in the photomicrograph in b. The entire axonal trajectory of this neuron is illustrated in red in a, with thick red areas indicating the location of terminal fields in GPe, GPi, and SNr. [Scale bars: b, 100 μm; c, 1 mm; g, 100 μm (also valid for d–f).] Abbreviations: a, anterior; ac, anterior commissure; CD, caudate nucleus; d, dorsal; ic, internal capsule; GPi-L, lateral portion of GPi; GPi-M, medial portion of GPi; ic, internal capsule; p, posterior; Put, putamen.
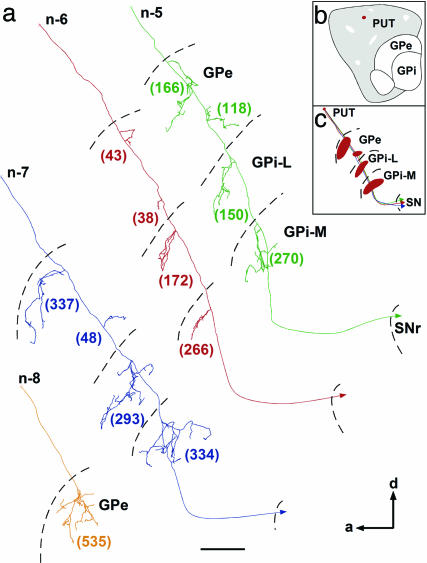
Fig. 2.
Reconstructions along the sagittal plane of four singly labeled axons that project to GPe, GPi, and SNr and whose cell body is located in the matrix compartment of the putamen. (a) Three of them innervate GPe, GPi, and SNr (n-5, n-6, and n-7), whereas the other arborizes within GPe only (n-8). Neurons projecting to the three striatal targets yield several distinct arborizations within each pallidal segment: they had one or two collaterals that arborize within both anterior and posterior halves of GPe and GPi. The total number of terminal boutons in each target structures is indicated in parentheses. The exact location of the injection site in putamen matrix compartment is indicated by the red spot in b, whereas the entire axonal trajectory of this neuron is illustrated in red in c, with thick red areas indicating the location of terminal fields in GPe and GPi. (Scale bar: 500 μm.) See the Fig. 1 legend for abbreviations.
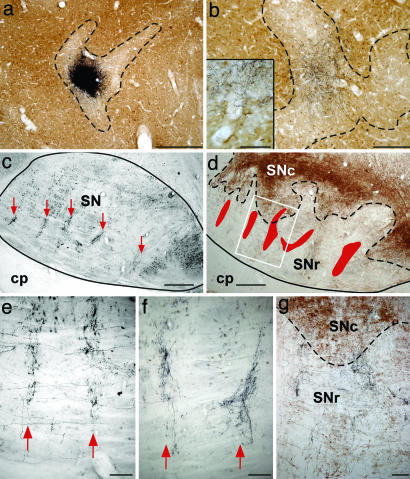
Fig. 3.
Striatofugal axon terminals organized in columns in the SNr after tracer injection in a single caudate striosome. (a) Photomicrograph showing the location of the injection site. (b) Local network of terminals in adjacent striosome, also shown at higher magnification (Inset). Dashed lines in a and b trace the border between striosome and matrix. (c) Low-magnification view of a sagittal section through the SN showing columns of axon terminals oriented dorsoventrally (red arrows). (d) Adjacent section immunostained to reveal the anterograde tracer and the enzyme tyrosine hydroxylase, a faithful marker of dopaminergic neurons. Red areas show the location of the columns, and the dashed line traces the limit between SNc (brown immunoprecipitate indicative of tyrosine hydroxylase+ neurons) and SNr. (e and f) Different columns formed by distinct axons of neurons located in the same striosome seen at a higher magnification. (g) Photomicrograph showing an area that corresponds to that indicated by the white rectangle in d. It shows that the vast majority of axon terminals of striosomal axons are confined to the SNr. (Scale bars: a, c, and d, 500 μm; b, 200 μm; b Inset and e–g, 100 μm.) cp, cerebral peduncle (see the Fig. 1 legend for other abbreviations).
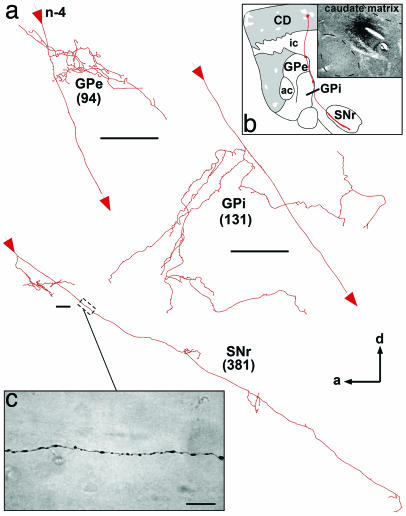
Caudate Matriceal Output. Nine axons from four injection sites have been traced from neurons located in the matrix compartment of the caudate nucleus. Seven of them branched in the GPe, GPi, and SNr (Fig. 4), whereas the two others innervated the GPe only. At the pallidal level, striatofugal axons branched in the dorsomedial part of GPe and GPi, and, along their course through the pallidum, the main axons exhibited large varicosities. At the nigral level, the axons coursed throughout the entire anterior–posterior axis of the SNr, leaving en passant numerous isolated varicosities (Fig. 4c). The axons also emitted some poorly branched and varicose collaterals that ran parallel to their main trajectory, without extending themselves much along the dorsoventral axis of the SNr. All axons that emerged from neurons located in the matrix compartment of the caudate nucleus arborized more profusely and yielded a larger number of terminal boutons in the SNr than in the two pallidal segments.
Fig. 4.
Camera lucida reconstruction from serial sagittal sections of a single axon (n-4) that projects to GPe, GPi, and SNr and whose cell body is located in the matrix compartment of the caudate nucleus. Detailed views of the arborization of this axon in GPe, GPi, and SNr are shown in a. The main axon crosses the internal capsule and enters through the GPe and GPi, where it leaves collaterals in the dorsomedial border of the nuclei. The main axon continues its course through the SNr, where it yields a large number of varicosities (c) and some short collaterals along its anterior–posterior trajectory. The total number of terminal boutons yielded by this axon into in each target structure is indicated in parentheses. The entire axonal trajectory of this neuron is illustrated in red in a, with thick red areas indicating the location of terminal fields in GPe, GPi, and SNr. The photomicrograph in b depicts the exact location of the injection site in the caudate matrix. (Scale bars: a, 100 μm; c, 10 μm.) See the Fig. 1 legend for abbreviations.
Caudate Striosomal Output. One of the five injection sites placed in the caudate nucleus of different monkeys was confined to a single striosome (Figs. 1b and 3a), and the axons of six labeled neurons located within this striosome were individually traced. All of these axons innervated the GPe, GPi, and SNr (Fig. 1 a and c–g). At the nigral level, each of them emitted four to six collaterals emerging at various points along the anterior–posterior axis of the SNr and forming ventrodorsally oriented columns (Figs. 1g, 3 c–f, and 5 a–d). All striosomal axons arborized within the SNr, but only two of them had thin collaterals that reached the ventral aspect of the SNc (Fig. 3 d and g). Like caudate matriceal neurons, the caudate striosomal neurons had an axon that arborized much more profusely at the nigral than at the pallidal level, as revealed by our quantitative estimates of terminal boutons displayed by both matriceal and striosomal neurons in each striatal target. These quantitative data (n = 15 axons) revealed that caudate neurons yielded a mean of 22% of their total number of terminal boutons in the GPe, 19% in the GPi, and 59% in the SNr.
Fig. 5.
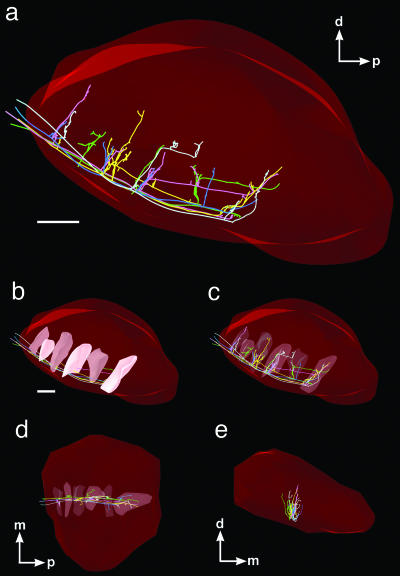
Three-dimensional reconstruction of five single axons originating from neurons located in the same caudate nucleus striosome. Each axon emits four to six collaterals arborizing in distinct columns scattered along the anterior–posterior axis of SNr (a). They all emit collaterals that form ventrodorsally oriented columns within the exact same regions of the SNr. Each axon is depicted according to a specific color, and the columns they formed are represented by modules within a large red structure that represents the SNr. The columns are represented by filled modules in the sagittal plane (b) and by transparent module in the sagittal (c), horizontal (d), and frontal (e) planes. [Scale bars in a and b: 500 μm (also valid for c–f).] m, medial (see the Fig. 1 legend for other abbreviations).
Putamen Matriceal Output. Twelve axons from seven injection sites have been traced from neurons located in the matrix compartment of the putamen. In contrast to caudatofugal axons, putamenofugal axons branched more profusely at the pallidal level than at the nigral level. Nevertheless, 10 of the 12 reconstructed axons branched in the three main striatal targets (GPe, GPi and SNr) (Figs. 2 and 6). The axonal branching pattern in the SNr was similar to that of axons originating from neurons in the caudate matrix, except that the number of axon terminals at the nigral level was much smaller than that provided by caudate matriceal neurons. Two putamenofugal axons branched only in the GPe. Their terminal arborization and the number of terminal varicosities they yielded at the GPe level were much larger than those of other putamenofugal axons that branched into GPe, GPi, and SNr (n-8; Fig. 2). Putamen neurons (n = 10) provided a mean of 28% of their total number of terminal boutons in the GPe, 55% in the GPi, and 17% in the SNr.
Fig. 6.
Camera lucida reconstruction from serial sagittal sections of a single axon (n-1) that projects to GPe, GPi, and SNr and whose cell body is located in the matrix compartment of the putamen. Detailed views of the arborization of this axon in GPe, GPi, and SNr are shown in c. The total number of terminal boutons yielded by this axon in each target structure is indicated in parentheses. The exact location of the injection site in the putamen matrix compartment is depicted in a Inset and in the photomicrograph in b. The entire axonal trajectory of this neuron is illustrated in red in a, with thick red areas indicating the location of terminal fields in GPe, GPi, and SNr. (Scale bars: b, 250 μm; c, 100 μm.) See the Fig. 1 legend for abbreviations.
Discussion
Direct and Indirect Pathways. The present single-axon tracing study provides a detailed comparison of the axonal branching pattern of single neurons located in striosomes and matrix compartments of primate striatum. Twenty-four of the 27 reconstructed axons arborized in all three striatal target structures (GPe, GPi, and SNr), revealing the highly collateralized and widely distributed aspect of the striatofugal system. This pattern of organization allows individual striatal neurons to send efferent copies of the same neural information to virtually all striatal targets. Our results are in agreement with single-cell labeling studies in rats (9, 10) and monkeys (13), which have reported that the vast majority of striatal neurons project to more than one striatal target site. However, striatofugal axons that branch into more than one target site appear more abundant in primates than in rodents; ≈90% of axons traced in the present primate study exhibited this feature, compared with 63.6% in rodents (9, 10). The results from single-axon tracing studies contradict the current concept of a dual (direct/indirect) striatal projection system (3, 14), which attributes the abnormal movements seen in Parkinson's and Huntington's diseases to an imbalance between the two pathways. However, the present study shows that virtually all striatal neurons project to GPe, which is considered a key relay center in the indirect pathway. Furthermore, our data reveal that neurons projecting solely to GPi or SNr or both, which would make up the direct pathway, do not appear to exist in primates.
Dopaminergic Receptors and Striatal Peptides. The fact that the majority of striatofugal axons arborize within the three major striatal target structures is at odds with the distribution of neuropeptides and dopamine receptors at the basal ganglia level. The current concept of direct and indirect striatal output systems stipulates that striatal neurons projecting to GPe express D2 receptor and release enkephalin, whereas neurons projecting to GPi and/or SNr exhibit D1 receptor and liberate substance P and/or dynorphin. However, there are many findings that do not support such a marked chemical segregation between the putative direct and indirect pathways. First, recent studies have detected a significant number of striatal neurons that express both D1 and D2 receptors (15–19), as well as enkephalin and substance P (20–22). Second, although GPe is markedly enriched in enkephalin, a significant amount of this neuroactive peptide also occurs in the SNr of primates (23, 24). Third, GPe neurons in humans have been shown to express neurokinin-1 receptor and to be closely contacted by substance P-positive fibers that arborize within the GPe (25), a finding that suggests functional interactions between substance P fibers and GPe neurons. One way of reconciling these various findings is to envisage the possibility that single striatal neurons might forward enkephalin in collaterals innervating GPe and substance P and/or dynorphin in collaterals branching in GPi and SNr. The mechanism whereby striatal neurons could send different peptides in distinct axonal branches is still unknown, but molecule trafficking studies have identified many mRNAs and proteins that are transported in a given neuronal process (26–29), so that striatal peptides or mRNAs could be conveyed differently among the diverse axonal branches of single axons, hence explaining the differential distribution of peptides in striatal target sites. Neuroactive peptides present at the basal ganglia level might also arise from nonstriatal sources, such as the dorsal raphe nucleus, which is known to project to pallidum and nigra (30–32) and to harbor a significant proportion of substance P-containing neurons (33). It is also worth noting that GPe contains an intrinsic population of enkephalin neurons (34, 35). These different sources of peptides could contribute to the differential peptide localization seen in GPe, GPi, and SNr.
Associative and Sensorimotor Striatal Territories. The quantitative estimate of the number of axon terminals yielded by each individual axons reveals that, although the great majority of striatal neurons arborize within the three striatal target nuclei, neurons located in the caudate nucleus project more abundantly to SNr, whereas those in the putamen aim chiefly at GPi. This organization is supported by results of retrograde cell-labeling studies in monkeys (36, 37), and it might have important functional implication. The fact that cortical inputs to caudate nucleus and putamen derives, respectively, from association and sensorimotor regions (13) suggests that GPi and SNr (the two major output structures of the basal ganglia) are involved in the processing of different functional modalities and, as such, cannot be considered equivalent to one another. Indeed, a recent electrophysiological study in primates has shown that, whereas GPi neurons are mainly concerned with the control of basic movement parameters, SNr neurons respond best to events related to memory, attention, or movement preparation (38). These functional differences are even more obvious in the parkinsonian state, where a remarkable neuronal hyperactivity occurs in GPi but not in SNr, as noted in patients suffering from idiopathic Parkinson's disease as well as in monkeys rendered parkinsonian after 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine injection (39–41). This finding explains why the GPi became the preferential target of stereotaxic surgical lesions to alleviate motor symptoms in Parkinson's disease (42). These results suggest that the cortico-caudato-nigral pathway is chiefly involved in the planning of motor acts, whereas the corticoputameno-pallidal pathway is principally concerned with their execution (Fig. 7).
Fig. 7.
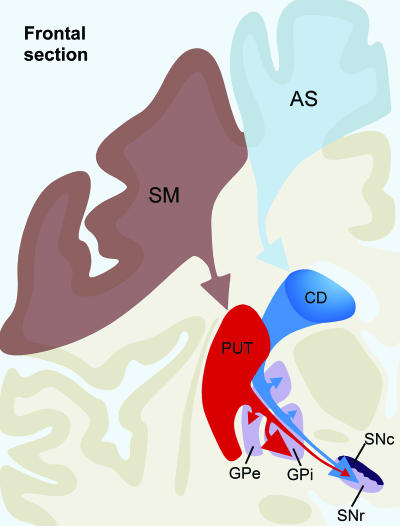
Schematic illustration of the overall organization of primate ganglia, as revealed by neuronal tracing studies. The cerebral cortex imposes functional subdivisions on the striatum: the head of the caudate nucleus, which receives inputs mainly from associative cortical areas, is considered the associative striatal territory (AS), whereas the putamen, which receives afferents principally from sensory and motor cortical areas, is regarded as the sensorimotor striatal territory (SM). Although the vast majority of neurons in either the caudate or the putamen project to the GPe, GPi, and SNr, those lying in AS (in blue) target more heavily the SN than the GPi, whereas neurons in SM (in red) arborize more profusely in GPi than in SN. The size of arrows indicates the relative importance of each type of input to the GPe, GPi, and SN. See the Fig. 1 legend for abbreviations.
Striatal Compartmentalization. Little is known about the efferent projections of neurons located within striosomes. Our current knowledge of the projections of striatal neurons located in matrix or striosomes derives from retrograde cell-labeling studies that were undertaken in rodents and that involved bulk injections of retrograde tracers in the various striatal target structures combined with histochemical visualization of the two striatal compartments (4, 7, 8). These investigations led to the concept that striosomal and matriceal neurons target SNc and SNr, respectively. The present study has provided the first description of the axonal projection of single striatal neurons located within striosomes. Although based on the injection of only one striosome located in the caudate nucleus, the detailed tracing of six individually labeled axons emerging from that striosome has indicated that, in contrast to current beliefs, striosomal neurons appear to project much more profusely to SNr than to SNc. Furthermore, axons of striosomal and matriceal origin were found to differ fundamentally from one another in the way they arborize within SN. Axons of striosomal neurons course along the entire anterior–posterior axis of SNr, giving off four to six collaterals oriented ventrodorsally. Each labeled neuron lying in the same striosome was found to contribute equally to these perpendicularly oriented columns. These results suggest that each neuron belonging to a single striosome has a multiple, modular representation in SNr. In contrast, axons of matriceal neurons yield several en passant terminals as they course through the SNr, a mode of organization that corresponds to the onion-like pattern of striatal input to SNr reported recently (43, 44). Dopamine D1 receptors are reportedly more abundant in striosomes than matrix, whereas the inverse is true for D2 receptors (45). Because activation of D1 and D2 receptors produces opposite effects, it is likely that dopaminergic modulation at the striatal level leads to a reverse action at the SNr level. Striosomal axons that form the typical “columns” at the SNr level might generate a robust inhibition after dopamine release, whereas the inverse could occur via the thin and elongated terminal field (“rods”) formed by matriceal neurons. The complex and varied axonal arborization patterns displayed by striatofugal axons at the SNr level might serve to mediate various types of neural information. Because striosomes are known to be innervated by cortical areas and subcortical structures related to the limbic system (7, 12, 46, 47), SNr could play a role in the integration of limbic, cognitive, and sensorimotor information. Furthermore, because SNr neurons exhibit extensive local axon collateral networks innervating dopaminergic SNc neurons (44, 48–50), they could thus contribute to an indirect nigrostriatal loop circuit through which the striatum might up-regulate its level of dopaminergic transmission via a disinhibition of nigrostriatal neurons. This idea is supported by physiological studies showing a disinhibition of dopaminergic neurons after striatal stimulation or GABA agonist in SNr (51, 52). It is clear that further anatomical and physiological data are required to confirm these various hypotheses. For example, the pattern of axonal projection of striosomal neurons reported here needs to be validated by injections in striosomes located in putamen and in ventral striatum.
Concluding Remarks. The present study leads to three major conclusions. First, the caudate nucleus, which processes chiefly neural information of the associative type, projects mainly to the SNr, whereas the postcommissural putamen, which represents the sensorimotor striatal territory, is largely oriented toward the GPi. Second, the striatofugal projections can no longer be viewed as a dual (direct and indirect) system, because the axon of most striatofugal neurons projects to the three major striatal targets (GPe, GPi, and SNr). Third, striosomes and matrix outputs are organized differently at the nigral level, but neurons in both compartments target GPe, GPi, and SNr.
Acknowledgments
This work was supported by Canadian Institutes for Health Research Grant MOP-5781 (to A.P.).
Author contributions: M.L. designed research; M.L. performed research; M.L. and A.P. analyzed data; and M.L. and A.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SN, substantia nigra; SNc, SN pars compacta; GPe, external segment of the globus pallidus; GPi, internal segment of the globus pallidus; SNr, SN pars reticulata; BDA, biotin dextran amine.
References
- 1.Ferrante, R. J., Kowall, N. W., Beal, M. F., Martin, J. B., Bird, E. D. & Richardson, E. P., Jr. (1987) J. Neuropathol. Exp. Neurol. 46, 12-27. [DOI] [PubMed] [Google Scholar]
- 2.Graveland, G. A., Williams, R. S. & DiFiglia, M. (1985) Science 227, 770-773. [DOI] [PubMed] [Google Scholar]
- 3.Wichmann, T. & DeLong, M. R. (1996) Curr. Opin. Neurobiol. 6, 751-758. [DOI] [PubMed] [Google Scholar]
- 4.Gerfen, C. R., Herkenham, M. & Thibault, J. (1987) J. Neurosci. 7, 3915-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerfen, C. R. (1992) Trends Neurosci. 15, 133-139. [DOI] [PubMed] [Google Scholar]
- 6.Graybiel, A. M. (1990) Trends Neurosci. 13, 244-254. [DOI] [PubMed] [Google Scholar]
- 7.Gerfen, C. R. (1984) Nature 311, 461-464. [DOI] [PubMed] [Google Scholar]
- 8.Gerfen, C. R. (1985) J. Comp. Neurol. 236, 454-476. [DOI] [PubMed] [Google Scholar]
- 9.Kawaguchi, Y., Wilson, C. J. & Emson, P. C. (1990) J. Neurosci. 10, 3421-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu, Y., Richard, S. & Parent, A. (2000) Neurosci. Res. 38, 49-62. [DOI] [PubMed] [Google Scholar]
- 11.Emmers, E. & Akert, K. (1963) Stereotaxic Atlas of the Brain of the Squirrel Monkey (Saimiri sciureus) (Univ. of Wisconsin Press, Madison).
- 12.Levesque, M. & Parent, A. (1998) Cereb. Cortex 8, 602-613. [DOI] [PubMed] [Google Scholar]
- 13.Parent, A. (1990) Trends Neurosci. 13, 254-258. [DOI] [PubMed] [Google Scholar]
- 14.DeLong, M. R. (1990) Trends Neurosci. 13, 281-285. [DOI] [PubMed] [Google Scholar]
- 15.Aizman, O., Brismar, H., Uhlen, P., Zettergren, E., Levey, A. I., Forssberg, H., Greengard, P. & Aperia, A. (2000) Nat. Neurosci. 3, 226-230. [DOI] [PubMed] [Google Scholar]
- 16.Akaike, A., Ohno, Y., Sasa, M. & Takaori, S. (1987) Brain Res. 418, 262-272. [DOI] [PubMed] [Google Scholar]
- 17.Ohno, Y., Sasa, M. & Takaori, S. (1987) Life Sci. 40, 1937-1945. [DOI] [PubMed] [Google Scholar]
- 18.Calabresi, P., Mercuri, N., Stanzione, P., Stefani, A. & Bernardi, G. (1987) Neuroscience 20, 757-771. [DOI] [PubMed] [Google Scholar]
- 19.Surmeier, D. J., Song, W. J. & Yan, Z. (1996) J. Neurosci. 16, 6579-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson, K. D. & Reiner, A. (1991) J. Comp. Neurol. 303, 658-673. [DOI] [PubMed] [Google Scholar]
- 21.Besson, M. J., Graybiel, A. M. & Quinn, B. (1990) Neuroscience 39, 33-58. [DOI] [PubMed] [Google Scholar]
- 22.Penny, G. R., Afsharpour, S. & Kitai, S. T. (1986) Neuroscience 17, 1011-1045. [DOI] [PubMed] [Google Scholar]
- 23.Reiner, A., Medina, L. & Haber, S. N. (1999) Neuroscience 88, 775-793. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki, S. & Parent, A. (1984) Brain Res. Bull. 13, 319-329. [DOI] [PubMed] [Google Scholar]
- 25.Mounir, S. & Parent, A. (2002) Neurosci. Res. 44, 71-81. [DOI] [PubMed] [Google Scholar]
- 26.Steward, O. (2002) Neuron 36, 338-340. [DOI] [PubMed] [Google Scholar]
- 27.Kindler, S. & Monshausen, M. (2002) Mol. Neurobiol. 25, 149-165. [DOI] [PubMed] [Google Scholar]
- 28.Kiebler, M. A. & DesGroseillers, L. (2000) Neuron 25, 19-28. [DOI] [PubMed] [Google Scholar]
- 29.Smith, R. (2004) Neuroscientist 10, 495-500. [DOI] [PubMed] [Google Scholar]
- 30.Charara, A. & Parent, A. (1998) J. Chem. Neuroanat. 15, 111-127. [DOI] [PubMed] [Google Scholar]
- 31.Charara, A. & Parent, A. (1994) Brain Res. 640, 155-170. [DOI] [PubMed] [Google Scholar]
- 32.Lavoie, B. & Parent, A. (1990) J. Comp. Neurol. 299, 1-16. [DOI] [PubMed] [Google Scholar]
- 33.Sergeyev, V., Hokfelt, T. & Hurd, Y. (1999) NeuroReport 10, 3967-3970. [DOI] [PubMed] [Google Scholar]
- 34.Martorana, A., Fusco, F. R., D'Angelo, V., Sancesario, G. & Bernardi, G. (2003) Exp. Neurol. 183, 311-319. [DOI] [PubMed] [Google Scholar]
- 35.Voorn, P., van de Witte, S., Tjon, G. & Jonker, A. J. (1999) Ann. N.Y. Acad. Sci. 877, 671-675. [DOI] [PubMed] [Google Scholar]
- 36.Feger, J. & Crossman, A. R. (1984) Neurosci. Lett. 49, 7-12. [DOI] [PubMed] [Google Scholar]
- 37.Parent, A., Bouchard, C. & Smith, Y. (1984) Brain Res. 303, 385-390. [DOI] [PubMed] [Google Scholar]
- 38.Wichmann, T. & Kliem, M. A. (2004) J. Neurophysiol. 91, 815-827. [DOI] [PubMed] [Google Scholar]
- 39.Merello, M., Balej, J., Delfino, M., Cammarota, A., Betti, O. & Leiguarda, R. (1999) Movement Disorders 14, 45-49. [DOI] [PubMed] [Google Scholar]
- 40.Hutchison, W. D., Lozano, A. M., Davis, K. D., Saint-Cyr, J. A., Lang, A. E. & Dostrovsky, J. O. (1994) NeuroReport 5, 1533-1537. [DOI] [PubMed] [Google Scholar]
- 41.Wichmann, T., Bergman, H., Starr, P. A., Subramanian, T., Watts, R. L. & DeLong, M. R. (1999) Exp. Brain Res. 125, 397-409. [DOI] [PubMed] [Google Scholar]
- 42.Okun, M. S. & Vitek, J. L. (2004) Movement Disorders 19, 375-389. [DOI] [PubMed] [Google Scholar]
- 43.Deniau, J. M., Menetrey, A. & Charpier, S. (1996) Neuroscience 73, 761-781. [DOI] [PubMed] [Google Scholar]
- 44.Mailly, P., Charpier, S., Menetrey, A. & Deniau, J. M. (2003) J. Neurosci. 23, 5247-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levey, A. I., Hersch, S. M., Rye, D. B., Sunahara, R. K., Niznik, H. B., Kitt, C. A., Price, D. L., Maggio, R., Brann, M. R., Ciliax, B. J., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 8861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerfen, C. R. (1989) Science 246, 385-388. [DOI] [PubMed] [Google Scholar]
- 47.Eblen, F. & Graybiel, A. M. (1995) J. Neurosci. 15, 5999-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karabelas, A. B. & Purpura, D. P. (1980) Brain Res. 200, 467-473. [DOI] [PubMed] [Google Scholar]
- 49.Grofova, I., Deniau, J. M. & Kitai, S. T. (1982) J. Comp. Neurol. 208, 352-368. [DOI] [PubMed] [Google Scholar]
- 50.Juraska, J. M., Wilson, C. J. & Groves, P. M. (1977) J. Comp. Neurol. 172, 585-600. [DOI] [PubMed] [Google Scholar]
- 51.Kalivas, P. W. (1993) Brain Res. Brain Res. Rev. 18, 75-113. [DOI] [PubMed] [Google Scholar]
- 52.Bunney, B. S., Chiodo, L. A. & Grace, A. A. (1991) Synapse 9, 79-94. [DOI] [PubMed] [Google Scholar]