Abstract
Seemingly contradicting results raised a debate over the ability of DNA to transport charge and the nature of the conduction mechanisms through it. We developed an experimental approach for measuring current through DNA molecules, chemically connected on both ends to a metal substrate and to a gold nanoparticle, by using a conductive atomic force microscope. Many samples could be made because of the experimental approach adopted here, which enabled us to obtain reproducible results with various samples, conditions, and measurement methods. We present multileveled evidence for charge transport through 26-bp-long dsDNA of a complex sequence, characterized by S-shaped current–voltage curves that show currents >220 nA at 2 V. This significant observation implies that a coherent or band transport mechanism takes over for bias potentials leading to high currents (>1 nA).
Keywords: molecular electronics, scanning probe microscopy, nanoelectronics
Experimental observations that seemed to be in dissonance raised a debate over the ability of DNA to transport charge and the nature of the conduction mechanisms through it (1–13). These conflicts stem from the variety of measurement approaches, sample preparations, experimental setups, and environmental conditions. The main factors that were difficult to control in those experiments were the interaction of DNA with the substrate (1, 13) and the contacts between the molecules and the electrodes (1, 14). Inspired by Cui et al. (14) and Xu et al. (15), we devised an experimental approach that overcomes these difficulties (16). Current passing through the dsDNA molecules is measured by using a metal-covered atomic force microscope (AFM) tip, while the molecules are chemically connected to a metal substrate at one end and to a gold nanoparticle (GNP) at the opposite end. Here we present multileveled evidence for charge transport through dsDNA molecules of a complex sequence, 26 bp long, characterized by S-shaped current–voltage (I–V) curves in which we measured currents >220 nA at 2 V. This significant observation is supported by topography–current maps, comparative I–V measurements, and 3D-mode (17) and current-stretching experiments. It implies that some coherent or band transport mechanism takes over for the higher currents (18).
Methods and Techniques
In the present study, ssDNA molecules (5′-CAT TAA TGC TAT GCA GAA AAT CTT AG-3′-C3H6-SH) are connected via a propyl-thiol end group to a gold surface, forming a packed monolayer. Complementary strands are connected via thiol groups to a 10-nm GNP. One or a few of them are then hybridized with the single strands that had been adsorbed on the surface to form a dsDNA molecule(s) connecting the GNP and the surface.
Full details of the sample preparation were reported recently (16). Briefly, the 3′-thiolated ssDNA is maintained protected in its oxidized form, (CH2)3-S-S-(CH2)3-OH, until usage. Before adsorption on the gold surface or on the GNP, the protecting group is removed by reduction with Tris(2-carboxyethyl) phosphine. The reduced DNA oligomer is then separated from all residues on a column (BioSpin 6, Bio-Rad) and immediately pipetted onto a clean, annealed gold surface. The reduced and purified thiolated complementary ssDNA (5′-CTA AGA TTT TCT GCA TAG CAT TAA TG-3′-C3H6-SH) oligomer is stirred with the GNP suspension (the GNP is rinsed in deionized water prior to the DNA adsorption to remove excess surfactant). The resulting ssDNA-GNP derivative is diluted with Tris·HCl buffer and pipetted onto the ssDNA monolayer. Incubation of the sample for 12 h at 100% relative humidity affords hybridized dsDNAs that are connected at both ends as described. The sample is rinsed with fresh buffer solution to remove nonspecifically bound ssDNA-GNP. Just before AFM measurements the sample is rinsed with deionized water to remove excess salts. Depending on the monolayer density, up to 10 parallel molecules can fit underneath the GNP.¶ Electrical measurements are then performed by using conductive AFM. A schematic of this configuration is shown in Fig. 1a.
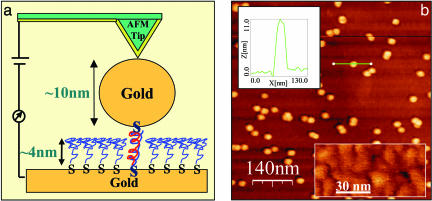
Fig. 1.
Sample layout and AFM image. (a) Schematic diagram: a smoothed gold substrate is covered by a packed monolayer of 26-bases-long ssDNA molecules of a complex sequence, ≈4 nm thick, connected to the substrate via (CH2)3 spacers and thiol end groups. Complementary strands, connected with a (CH2)3 spacer and a thiol end group to a 10-nm GNP, are hybridized with the monolayer single strands. Hence, the dsDNA molecules are connected chemically to the metal substrate and to the GNP only at their ends and do not come in direct contact with the metal substrate. The gold particles are contacted by means of a metal-covered AFM tip to close the electrical circuit. (b) An AFM image of the sample. The GNPs that are connected to the substrate through the dsDNA molecules and partially cover the surface are evident on the background of the ssDNA monolayer. (Upper Inset) A cross section over one of the GNPs (green line) is shown. The height of the GNP is ≈10 nm over the monolayer. (Lower Inset) A90 × 50-nm2 area of the ssDNA monolayer (in a better color contrast) is shown. The observed features are composed of ssDNA.
Images were obtained with a commercial AFM (Nanotec Electronica, Madrid) in dynamic mode. Soft cantilevers (OMCL-RC800PSA, Olympus Optical Co., Ltd., Tokyo) with a nominal force constant of 0.3 Nm–1, resonance frequency of 75–80 kHz, and tip radius of 15–20 nm were used (the tip radius was measured with a scanning electron microscope). For the electrical measurements, the same tips are covered with ≈3 nm of Cr and ≈15–30 nm of evaporated gold or directly sputtered gold/palladium. Consequently, the tip radius grows to 25–40 nm, the force constant increases to 1 Nm–1, and the resonance frequency drops to 50–70 kHz after coverage. The cantilever is oscillated close to its resonance frequency, and the amplitude and the phase, relative to the oscillatory driving force, are measured through the tip deflection signal as detected by the photodetector. The feedback is performed on the amplitude signal channel.
The tip is typically a few to tens of nanometers above the surface, and the oscillation amplitude for imaging is 5–50 nm. The surface morphology of the samples, characterized by using AFM, is shown in Fig. 1b. The surface of the single-stranded monolayer appeared rather smooth (<1 nm roughness), although the “bulges” of the ssDNA molecules can be observed (Fig. 1b, Lower Inset). The 10-nm GNPs, which are connected to the dsDNA, appear very clear on the background of this monolayer (Fig. 1b).∥ The measured height of the GNP above the monolayer surface is ≈10 nm (Fig. 1b, Upper Inset), which means that the double strands formed do not protrude above the thickness of the monolayer, which is ≈4 nm. Because the length of the dsDNA is ≈9 nm, this difference implies that the dsDNA molecules are tilted relative to the surface normal, as expected for thiols (see also scheme in Fig. 4 Inset) (19, 20). Note that when the noncomplementary ssDNA oligomers are attached to the GNP, the number of particles observed on the surface is 2 orders of magnitude smaller (16).
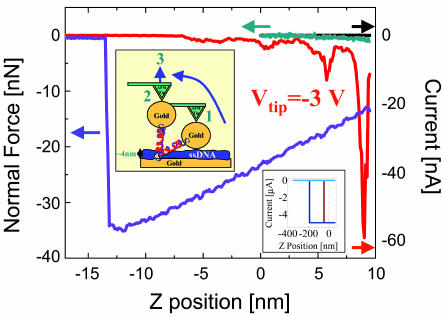
Fig. 4.
Current–distance (black, forward; red, backward) and force–distance (green, forward; purple, backward) curves measured simultaneously. A voltage of –3 V was applied to the tip during retraction. The current rise (a negative value for negative voltage) that we observed during retraction of the tip is interpreted as being caused by a more relaxed configuration of the molecule when the tilt of the molecule is first recovered (see scheme in the Inset). The current then drops rapidly with the stretching. The bumps in the current may be related to sequential stretching steps or any other structural effect. Note that the length calibration is correct for the first few nanometers, but when the molecule is strongly stretched, part of the tip motion is incorporated in cantilever bending, and one can assume that the distance from +5 to –5 nm (zero is the tip distance above the surface before approach, the starting position depending on the set point) should be somewhat scaled down. (Inset) Similar measurements performed on a GNP that is connected to noncomplementary strands (no current), on the ssDNA monolayer (no current), on a GNP laid on the bare gold (brown), and directly on the bare gold (blue). The latter two measurements show very high currents (>5 μA) that last over long stretching distances (50 and 100 nm, mostly cantilever deflection), probably because of the metals melting together. These measurements prove that the currents that we measured cannot be attributed to direct GNP-surface contact.
Results
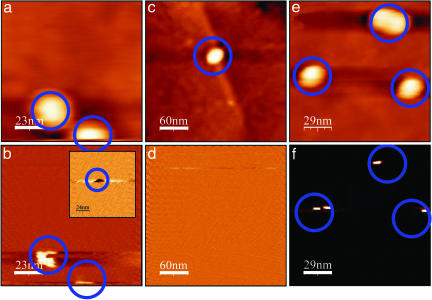
The conductive properties of the dsDNA molecules attached to the GNP and the insulating behavior of the ssDNA layer can be demonstrated qualitatively by comparing the topography and the current images, which are measured simultaneously. Fig. 2 shows alternating measurements on complementary (a, b, e, and f) and noncomplementary (c and d) strands connected to the GNPs, such that with the complementary strands dsDNA are formed and with the noncomplementary strands no dsDNA is formed. The topography images are shown in a, c, and e. No current is observed in current images measured simultaneously with the topography images at zero voltage (as in the image in d). b, d, and f were measured together with the following topography image at a 4-V bias voltage. After the 4-V application current signal was observed only for dsDNA at the position of the GNPs. Four volts is larger than the maximum voltage used in the I–V curves shown later and therefore demonstrates that even at this high voltage no leakage current is observed on the ssDNA monolayer. The neutral background indicates that the ssDNA monolayer insulates even at a high bias voltage. This measurement confirms that current flows only when the GNPs connected to the dsDNA are contacted and not through the ssDNA monolayer.
Fig. 2.
Simultaneously acquired topography and current maps. Shown are measurements on a sample with GNPs connected to dsDNA (a and b; 115 × 115 nm2), on a sample with GNPs connected to noncomplementary strands (the same strand as on the surface) (c and d; 300 × 300 nm2), and immediately after on a sample with GNPs connected to a dsDNA (to prove that the tip is still conductive) (e and f; 170 × 170 nm2). a, c, and e show topography images. No current is observed for current maps recorded simultaneously with a, c, and e with zero bias voltage on the tip (V = 0). b, d, and f present current maps measured simultaneously with the next topography images (not shown), for which 4 V were applied to the tip. A current signal is clearly observed at the positions corresponding to the positions of GNPs connected to dsDNA (blue circles), and no current is observed for the noncomplementary ones. An opposite signal is measured for –4V (b Inset). No current is measured in any other position on the ssDNA in all cases.
More quantitative and detailed information is obtained from the electrical transport measurements presented in Fig. 3, which shows I–V curves measured between the metallized tip and the substrate in various configurations. The AFM software was especially modified to have a good control over the applied force and the tip position relative to the sample while performing the electrical measurements. The distance between the tip and the particle, or the surface under the tip, is reduced while recording the tip deflection (green curves in the insets). An I–V curve is acquired at a predefined distance (presented in the insets together with the measurements illustrated). The measurements were done at various humidity conditions ranging from ambient (relative humidity of ≈50–85%) to N2 atmosphere (relative humidity of ≈15%) with no observable difference. When performing a measurement in a nonvacuum or nonliquid environment, as are all of the measurements reported here, the tip–surface distance is reduced until “jump to contact” (JTC) between the tip and the surface occurs (green curves). Then, the tip and the surface underneath move together while the AFM cantilever is deflected further (Fig. 3d, Inset). To avoid tip-loading force on the particle or the monolayer, the approach can be stopped just before the JTC (Fig. 3 a–c, Insets) by either limiting the approach distance or limiting the approach by using a feedback from the amplitude reduction. When the tip is retracted (red curves), a hysteresis is observed as a result of remaining tip–surface contact caused by adhesion and capillary force resulting from a water meniscus located between the tip and the surface (forces in the nanonewton range). The tip–particle (or tip–ssDNA monolayer) contact interaction has the magnitude of the adhesion force alone with no apparent contribution of the tip load. “Snap off contact” (SOC) occurs at a distance farther away from the surface than the JTC distance. The adhesion indicates that good contact was established between the tip and the particle or the surface underneath. Before the JTC and after the SOC, the tip oscillates near its resonance frequency to improve its stability** (as seen in all of the insets).
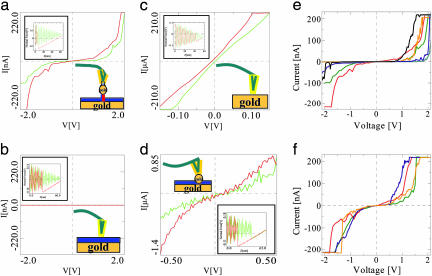
Fig. 3.
I–V curves that were measured at a preset height above the surface at the closest point to the surface during force–distance measurements (shown in the insets). The approach in measurements a–c was halted just before the JTC of the tip to avoid pressure on the surface or the particle underneath. The hysteresis in the backward curves in the insets is caused by adhesion and indicates that good contact was established with the surface or the particle underneath. The green curves are the “forward” and were measured first; the red curves are the “backward” and were measured after the green ones. It can be observed that the current values in the red curves are usually higher than those of the green curves. This is probably because of the formation of improved contact between the metal tip and the particle or the surface during the first measurement. (a) This measurement was performed on a metal particle without pressing on it, as seen in the inset. The current measured is >220 nA at 2 V. (b) I–V curve, measured on the ssDNA monolayer without pressing it. Negligible current was observed here, indicating that the monolayer is insulating. (c) A measurement that was taken on a bare gold surface showing ohmic behavior, as expected from metal–metal contact. The resistance here is ≈500 Ω. (d) I–V curve measured on a metal particle, connected to the surface through a dsDNA while pressing it strongly to the metal substrate, as seen in the force–distance curve in the inset. This curve also shows a nearly linear behavior. (e and f) Two sets of I–V curves that were measured on different GNPs on different samples, tips, and dates, which demonstrate the variation in the data. Lower maximal currents were measured as well in many cases (not shown). e emphasizes curves with a gap, whereas f emphasizes curves with a nonzero or small gap.
Fig. 3a shows a typical I–V curve that was measured when the metal tip was brought in contact with one of the GNPs without pressing it (see Inset). Typically, when the voltage exceeds ≈1V, a gradual rise in the current is observed. The current can reach values of up to 220 nA at 2 V. Beyond this value the preamplifier is saturated, and therefore higher values cannot be recorded. The curves are generally not symmetric, probably because of asymmetry in the contacts and the molecule itself. They have an S-type shape, when typically the resistance measured between –1 and 1 V is ≈60 MΩ. At ≈2 V the resistance is of the order of 2 MΩ. The reproducibility of the general curve shapes in consecutive sets of measurements indicates their stability at these voltages.
The flat curve in Fig. 3b was measured on the ssDNA monolayer without pressing it (the applied force is similar to that applied for Fig. 3 a and c), as shown in the inset. Again, the hysteresis in the retraction force–distance curve indicates that good contact was established between the tip and the surface. This curve is representative of many measurements that were performed in many positions on the monolayer.†† Similar measurements were done on GNPs labeled with noncomplementary ssDNA with no current rise. All these curves indicate insulating behavior. Fig. 3c shows an I–V curve that was measured on a bare gold surface to check the reliability of the measurement method. An ohmic curve was obtained (before JTC), showing resistance of ≈500 Ω (the resistance in these measurements at different positions spans 4–500 Ω), as expected from contact between a metal tip and a metal substrate. Finally, Fig. 3d presents a curve that was measured on one of the GNPs while pressing it strongly to the surface (see Inset). The measured current (order of 1 μA) results from a contact with the substrate through the pressed ssDNA monolayer. Fig. 3 e and f show two collections of curves that were measured on various GNPs and different samples with different tips. Note that the maximal Coulomb blockade signature of a completely isolated 10-nm GNP is ≈0.2 eV, and therefore its contribution to the shape of the I–V curve is minimal.
To further verify that the high current does not originate from any direct contact between the GNP and the metal surface, we monitored the current at a fixed voltage, –3 V, applied to the tip while retracting the tip and monitoring the force vs. distance. Fig. 4 presents the results obtained from this procedure; both the force–distance and the current–distance curves, measured simultaneously, are shown. The current value (negative because of the voltage sign) increases at the beginning of the retraction (red curve, right to left) and then decreases, first sharply and then in jumps that can be interpreted in several ways. The initial increase in the current is possibly caused by the structural relaxation of the dsDNA after straightening of the propyl group, through which the DNA is attached to the surface. This alkane chain is most probably initially not in its “all-trans” configuration. When the chain is stretched by the retraction of the tip, the propyl group is transformed to the all-trans configuration, which leads to better conductivity (19). The adhesion forces between the tip and the particle are in the nanonewton range, whereas the interplanar forces in the DNA are in the 10- to 100-pN range, indicating that the molecule is stretched when the particle is still attached to the tip, and only then is the meniscus also stretched and SOC occurs. These measurements show that the current falls off over a few nanometers, in accordance with current flowing only through the molecule. Fig. 4 Inset shows similar stretching measurements done on GNP connected to a noncomplementary strand (no current), on the ssDNA (no current), on a GNP laid directly on the bare gold, and directly on the bare gold. The last two measurements show high current (>5 μA) over a long stretching distance (50 and 100 nm), probably caused by the gold melting and a sharp SOC followed by an immediate current suppression. This clear difference in behavior between the control experiments and the dsDNA-bound GNP verifies that current flows only through the dsDNA and not through any direct contact of the GNP with the metal surface.
The water layer and existing counter ions around the DNA do affect the DNA structure and therefore, in principle, may affect the results. Their existence, however, cannot explain by itself the observations, because also in the case of ssDNA the water and the counter ions exist, but no current was monitored. However, one should realize that in the case of ssDNA, no GNP-DNA-substrate junction is present.
Discussion
The reported measurements clearly demonstrate the ability of short dsDNA molecules to transport relatively high currents when the potential across the molecule exceeds ≈ 1 V. The current, measured through a complex sequence in the present studies (>220 nA at 2 V), is at least 22 nA per molecule at 2 V and is considerably higher than measured before in “dry” DNA (6, 7, 10–12). It has already been shown for other molecules that in several cases the current is increased by ≈3 orders of magnitude when the molecule is bonded chemically to the electrodes (21). Hence, we attribute our observation to the fact that on one hand the dsDNA is chemically bonded to the contact electrodes, and on the other hand it is not interacting with the substrate along its length. Furthermore, it was shown that control over the vertical applied force when performing the I–V curves is of major importance for the stability of the measurements (17, 22). The controlled AFM-measurement method developed here significantly contributes to the stability and reproducibility of the electrical measurements. In addition, the stretching measurement (measuring the current at a constant bias while extending the tip substrate separation) showed that the current amplitude depends strongly on the dsDNA distortion and therefore on the applied force. Consequently, the control over the force enables us to observe the high current measured here.
Several mechanisms have been suggested for charge transport in DNA (1, 3, 4, 18). Our measurements may be consistent with the mechanisms of incoherent hopping for the low currents (<1 nA) but clearly imply the existence of a faster conduction mechanism (e.g., coherent or band transport for the higher bias potential at which the current rises). This mechanism may be of importance at least at room temperature, at which phononassisted interactions may take place (18). The high measured current, although surprising, does not necessarily contradict previous reported results (1–13), in which the currents may have been limited by the attachment to the surface or by noncovalent bonding to the metal electrodes. It diverges, however, from the mechanisms that are suggested to account for the kinetic measurements in solution. The maximal possible currents expected according to this mechanism is well below 1 nA (18). In the kinetic measurements a donor and an acceptor are attached to the dsDNA at a distance; after photoexcitation, a single charge carrier (usually a hole) is injected into the chain and travels the distance, and the process is followed by a recombination event (23). In the present study, as well as in the experiment performed by Xu et al. (15), the dsDNA molecule is covalently attached to two metal electrodes. As a result, the electrochemical potentials (Fermi levels) of the molecule and the electrodes must equalize. This process involves either significant charge reorganization on the molecule or charge transfer between the metal electrodes and the molecule. This equalization process changes substantially the energy levels of the DNA. Hence, in this case, the charge transport mechanisms can differ significantly from the incoherent hopping process, commonly accepted for the kinetic measurements. We have no sufficient experimental data to clearly identify the specific mechanism(s) acting in our measurement, but possible ones could include multicharge injection into the chain (limited by polaron formation), fast hopping with reduced loss of phase, and band transport. The latter is inconsistent with previous calculations that report a wide band gap and a narrow band width (e.g., by de Pablo et al. in ref. 8). In none of these calculations, however, was the covalent attachment to the electrodes accompanied by charge injection and band bending taken into account (as well as the backbone). Therefore, the band-conduction picture may differ from those calculations, and band conduction that allows for high currents should still be considered.
Generally, our results are consistent with scanning tunneling microscopy measurements reported by Xu et al. (15), in which large statistics on the conductivity of dsDNA covalently connected to the tip and the substrate on opposite ends was monitored in aqueous solution, in which the B-form DNA structure is apparently preserved. They measured currents of similar magnitude for shorter, 8- to 14-bp-long dsDNA, however, with no gap in the I–V curves, as observed in some of our measurements. The similarity in the results of both experiments indicates that the electrode–bridge–electrode configuration vs. donor–bridge–acceptor configuration is much more important than the difference between aqueous and ambient environments.
Acknowledgments
We thank Shirley Daube, Ignacio Horcas, Noa Lapidot, Julio Gomez, and Igor Brodsky for assistance with sample preparation and measurements and for fruitful discussions. The research was supported by the FIRST foundation, the Israeli Academy of Sciences and Humanities, and the German Israel Foundation and by European grant for Future and Emerging Technologies IST-2001-38951.
Author contributions: H.C., C.N., R.N., and D.P. designed research; H.C. and C.N. performed research; H.C., C.N., R.N., and D.P. analyzed data; H.C., C.N., R.N., and D.P. wrote the paper; H.C. developed the controlled measurement methods, prepared some of the samples, and performed the electrical measurements; C.N. developed the sample configuration and performed the sample preparation; R.N. was the group leader for sample preparation; and D.P. was the group leader for electrical measurements.
Abbreviations: AFM, atomic force microscope/microscopy; GNP, gold nanoparticle; I–V, current–voltage; JTC, jump to contact; SOC, snap off contact.
Footnotes
The surface area of the GNP is 300 nm2. If 25% of the bottom is available for dsDNA attachment and the surface area per ssDNA is ≈2.5 × 2.5 nm2, then there is room for ≈10 molecules at most. This value is in accord with the ssDNA coverage density, ≈3 × 1013 cm2 (as found from radioactive labeling of the ssDNA). The coverage of dsDNA, however, is likely to be smaller than 100%; thus, probably <10 molecules are connecting the GNP and the substrate.
The ssDNA monolayer thickness was characterized by both AFM (by “shaving” a square on the surface) and ellipsometry and was found to be ≈4 nm. The GNP coverage was measured by using AFM and was found to be ≈500 GNPs/μm2 for complementary strands (dsDNA) and ≈5 GNPs/μm2 for noncomplementary strands (same strand as the one connected to the surface).
The observed oscillation is caused by the driving oscillation but appears with a lower frequency because of aliasing.
Additional measurements, performed in 3D mode on the ssDNA monolayer (22), verify that the tip deflection after voltage application is <2 Å and does not affect the I–V results. In addition, the current was monitored while the tip penetrated ≈4 nm into the ssDNA monolayer, and it was found that current rises only when good contact is established between the tip and the metal surface.
References
- 1.Porath, D., Cuniberti, G. & Di Felice, R. (2003) in Topics in Current Chemistry, ed. Shuster G. (Springer, Berlin), Vol. 237, pp. 183–227. [Google Scholar]
- 2.Dekker, C. & Ratner, M. (2001) Phys. World 14, 29–33. [Google Scholar]
- 3.Endres, R. G., Cox, D. L. & Singh, R.R.P. (2004) Rev. Mod. Phys. 76, 195–214. [Google Scholar]
- 4.Di Ventra, M. & Zwolak, M. (2003) in DNA Electronics: Encyclopedia of Nanoscience and Nanotechnology, ed. Singh Nalwa, H. (American Scientific Publishers, Stevenson Ranch, CA), pp. 475–493.
- 5.Braun, E., Eichen, Y, Sivan, U. & Ben-Yoseph, G. (1998) Nature 391, 775–778. [DOI] [PubMed] [Google Scholar]
- 6.Fink, H. W. & Schönenberger, C. (1999) Nature 398, 407–410. [DOI] [PubMed] [Google Scholar]
- 7.Porath, D., Bezryadin, A., de Vries, S. & Dekker, C. (2000) Nature 403, 635–638. [DOI] [PubMed] [Google Scholar]
- 8.de Pablo, P. J., Moreno-Herrero, F., Colchero, J., Gomez-Herrero, J., Herrero, P., Baro, A. M., Ordejon, P., Soler, J. M. & Artacho., A. (2000) Phys. Rev. Lett. 85, 4992–4995. [DOI] [PubMed] [Google Scholar]
- 9.Storm, A. J., van Noort, J., de Vries, S. & Dekker, C. (2001) Appl. Phys. Lett. 79, 3881–3883. [Google Scholar]
- 10.Watanabe, H., Manabe, C., Shigematsu, T., Shimotani, K. & Shimizu, M. (2001) Appl. Phys. Lett. 79, 2462–2464. [Google Scholar]
- 11.Zhang, Y., Austin, R. H., Kraeft, J., Cox, E. C. & Ong, N. P. (2002) Phys. Rev. Lett. 89, 198102. [DOI] [PubMed] [Google Scholar]
- 12.Shigematsu, T., Shimotani, K., Manabe, C., Watanabe, H. & Shimizu, M. (2003) J. Chem. Phys. 118, 4245–4252. [Google Scholar]
- 13.Kasumov, A. Y., Klinov, D. V., Roche, P.-E., Gueron S. & Bouchiat, H. (2004) Appl. Phys. Lett. 84, 1007–1009. [Google Scholar]
- 14.Cui, X. D., Primak, A., Zarate, X., Tomfohr, J., Sankey, O. F., Moore, A. L., Moore, T. A., Gust, D., Harris, G. & Lindsay, S. M. (2001) Science 294, 571–574. [DOI] [PubMed] [Google Scholar]
- 15.Xu, B., Zhang, P., Li, X. & Tao, N. (2004) Nano Lett. 4 (6), 1105–1108. [Google Scholar]
- 16.Nogues, C., Cohen, S. R., Daube, S. S. & Naaman, R. (2004) Phys. Chem. Chem. Phys. 6, 4459–4466. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Navarro, C., Gil, A., Alvarez, M., de Pablo, P. J., Moreno-Herrero, F., Horcas, I., Fernandez-Sanchez, R., Colchero, J., Gomez-Herrero, J. & Baro, A. M. (2002) Nanotechnology 13, 314–317. [Google Scholar]
- 18.Bixon, M. & Jortner, J. (1999) Adv. Chem. Phys. 106, 35–203. [Google Scholar]
- 19.Haran, A., Waldeck, D. H., Naaman, R., Moons, E. & Cahen, D. (1994) Science 263, 948–950. [DOI] [PubMed] [Google Scholar]
- 20.Kelley, S. O., Barton, J. K., Jackson, N. M. & Hill, M. G. (1997) Bioconjugate Chem. 8, 31–37. [DOI] [PubMed] [Google Scholar]
- 21.Salomon, A., Cahen, D., Lindsay, S., Tomfohr, J., Engelkes, V. B. & Frisbie, C. D. (2003) Adv. Mater. 15, 1881–1890. [Google Scholar]
- 22.Cohen, H., Nogues, C., Ullien, D., Daube, S., Naaman, R. & Porath, D. (2005) Faraday Discuss., in press. [DOI] [PubMed]
- 23.Murphy, C. J., Arkin, M. R., Jenkins, Y., Ghatlia, N. D., Bossmann, S. H., Turro, N. J. & Barton, J. K. (1993) Science 262, 1025–1029. [DOI] [PubMed] [Google Scholar]