Abstract
Regeneration of plant organs is often the essential step in genetic transformation; however, the regeneration ability of a plant varies depending on the genetic background. By conventional crosses of low-regeneration rice strain Koshihikari with high-regeneration rice strain Kasalath, we identified some quantitative trait loci, which control the regeneration ability in rice. Using a map-based cloning strategy, we isolated a main quantitative trait loci gene encoding ferredoxin-nitrite reductase (NiR) that determines regeneration ability in rice. Molecular analyses revealed that the poor regeneration ability of Koshihikari is caused by lower expression than in Kasalath and the specific activity of NiR. Using the NiR gene as a selection marker, we succeeded in selectively transforming a foreign gene into rice without exogenous marker genes. Our results demonstrate that nitrate assimilation is an important process in rice regeneration and also provide an additional selectable marker for rice transformation.
Keywords: regeneration ability, ferredoxin-nitrate reductase, selectable marker
Regeneration of plants from cell culture is a critical step in the production of novel varieties of plants. Generally, it is not easy to culture and regenerate monocot plants, including agronomically important crops such as rice, wheat, and maize. In rice, an efficient culture system using mature seeds has been established based on research with model varieties such as Nipponbare (Japonica) and Kasalath (Indica). However, many leading varieties used for food production, such as Koshihikari in Japan and IR64 in tropical countries, have low regeneration ability in the mature seed culture system, resulting in a serious obstacle to efficient production of transgenic plants. It has been indicated that regeneration ability depends mainly on a few key genes (1–7), but no gene has yet been identified in any plant species. To understand the regeneration process and resolve the low regeneration ability of a leading Japanese variety, Koshihikari, we have attempted to isolate major quantitative trait loci (QTL), which would increase the regeneration ability of Koshihikari.
Materials and Methods
Culture Conditions and Regeneration Test. Mature seeds were dehusked and surface-sterilized in 70% ethanol for 30 s, vigorously shaken in 1.5% sodium hypochlorite for 30 min, and rinsed five times in sterilized water. For the induction of calli, sterilized seeds were placed on the surface of an agar medium containing CHU (N6) basal salt mixture (Sigma), 2 mg/liter glycine, 0.5 mg/liter nicotinic acid, 0.5 mg/liter pyridoxine-HCl, 1 mg/liter thiamine-HCl, 0.1 g/liter myo-inositol, 0.3 g/liter casamino acid, 2.878 g/liter proline, 2 mg/liter 2,4-dichlorophenoxyacetic acid, 30 g/liter sucrose, and 3 g/liter gelrite. The medium was adjusted to pH 5.8. Seeds were incubated in the medium at 29.5°C. Four weeks after inoculation calli formed from seeds were transferred onto regeneration medium containing MS plant salt mixture (Wako), 5 mg/liter nicotinic acid, 10 mg/liter pyridoxine-HCl, 10 mg/liter thiamine-HCl, 0.1 g/liter myo-inositol, 2 g/liter casamino acid, 0.2 mg/liter naphthalene acetic acid, 2 mg/liter kinetin, 30 g/liter sorbitol, 30 g/liter sucrose, and 3 g/liter gelrite at pH 5.8. After 4 weeks, regenerated shoots were counted. Regeneration ability is given as number of shoots per seed.
Bacterial Artificial Chromosome (BAC) Library. A Kasalath BAC library containing 18,816 clones with an average insert size of 120 kb was constructed with the CopyControl pCC1BAC vector (Epicentre Technologies, Madison, WI). We combined 384 clones into 49 pools for PCR-based screening with DNA markers P121 and P182.
DNA Construction for Transgenic Analyses. For complementation tests, three genomic fragments of Kasalath containing each candidate gene region were subcloned into the binary vector pBI101-Hm3 (8). For selection marker tests, we generated five constructs based on pBI101 as a vector backbone (Clontech). The NheI–HindIII fragment containing nptII was excised from pBI101 and self-ligated (pBI101 minus nptII). The cauliflower mosaic virus 35S promoter from pBI221 (Clontech) was introduced into the upstream site of the β-glucronidase (GUS) gene of pBI101 minus nptII to produce a plasmid designated 35SGUS. The empty vector was generated by removing the GUS-NOS terminator of pBI101-nptII. The 12.2-kb XhoI–StuI genome fragment of Kasalath containing ferredoxin-nitrite reductase (NiR), used for complementation tests, was introduced into the 35SGUS plasmid at the SalI site and designated NiR genome+35SGUS. The 5′ sequence of the NiR gene, containing the 5′ flanking and 5′ noncoding regions, was amplified from Kasalath genomic DNA with a 5′ primer containing a NheI linker that annealed 1.3 kb upstream of the translation initiation site and a 3′ primer with a HindIII linker that annealed just upstream of the translation initiation site. PCR products were cloned into pBI101 at the NheI–HindIII site where nptII was excised, and the construct was labeled pNiR+GUS. NiR cDNA was synthesized by RT-PCR from Kasalath callus RNA by using one primer complementary to the translation initiation site and the other complementary to the translation stop site. Kasalath NiR cDNA was cloned into pCR4 Blunt-TOPO vector (Invitrogen), and the NiR terminator region was joined downstream of the NiR cDNA (NiRcDNA::NiRt). This NiR cDNA::NiR terminator fragment was introduced into the downstream site of the NiR promoter in pNiR+GUS. The pNiR::NiRcDNA::NiRt+35SGUS plasmid was constructed to introduce a cauliflower mosaic virus 35S promoter fragment into pNiR::NiRcDNA::NiRt+GUS. Rice actin 1 promoter, obtained from pBIActnos (9), was introduced into the upstream region of NiR cDNA in the NiRcDNA::NiRt construct. The NiRcDNA::NiRt fragment was cloned into empty pBI101 vector, and then the 35SGUS fragment was introduced to construct pAct::NiRcDNA::NiRt+35SGUS.
RT-PCR. Complementary DNA was synthesized from 2 μg of total RNA with Omniscript Reverse Transcriptase (Qiagen, Valencia, CA) with oligo(dT) primers. Semiquantitative PCR was performed with gene-specific primers A and A′ (see Fig. 3a), and saturated RT-PCR was performed with gene-specific primers B and B′ (see Fig. 3a). Primer sequences and PCR conditions are available on request. Real-time RT-PCR was performed with a SybrGreen detection system on a Light Cycler (Roche Diagnostics). RNA expression data were normalized to the levels of polyubiquitin (GenBank accession no. U37687) RNA.
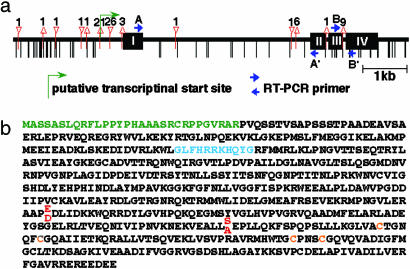
Fig. 3.
Primary structure of PSR1. (a) Comparison of the NiR sequence in Koshihikari and Kasalath. Differences of the Kasalath allele compared with Koshihikari are shown. Black boxes represent the coding regions, and vertical bars indicate single nucleotide substitutions. Triangles and inverted triangles indicate deletion and insertion sites, respectively, and numbers above the triangles represent the numbers of deleted or inserted nucleotides. Arrows indicate RT-PCR primers used in expression analyses. (b) Amino acid sequence of NiR. Three putative functional regions, chloroplast transit peptide region (green), ferredoxin-binding region (blue), and 4Fe-4S cluster (orange), are indicated. These domain features are from the Protein Information Resource database (accession no. JC4395). Two amino acid differences were found between Koshihikari and Kasalath (stacked red letters).
Immunoblot Analysis. Calli were homogenized in extraction buffer (50 mM Tris·HCl, pH 7.9/2 mM EDTA/5 mM cystein). For immunoblotting, 25 μg of protein was separated by electrophoresis on a 7.5% SDS/polyacrylamide gel and transferred to a nitrocellulose membrane (Amersham Pharmacia). Western blot assays were performed as described (10).
NiR Enzyme Assay. Koshihikari and Kasalath cDNAs were amplified by RT-PCR and subcloned into the bacterial expression vector pET-32a (Novagen). Recombinant proteins were induced by 0.2 mM isopropyl β-d-thiogalactoside in 20°C culture and extracted by sonication. Protein purification was performed by using TALON metal affinity resin (BD Biosciences, Franklin Lakes, NJ), and the concentrations were determined with a protein assay kit (Bio-Rad). For in vitro assays of NiR, 50 μl of crude extracts of calli or purified recombinant proteins was added to an 850-μl assay mixture (50 mM Tris·HCl, pH 7.5/0.5 mM NaNO2/1 mM methyl viologen), and the enzyme reaction was initiated by the addition of 100 μl of 0.12 M Na2S2O4 dissolved in 0.2 M NaHCO3. After incubation at 30°C for 60 min, the reaction was terminated by vigorous vortexing until the color of methyl viologen disappeared completely. The concentration of reduced nitrite ions was determined with a 2,3-diaminonaphthalene kit (Dojindo Laboratories, Kumamoto, Japan).
Results
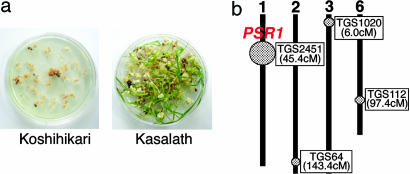
Identification of a Regeneration-Related Gene. In preliminary studies, Koshihikari calli turned brown, and no green tissue was obtained when mature seed-derived calli were used for regeneration, whereas Kasalath calli were white or yellow and formed shoots with high frequency (Fig. 1a). We therefore chose Kasalath (Indica) as a high-regenerative variety to cross with Koshihikari.
Fig. 1.
QTL analysis for regeneration ability in rice. (a) Comparison of the regeneration ability between Koshihikari (Japonica, Left) and Kasalath (Indica, Right). (b) Linkage map and positions of QTL for regeneration ability identified in crosses between Koshihikari and Kasalath. Circles and their diameters indicate the position and efficiency of each QTL, respectively. The closest PCR land markers (TG marker) and their positions are also shown.
The Koshihikari × Kasalath F1 was backcrossed with Koshihikari to produce 99 BC1F1 plants from which BC1F2 seeds were harvested. For QTL mapping, the genotypes of the 99 BC1F1 plants were determined by using 262 PCR markers, which were almost equally dispersed on the 12 rice chromosomes. Twenty BC1F2 seeds from each BC1F1 line were tested for regeneration ability. We then performed QTL analysis with the qgene computer program (11) and found four putative QTL located on chromosomes 1, 2, 3, and 6 in which Kasalath alleles were correlated with a positive effect on regeneration ability. The QTL located on the short arm of chromosome 1 near PCR marker TGS2451 had the largest effect and was designated Promoter of Shoot Regeneration 1 (PSR1) (Fig. 1b).
To roughly locate the map position of PSR1, we selected 30 BC2F1 plants that had Kasalath alleles flanking PSR1 and induced calli from 10 BC2F2 seeds of each BC2F1 line. On the basis of the genotypes and phenotypes of BC2F2 plants, PSR1 was mapped to ≈45.4 cM on chromosome 1.
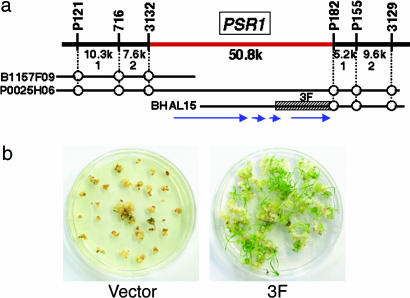
For high-resolution mapping, ≈3,800 BC3F2 seeds from selected BC3F1 plants containing a small heterozygous Kasalath chromosomal segment around PSR1, but with the homozygous Koshihikari genotype around the other three QTL regions, were analyzed. Fine-mapping analysis revealed that PSR1 lies in the interval between two molecular markers, 3132 and P182, with a distance of 50.8 kb (Fig. 2a). In this region, four putative genes were predicted by RiceGAAS analyses (http://RiceGAAS.dna.affrc.go.jp). To identify the PSR1 gene, we constructed a Kasalath BAC library containing ≈2 × 104 clones with an average insert size of 120 kb and isolated a BAC clone, BHAL15, that included most of the PSR1 candidate region (Fig. 2a). Several fragments covering each candidate gene were subcloned from BHAL15 and introduced into Koshihikari calli. Among them, one 12.2-kb fragment (Fig. 2a) covering the entire region of a putative NiR gene provided the regeneration ability of Koshihikari calli (Fig. 2b), indicating that PSR1 may be a NiR gene.
Fig. 2.
Map-based cloning of the strongest QTL, PSR1. (a) Genetic and physical map of the PSR1 region. The vertical bars represent molecular markers. The physical distances between adjacent markers and the numbers of recombinants are shown under the uppermost horizontal line. B1157F09 (BAC) and P0025H06 (P1 artificial chromosome) are Nipponbare clones of the Rice Genome Research Program. The hatched box region of Kasalath BHAL15 (3F) was used for complementation tests. Circles indicate the positions of each marker. Arrows indicate the position of predicted genes. (b) Complementation test. Koshihikari calli introduced an empty vector (Left) and the 3F fragment (Right).
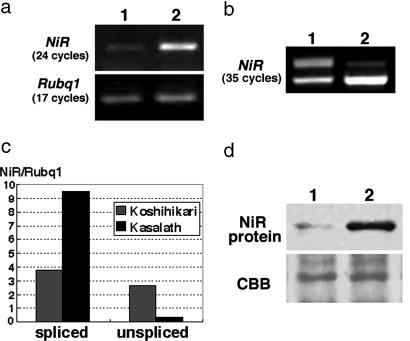
Sequence and Expression Analyses of the PSR1 Gene. We compared the NiR sequences of Koshihikari and Kasalath and found many single nucleotide polymorphisms and nucleotide deletions and insertions, especially in the promoter regions (Fig. 3a). However, these polymorphisms produce only two amino acid changes: E375D and S453A (Fig. 3b). Because sequence comparisons suggested the possibility that the expression level of NiR is different in these two plants, we examined NiR expression in callus by semiquantitative RT-PCR. NiR expression was detected in both Koshihikari and Kasalath, but the expression level was much lower in Koshihikari when ubiquitin expression was used as a control (Fig. 4a). We also detected two transcripts with different sizes in Koshihikari, the unspliced transcript retaining the third intron and the correctly spliced transcript (Fig. 4 b and c). We also examined the amount of NiR protein by immunoblot assay. The NiR protein was detected at a much higher level in Kasalath than in Koshihikari (Fig. 4d).
Fig. 4.
Comparison of NiR expression. (a) Semiquantitative RT-PCR analysis of NiR expression performed with primers A and A′ indicated in Fig. 3a. NiR was detected at 24 cycles. Rubq1, detected at 17 cycles, was used as a control. Lane 1, Koshihikari; lane 2, Kasalath. (b) Saturated RT-PCR analysis of NiR expression performed with primers B and B′ indicated in Fig. 3a. NiR was detected at 35 cycles. Lane 1, Koshihikari; lane 2, Kasalath. (Upper) The unspliced transcript. (Lower) The correctly spliced transcript. (c) Real-time PCR analysis of NiR expression. Expression levels were normalized by using Rubq1. (d) Immunoblot analysis of the crude extract from Koshihikari (lane 1) and Kasalath (lane 2) calli with antibody against NiR. The Coomassie brilliant blue (CBB)-stained RbcS protein was used as a loading control.
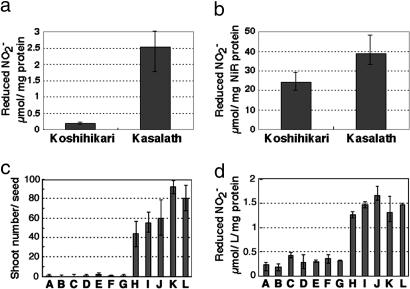
NiR Activity. The lower level of NiR expression in Koshihikari may result in lower enzymatic activity. This hypothesis was confirmed with nitrite reductase assays using crude extracts. NiR activity in extracts of Koshihikari calli was ≈22 times lower than that of Kasalath (Fig. 5a). Lower NiR activity in Koshihikari is caused not only by lower expression of NiR protein but also by lower specific activity of the NiR enzyme in Koshihikari. When we examined recombinant proteins produced in Escherichia coli, the activity of the Kasalath NiR was 1.6 times higher than the Koshihikari NiR (Fig. 5b), suggesting that amino acid differences also caused the differences in enzyme activity. We also examined the relationship between NiR activity in calli and regeneration ability in various kinds of Japonica rice varieties and found that the varieties with high regeneration ability always had high NiR activity and vice versa (Fig. 5 c and d). This finding clearly demonstrates that NiR activity is a determining factor for the regeneration ability of Japonica rice varieties.
Fig. 5.
Comparison of NiR enzyme activity. (a) NiR activity in the crude extract from Koshihikari and Kasalath calli. Enzyme activity was measured by determining the number of reduced nitrite ions per 1 mg of total protein for 1 h. (b) NiR enzyme activity of the recombinant protein produced in E. coli. Enzyme activity was measured by determining the number of reduced nitrite ions per 1 mg of purified NiR protein for 1 h. (c and d) Regeneration ability (c) and NiR enzyme activity (d) in various Japonica varieties: A, Koshihikari; B, Ginbouzu; C, Moritawase; D, Norin1; E, Milky-queen; F, Hitomebore; G, Akitakomachi; H, Nipponbare; I, Norin22; J, Norin8; K, Sasanishiki; L, Taichung65.
NiR as a Selection Marker. NiR catalyzes the reduction of nitrite to ammonium and functions as a key enzyme in nitrate assimilation. Nitrate is commonly used as a nitrogen source for plant cell culture, but its metabolite, nitrite, has a toxic effect on plant cell growth (12, 13). Thus, rapid metabolism of nitrite is crucial for plant cell growth and regeneration. In this context, Kasalath calli, having higher NiR activity, may rapidly metabolize the nitrite, whereas Koshihikari calli may not. To test this hypothesis we measured the accumulation of nitrite in callus culture medium and found that there is appreciable accumulation of nitrite ions in the Koshihikari culture medium, but it was not detected in the Kasalath medium (Table 1).
Table 1. Accumulation of nitrite and ammonium ions in the regeneration medium.
| Rice strain | NO2– | NH4+ |
|---|---|---|
| Koshihikari | 1.2 | 14 |
| Kasalath | <0.03* | 17 |
Each content was determined by capillary electrophoresis, which is represented by μmol per g of medium.
Assay limit is 0.03
Genes encoding antibiotic and herbicide resistance are widely used as selection markers in plant transformation (13). Recently, a positive selection system based on the E. coli phosphomannos-isomerase (pmi) gene as selection marker was developed in various plants, including rice (14). These selection markers are exogenous for plants. Moreover, several methods have been reported to generate selection marker-free transgenic plants, including site-specific recombination and intrachromosomal recombination to remove the selection marker and cotransformation and transposable elements to segregate the selection marker (15, 16). In the chloroplast transformation system, a method that did not use an antibiotic resistance gene was established (17). However, such a method is generally time-consuming and inefficient. As already shown, the introduction of the Kasalath NiR genome sequence conferred enhanced regeneration ability on Koshihikari (Fig. 2b). This result suggests that the Kasalath NiR gene could be used as a selection marker for gene transformation into recalcitrant strains of rice without any additional selective agents.
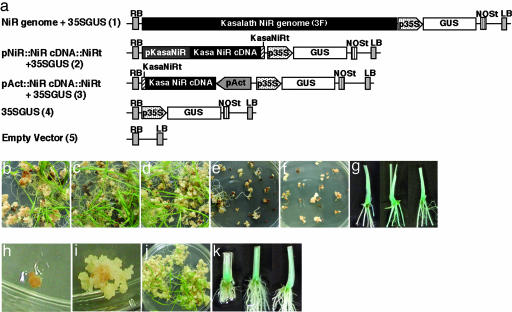
To test this idea, we constructed five vectors carrying the GUS gene (GUS) as a reporter: Kasalath NiR genome + 35S promoter::GUS (plasmid 1 in Fig. 6a), Kasalath NiR promoter::NiR cDNA::NiR terminator + 35S promoter::GUS (plasmid 2 in Fig. 6a), rice Actin1 promoter::NiR cDNA::NiR terminator + 35S promoter GUS (plasmid 3 in Fig. 6a), and two control vectors not including NiR (plasmids 4 and 5 in Fig. 6a). These constructs were transformed into Koshihikari calli by Agrobacterium tumefciens-mediated transformation by standard methods (18) and cultured on medium containing no chemicals for selection. Calli transformed with the three constructs carrying the Kasalath NiR formed many shoots (Fig. 6 b–d) with GUS activity (Fig. 6g). Calli transformed with constructs lacking Kasalath NiR formed no or few shoots (Fig. 6 e and f). These calli did not express GUS activity (data not shown). About 54% of calli transformed with Kasalath NiR as a selection marker regenerated into plants. Of these, >80% stained for GUS activity. This result shows that the Kasalath NiR gene is useful as a selection marker for transformation of Koshihikari rice.
Fig. 6.
Transformation using the NiR gene as a selection marker. (a) Schematic representation of the T-DNA region of the constructs tested. RB, right border; LB, left border; p35S, cauliflower mosaic virus 35S promoter; NOSt, 3′ signal of nopaline synthase; pKasaNiR, Kasalath NiR promoter; KasaNiRt, 3′ signal of Kasalath NiR; pAct, rice Actin1 promoter. (b–f) Transformed Koshihikari calli carrying plasmids 1–5, respectively. (g) GUS expression in regenerated plants carrying plasmid 1 (Left),2(Center), and 3 (Right). (h and i) Transformed Nipponbare calli overexpressing NiR on 1,000 mg/liter NaNO2 selection medium: nontransformant (h) and transformant (i). (j) Regenerated Nipponbare plants from selected calli on 500 mg/liter NaNO2 regeneration medium. (k) GUS expression of regenerated Nipponbare plants. (Left) Plasmid 1. (Center) Plasmid 2. (Right) Plasmid 3.
We also attempted to adopt the Kasalath NiR selection system for high regenerative varieties. Because nitrite is toxic to plants, the addition of excess nitrite in the culture medium causes the growth inhibition of calli regardless of their regeneration ability. For example, variety Nipponbare cells cannot grow under high-nitrite conditions (Fig. 6h). However, when Nipponbare cells were transformed with the NiR overexpression construct (plasmid 3 in Fig. 6a), the cells grew normally (Fig. 6 i and j) and showed GUS activity (Fig. 6k). This result demonstrates that NiR can be used as a selection marker for transformation even in rice plants with high regeneration ability.
Discussion
Under certain conditions, differentiated plant tissues can revert to a dedifferentiated state and through additional cell divisions form unorganized cell aggregates or calli. Calli can regenerate into whole plants in response to plant hormones that stimulate root and shoot tissue formation. The competence of whole plant regeneration depends on totipotency, or the genetic potential that makes plant tissue culture possible. Plant culture systems have been applied to many areas of plant science and crop improvement, making it an essential technique, particularly in plant transformation.
It is possible that all plant species are totipotent, but it can be difficult to identify the culture conditions and stimuli required to express totipotency. A potential solution to this problem is to identify the QTL genes associated with callus formation and plant regeneration and transfer the high-regeneration ability QTL gene(s) into low-regeneration varieties. Several studies have described the improvement of regeneration ability by introgressing the QTL gene through conventional crosses (1, 19). However, this method is very laborious and gives no understanding of the molecular mechanisms of plant regeneration. In this study, we used a gene identified by QTL analysis to overcome a genetic barrier to plant regeneration through isolation and characterization of a nitrite reductase gene. Complementation of low-regeneration variety Koshihikari with the NiR gene from high-regeneration variety Kasalath is sufficient for establishing a practical cell culture system.
It is possible that other NiR or sulfite reductase (SiR) genes are involved in the same step of nitrite reduction. Rice has at least two candidate genes encoding NiR (PSR1/NiR and another putative NiR) and one SiR (data not shown). An additional candidate NiR may be a pseudogene because it has a stop codon in the coding region. The expression level of SiR is almost the same in Koshihikari and Kasalath calli and much lower than PSR1/NiR (data not shown), indicating that nitrite reduction in rice calli depends mainly on PSR1/NiR. Therefore, PSR1/NiR has a critical function in the assimilation of nitrogen in rice cell culture conditions. Actually, all of the Japonica rice varieties with low regeneration ability we examined showed low NiR activity (Fig. 5 c and d). It is likely, then, that the introduction of PSR1/NiR into these varieties would result in an increase of regeneration ability as in the case of Koshihikari.
Nitrate is the major source of nitrogen in most plants. In the assimilation pathway, nitrate is reduced to nitrite by nitrate reductase (NR), and, subsequently, NiR reduces nitrite to ammonium. Ammonium is then used for the synthesis of N-metabolites. Mutants impaired in nitrate assimilation have been isolated by various criteria such as chlorate resistance (20), absence of NR activity (21), accumulation of nitrite (22), and inability to grow on nitrate as sole nitrogen source (23). Some putative NiR mutants accumulating nitrite have been isolated from barley, and one of them was confirmed as a NiR-deficient mutant (24). However, any other NiR mutants in higher plants have been obtained by above screenings. In Arabidopsis, some T-DNA insertion mutants of the NiR gene have been detected but have no observable phenotype (http://arabidopsis.orghttp://arabidopsis.org). In this study, we found the deficiency of NiR activity in Koshihikari led to the low regeneration frequency. From this viewpoint, Koshihikari can be considered to be a NiR-deficient mutant. Such low NiR activity also was observed in shoot and root of Koshihikari as well as in callus (data not shown), and therefore Koshihikari may be available as a plant material to investigate the function and regulation of NiR in the nitrate assimilation pathway.
On the basis of the differences of NiR activity among rice varieties, we established a gene transformation method with NiR as a selection marker. A major advantage of using NiR for selection is that it is an endogenous rice gene that does not confer a selective advantage beyond that found widely within the rice species. Consequently, this transformation system using the NiR gene could ease some public concerns about genetically modified crops.
Acknowledgments
We thank Miyako Ueguchi-Tanaka for excellent technical advice in protein expression analyses and Ikuko Aichi, Masako Hattori, and Sakimi Watanabe for excellent technical assistance.
Author contributions: A.N., M.A., and M.M. designed research; A.N. performed research; M.A., S.L., T.T., E.R.A., and T.Y. contributed new reagents/analytic tools; A.N. and S.L. analyzed data; and A.N. and M.M. wrote the paper.
Abbreviations: QTL, quantitative trait loci; NiR, ferredoxin-nitrite reductase; PSR1, Promoter of Shoot Regeneration 1; BAC, bacterial artificial chromosome; GUS, β-glucronidase.
References
- 1.Armstrong, C. L., Romero-Severson, J. & Hodges, T. K. (1992) Theor. Appl. Genet. 84, 755-762. [DOI] [PubMed] [Google Scholar]
- 2.Koornneef, M., Bade, J., Hanhart, C., Horsman, K., Schel, J., Soppe, W., Verkerk, R. & Zabel, P. (1993) Plant J. 3, 131-141. [Google Scholar]
- 3.Yu, K. & Pauls, K. (1993) Plant Mol. Biol. 22, 269-277. [DOI] [PubMed] [Google Scholar]
- 4.Mano, Y., Takahashi, H., Sato, K. & Takeda, K. (1996) Breed. Sci. 46, 137-142. [Google Scholar]
- 5.Taguchi-Shiobara, F., Lin, S. Y., Tanno, K., Komatsuda, T., Yano, M., Sasaki, T. & Oka, S. (1997) Theor. Appl. Genet. 95, 828-833. [Google Scholar]
- 6.Takeuchi, Y., Abe, T. & Sasahara, T. (2000) Crop Sci. 40, 245-247. [Google Scholar]
- 7.Kwon, Y. S., Kim, K. M., Eun, M. Y. & Sohn, J. K. (2001) Mol. Cells 11, 64-67. [PubMed] [Google Scholar]
- 8.Sato, Y., Sentoku, N., Miura, Y., Hirichika, H., Kitano, H. & Matsuoka, M. (1999) EMBO J. 18, 992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimura, A., Ito, M., Kamiya, N., Sato, Y. & Matsuoka, M. (2002) Plant J. 30, 189-201. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura, A., Tamaoki, M., Sakamoto, T. & Matsuoka, M. (2000) Plant Cell Physiol. 41, 583-590. [DOI] [PubMed] [Google Scholar]
- 11.Nelson, J. C. (1997) Mol. Breed. 3, 239-245. [Google Scholar]
- 12.Hamilton, J. T. & Lowe, R. H. (1981) Agron. J. 73, 787-790. [Google Scholar]
- 13.Hallens, R., Mullineaux, P. & Klee, H. (2000) Trends Plant Sci. 5, 446-451. [DOI] [PubMed] [Google Scholar]
- 14.Datta, K., Baisakh, N., Oliva, N., Torrizo, L., Abrigo, E., Tan, J., Rai, M., Rehana, S., Al-Babili, A., Beyer, P., et al. (2003) Plant Biotechnol. J. 1, 81-90. [DOI] [PubMed] [Google Scholar]
- 15.Ebinuma, H., Sugita, K., Matsunaga, E., Endo, S., Yamada, K. & Komamine, A. (2001) Plant Cell Rep. 20, 383-392. [DOI] [PubMed] [Google Scholar]
- 16.Tu, J., Datta, K., Oliva, N., Zhang, G., Xu, C., Khush, G. S., Zhang, Q. & Datta, S. K. (2003) Plant Biotechnol. J. 1, 155-165. [DOI] [PubMed] [Google Scholar]
- 17.Daniell, H., Wiebe, P. O. & Fernandez-San Millan, A. (2001) Trends Plant Sci. 6, 237-239. [DOI] [PubMed] [Google Scholar]
- 18.Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. (1994) Plant J. 6, 271-282. [DOI] [PubMed] [Google Scholar]
- 19.Koornneef, M., Hanhart, C., Jongsma, M., Toma, I., Weide, R., Zabel, P. & Hille, J. (1986) Plant Sci. 45, 201-208. [Google Scholar]
- 20.Maliga, P. (1984) Ann. Rev. Plant Physiol. 35, 519-542. [Google Scholar]
- 21.Kleinhofs, A., Kuo, T. & Warner, R. (1980) Mol. Gen. Genet. 177, 421-425. [Google Scholar]
- 22.Duncanson, E. Gilkes, A. F, Kirk, D. W. & Wray, J. L. (1993) Mol. Gen. Genet. 236, 275-282. [DOI] [PubMed] [Google Scholar]
- 23.Pelsy, F., Kronenberger, J., Pollien, J. M. & Caboche, M. (1991) Plant Sci. 76, 109-114. [Google Scholar]
- 24.Ward, M. P., Abberton, M. T., Forde, B. G., Sherman, A. & Thomas, W. T. (1995) Mol. Gen. Genet. 247, 579-582. [DOI] [PubMed] [Google Scholar]