Centromere specification has been a topic of intense interest since it became clear that, under the right conditions, human centromeres can form over noncentromeric sites. At least 60 human “neocentromeres” have been described that retain no vestige of the familiar α-satellites and that map to apparently random locations in the genome (1). At the functional and cell biological levels (e.g., association with kinetochore proteins), not a single difference has been detected between standard human centromeres and neocentromeres. These and similar data from Drosophila suggest that animal centromeres are initiated in large part by epigenetic mechanisms (2). In this issue and a recent issue of PNAS, two new articles (3, 4) extend these observations to plants. Lee et al. (3) provide a new evolutionary perspective on centromere evolution, demonstrating that satellite repeats are gained and lost at astonishing rates. Nasuda et al. (4) take the story a step further to show that barley centromeres can move to new positions and that satellite DNA is not necessary for efficient centromere formation.
An Ancient Centromere Repeat Has Spawned Multiple Variants in the Grass Family
In both plants and animals, the major centromeric DNAs are small 100- to 200-bp satellite repeats that usually are organized in very long arrays. In rice, the major repeat is CentO, and in maize it is CentC. Both are known to interact with the key centromere protein Centromeric Histone H3 (CENH3) (5, 6). Grass centromeres also contain a specialized class of centromeric retroelements (CR elements) that bind to CENH3 and are thoroughly interspersed with the satellites (5). The presumption is that satellite arrays are the primary centromere repeats (5–7), whereas CR elements are either efficient centromere parasites (8) or facilitate the establishment of a centromeric state (9).
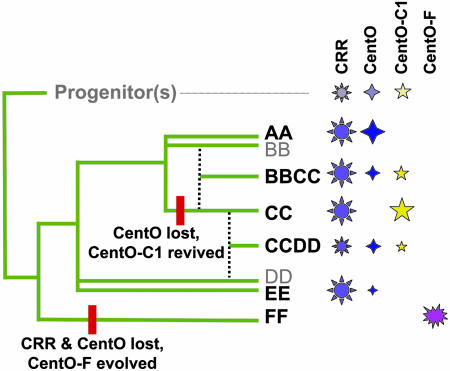
A central issue in centromere research is the “centromere paradox” (10), the apparent conflict between the importance of centromeric DNA and the fact that it evolves so quickly that identifying conserved sequences is often impossible. An answer to the paradox will require, in part, a better understanding of the variation that is present within large clades of related species. Toward this end, Lee et al. (3) took a biochemical approach to identify centromere repeats in two understudied rice species, Oryza rhizomatis and Oryza brachyantha. There are numerous allotetraploids in the Oryza genus, and the species are often referred to by the genomes they contain (e.g., BB, CC, BBCC, etc.). As shown in Fig. 1, O. rhizomatis is a diploid with a CC genome, and O. brachyantha is a diploid with a FF genome. Both species had been previously shown to lack CentO (11). Lee et al. (3) used chromatin immunoprecipitation to partially purify centromeric nucleosomes, extracted the associated DNA, and prepared small-insert libraries for sequencing. In O. rhizomatis (CC), the major repeats turned out to be variants of CentO named CentO-C1 and CentO-C2. CentO-C2 is present at centromeres and telomeres, and CentO-C1 is a centromere-specific sequence.
Fig. 1.
The distribution of centromere repeats in several rice genomes as described by Lee et al. (3). The phylogeny provided by Ge et al. (18) was used as a template to show evolutionary relationships. Cultivated rice (O. sativa) contains the AA genome, O. rhizomatis contains the CC genome, and O. brachyantha contains the FF genome. The major repeats are indicated with symbols; the size of symbol provides a rough indicator of how prevalent the repeat is. Major events in centromere evolution are indicated with red bars.
CentO-C1 shares sequence homology over an 80-bp region that also is conserved between rice CentO and maize CentC. Phylogenetic analysis suggests that CentC, CentO, and CentO-C1 diverged at roughly the same time several million years before the emergence of Oryza (3) (Fig. 1). By using pattern-matching software, Lee et al. also were able to identify significant homology between the conserved CentO/CentC/CentO-C1 domain and a known centromere repeat in pearl millet. These data provide strong evidence for an ancestral centromere repeat and a conserved sequence motif within it. It is unclear what function such a motif might provide. It is possible that the motif represents a conserved protein-binding site; however, no sequence-specific centromere binding proteins have been identified in plants. A second possibility, preferred by the authors, is that the motif confers structural stability to the repeat (3).
Genomewide Centromere Replacement in a Short Time Frame: Evidence for Centromere Drive?
A recently proposed model for centromere evolution (and potential solution to the centromere paradox) supposes that centromere repeats can influence how frequently they are recovered in progeny and evolve accordingly (12). Under this view, known as the centromere drive hypothesis, centromeres that are particularly good at chromosome segregation will arrive in reproductive cells more often than those that are not. Assuming that centromere function is at least partially determined by DNA sequence, it stands to reason that sequences that can confer segregation advantages will increase in number (13). It is an appealing idea, but like many evolutionary models it is difficult to test. For instance, the prevalence of CentO-C1 in O. rhizomatis suggests that it may have had a segregation advantage over other repeats in this lineage. However, whether it was sequence-based adaptive evolution or genetic drift is difficult to determine because both CentO and CentO-C1 preexisted in the progenitor species. More to the point would be a new centromere repeat that arises in a single species and sweeps through the genome.
As a part of their chromatin immunoprecipitation cloning study, Lee et al. (3) cloned centromeric DNA from an African rice species known as O. brachyantha (FF). Surprisingly, none of the 96 repeats sequenced proved to be CentO, Cento-C1, or CentO-C2. Instead, they found CentO-F, which lacks any detectable homology to other centromere repeats. CentO-F is found at every centromere in O. brachyantha and has a copy number that is similar to that of CentO in cultivated rice (≈20,000 per haploid genome). It is possible that CentO-F existed in the Oryza progenitor(s), but it seems unlikely, because there was no evidence of the repeat in any of the other 16 species analyzed.
Another remarkable characteristic of O. brachyantha (FF) is that it contains few if any canonical CR elements (known as CRR in rice). CR elements are the most conserved and reliable markers of cereal centromeres and interact with CENH3 as efficiently as satellite DNA (5, 6). CentO-F may have usurped the centromere functions that CR elements normally provide (9) or may simply be spreading so fast that the CR elements cannot keep pace (3). It is difficult to estimate how quickly it occurred, but a reasonable guess would be that CentO-F evolved and drove out CentO, CentO-C1, and CRR in a period of 7 million to 9 million years (approximate age of the FF genome; E. Kellogg, personal communication). By comparison, the α-satellites of New and Old World monkeys retain ≈64% sequence homology, even though the two superfamilies diverged ≈35 million years ago (13, 14). With respect to rapid evolution and shear aggressiveness, the behavior of rice CentO-F is consistent with the centromere drive model (3, 12).
Plant Centromeres Can Function Without Canonical Satellite Repeats
The concept of the epigenetically determined centromere became firmly entrenched when the first human neocentromere was identified and carefully documented (1, 2). Facultative neocentromeres also had been identified in maize and rye, but unlike animal neocentromeres, classic plant neocentromeres are meiosis-specific and lack kinetochores (15). Thus, until the work of Nasuda et al. (4), it remained possible that fully functional neocentromeres and the implied centromere plasticity were unique to animals.
Several “gametocidal” chromosomes from Aegilops (a wheat relative) have the unique property of inducing breakage on other chromosomes when introduced into wheat. When chromosomes from barley are introduced into such a wheat–Aegilops addition line, they too are subject to breakage. It was in such a gametocidal background that a series of barley translocation and deletion chromosomes were identified (Fig. 2). The two chromosomes analyzed in the most detail were telosomes containing a single arm of barley chromosome 7H. One of the telosomes had lost the original centromere (7HS*) and another had lost the centromere and at least a portion of the flanking pericentromeric DNA (7HS**). Importantly, all known centromere repeats and CR elements were absent on 7HS* and 7HS**. Nevertheless, both truncated chromosomes were completely stable in genetic crosses, and 7HS* reacted strongly with antisera to at least four known kinetochore proteins.
Fig. 2.
The origin of barley neocentromeres as described by Nasuda et al. (4). The study began with a translocation between barley chromosome 7HS and an unidentified wheat chromosome. An isochromosome derivative of the translocation was identified that had lost the wheat half of the chromosome as well as any evidence of the barley centromere. From the isochromosome, two telosomic derivatives were produced that appeared to have lost even larger segments of the original centromere. The telosomes lacked known centromere repeats from either barley or wheat yet were mitotically and meiotically transmissible.
The data are very similar to prior observations in Drosophila that suggest centromeres can spread into flanking chromatin (16). The primary difference is that the Drosophila centromere moved only into euchromatin, whereas barley centromere 7H appeared to move into (or over) heterochromatin. It is possible that in both cases the neocentromeres were close enough to an established centromere that small amounts of CENH3 and associated proteins were carried over (4, 16). The centromeric chromatin may then have spread and adopted a domain similar in size to the original centromere but with few or none of the original DNA sequences.
Nasuda et al. (4) have demonstrated convincingly that plant centromeres do not require satellite repeats to function normally and provided new support for the view that neocentromeres are established epigenetically. Indeed, although tandem repeat arrays can initiate new centromeres under experimental conditions (17), there is little evidence that it occurs naturally. It appears that centromere repeats normally accumulate after a new centromere has been established, selfishly and (at least initially) without regard to organismal fitness (6, 10, 12, 15). Despite this unlikely mode of evolution, simple repeats have emerged as dominant centromeric sequences in nearly all plants and animals. Future work is likely to focus on the proteins involved in the earliest stages of (neo)centromere formation as well as these still-enigmatic repeats, including what sequence features allow them to contribute to centromere formation and what proteins facilitate their interactions with the kinetochore.
Author contributions: R.K.D. wrote the paper.
See companion articles on pages 11793 in this issue and 9842 in issue 28 of volume 102.
References
- 1.Choo, K. H. A. (2001) Dev. Cell 1, 165–177. [DOI] [PubMed] [Google Scholar]
- 2.Karpen, G. H. & Allshire, R. C. (1997) Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- 3.Lee, H.-R., Zhang, W., Langdon, T., Jin, W., Yan, H., Cheng, Z. & Jiang, J. (2005) Proc. Natl. Acad. Sci. USA 102, 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasuda, S., Hudakova, S., Schubert, I., Houben, A. & Endo, T. R. (2005) Proc. Natl. Acad. Sci. USA 102, 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang, J., Birchler J. A., Parrott W. A. & Dawe, R. K. (2003) Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- 6.Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R. & Jiang, J. (2004) Nat. Genet. 36, 138–145. [DOI] [PubMed] [Google Scholar]
- 7.Houben, A. & Schubert, I. (2003) Curr. Opin. Plant Biol. 6, 554–560. [DOI] [PubMed] [Google Scholar]
- 8.Langdon, T., Seago, C., Mende, M., Leggett, M., Thomas, H., Forster, J., Jones, R. & Jenkins, G. (2000) Genetics 156, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topp, C. N., Zhong, C. X. & Dawe, R. K. (2004) Proc. Natl. Acad. Sci. USA 101, 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 11.Hass, B. L., Pires, J. C., Porter, R., Phillips, R. L. & Jackson, S. A. (2003) Theor. Appl. Genet. 107, 773–782. [DOI] [PubMed] [Google Scholar]
- 12.Malik, H. S. & Henikoff, S. (2002) Curr. Opin. Genet. Dev. 12, 711–718. [DOI] [PubMed] [Google Scholar]
- 13.Schrago, C. G. & Russo, C. A. (2003) Mol. Biol. Evol. 20, 1620–1625. [DOI] [PubMed] [Google Scholar]
- 14.Alves, G., Seuanez, H. N. & Fanning, T. (1994) Chromosoma 103, 262–267. [DOI] [PubMed] [Google Scholar]
- 15.Dawe, R. K. & Hiatt, E. N. (2004) Chromosome Res. 12, 655–669. [DOI] [PubMed] [Google Scholar]
- 16.Maggert, K. & Karpen, G. (2001) Genetics 158, 1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu, J., Stromberg G., Compitello G., Willard H. F. & Van Bokkelen G. (2005) Nucleic Acids Res. 33, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge, S., San, T., Lu, B.-R. & Hong, D.-Y. (1999) Proc. Natl. Acad. Sci. USA 96, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]