Abstract
The DNA of functional cis-regulatory modules displays extensive sequence conservation in comparisons of genomes from modestly distant species. Patches of sequence that are several hundred base pairs in length within these modules are often seen to be 80–95% identical, although the flanking sequence cannot even be aligned. However, it is unlikely that base pairs located between the transcription factor target sites of cis-regulatory modules have sequence-dependent function, and the mechanism that constrains evolutionary change within cis-regulatory modules is incompletely understood. We chose five functionally characterized cis-regulatory modules from the Strongylocentrotus purpuratus (sea urchin) genome and obtained orthologous regulatory and flanking sequences from a bacterial artificial chromosome genome library of a congener, Strongylocentrotus franciscanus. As expected, single-nucleotide substitutions and small indels occur freely at many positions within the regulatory modules of these two species, as they do outside the regulatory modules. However, large indels (>20 bp) are statistically almost absent within the regulatory modules, although they are common in flanking intergenic or intronic sequence. The result helps to explain the patterns of evolutionary sequence divergence characteristic of cis-regulatory DNA.
Keywords: genomic sequence conservation, indels, regulatory evolution
In the general case where the transcription factor target sites are not known in advance, interspecific sequence comparison is now the method of choice for physically identifying putative cis-regulatory modules in the intronic or intergenic DNA sequence of given animal genes. As has long seemed reasonable to assume on the grounds that they are functionally essential (1), these key regulatory units of the genome are evolutionarily conserved relative to flanking sequence. Thus, cis-regulatory modules can be detected computationally by interspecific comparison of the sequence surrounding the gene of interest, recognized as a block of sequence that has remained relatively similar between the two species, excised by PCR and incorporated in an expression vector. Their function can then be studied by direct gene transfer methods (e.g., refs. 2–12). The appropriate evolutionary species distance must be chosen: that is, not so close that unselected (i.e., “background”) sequence has not had time to diverge but not so far that the pattern of conservation has been lost by too much divergence. But at the “right” distance, cis-regulatory modules stand out from the immediately flanking background as patches of well conserved sequence that are usually several hundred base pairs in length and terminated at their boundaries by abrupt transitions to sequence that has diverged too greatly for easy computational alignment.
Despite its conventional rationale and remarkable practical usefulness, there has remained a deeply problematic aspect of the conservation of cis-regulatory module DNA. Cis-regulatory modules consist of clusters of transcription factor target sites, always of several different types, and some represented multiple times (for a review, see refs. 13 and 14). But as detailed functional studies have revealed the internal structure of some cis-regulatory modules, it has become clear that much of the sequence length that is included in the relatively conserved sequence patches identified by interspecific comparison must actually be located between, not within, the known transcription factor target sites. Why is this sequence conserved with respect to the external sequence flanking the module? Are we in fact missing a large fraction of the sequence-specific DNA–protein interactions? Is there some other function associated with cis-regulatory modular DNA that is sequence-dependent? Or do the true target sites somehow cast a “conservation shadow” about themselves and, if so, by what mechanism?
To address these questions, we examined five specific cis-regulatory modules from Strongylocentrotus purpuratus that were already known from earlier studies (15, 16) or are included in an ongoing analysis of an embryonic gene regulatory network (17, 18). The genes were otx, delta, gatae, wnt8, and brachyury. Each of these previously characterized cis-regulatory modules was known to be conserved to the typical extent over a stretch of several hundred base pairs in the DNA of another sea urchin species, Lytechinus variegatus. Outside the regulatory modules or protein coding exons, the genomic sequence of these two species is so divergent that it cannot reliably be aligned (19, 20). Therefore, we obtained the orthologous cis-regulatory modules and samples of the flanking sequence from the genome of a congener of S. purpuratus, Strongylocentrotus franciscanus. This strategy made possible a direct comparison of the modes of divergence, within and outside the cis-regulatory module sequences. In addition, an analysis of polymorphism within S. purpuratus in regulatory regions of the endo16 gene (21) proved directly relevant, and the results of that study were incorporated as well.
Materials and Methods
Genes and Cis-Regulatory Modules. The five cis-regulatory modules derive from three genes that encode transcription factors [namely, gatae (22), brachyury, and otx (23)] and two genes that encode signaling ligands [namely, delta (24) and wnt8 (25)]. The endo16 gene, also included in the analysis, encodes a terminal differentiation protein of the endoderm (26). The relevant cis-regulatory modules have been described (15, 16, 27, 28). Ordered and oriented bacterial artificial chromosome (BAC) sequence for all six genes is available from the authors upon request, for both S. purpuratus and L. variegatus; GenBank accession numbers are listed in Table 2, which is published as supporting information on the PNAS web site. A preliminary analysis of another transcription factor, gcm (29), was also undertaken (Fig. 4, which is published as supporting information on the PNAS web site).
The sequences used to perform an intraspecific comparison for the endo16 gene of S. purpuratus were collected from several sources: (i) those determined in the original study of the cis-regulatory modules controlling this gene (15, 16), (ii) a previously sequenced BAC insert, (iii) a contig from the whole-genome shotgun assembly (GenBank accession no. AAGJ00000000), and (iv) the sequences determined in the previous polymorphism study (21). Three active regions and nine flanking regions lying within the region 5′ of the conserved A and B modules of endo16 were analyzed.
Primer Design and Sequencing from S. franciscanus BACs. To obtain the tracts of sequence from the genomic regions surrounding the relevant cis-regulatory modules in S. franciscanus, primers that lie outside the highly conserved protein coding regions were required. For each gene, alignments between the S. purpuratus and L. variegatus BAC inserts had been previously performed in the course of the cis-regulatory analyses (20, 27, 28, 30, 31). To find suitable conserved regions for primer design, we also used blastn (32) and additional family relations (fr) analyses (20). For example, at a window size of 10 bp and a similarity of 90%, fr reveals tracts of conserved sequence easily seen in dot plots (28). Such highly conserved regions were taken as likely primer targets in the S. franciscanus sequence, because it is much less diverged from S. purpuratus than is L. variegatus (19). The fr routine produces a machine-readable XML file, which was used directly for computation of sets of PCR primer pairs, each of which lies in a conserved region. Primers were designed on the S. purpuratus sequence by using eprimer3 (33), and primer pairs were selected to yield overlapping products for sequencing. Appropriate BAC inserts from S. franciscanus served as templates in standard PCRs. For sequencing reactions, the amplified products were gel-purified, and the PCR primers were used as sequencing primers in standard Applied Biosystems Big Dye sequencing reactions, which were read on a 3730 DNA Sequencer (Applied Biosystems). Sequencing reads were assembled with the phred-phrap-consed package (34, 35) and mapped onto the S. purpuratus sequence with crossmatch (36). The crossmatch output was translated into XML and viewed in family relations. The assembled S. franciscanus sequences were preliminarily aligned to the S. purpuratus BAC sequences by using blastn to choose suitable regions for alignment with clustalw (37). Regions marked by long indels were examined by hand to confirm proper alignment. Identities, single base pair substitutions, and number and size of gaps were tabulated from the clustalw output. Approximately 30 kb of sequence was obtained by this method in the absence of any previously known tracts of S. franciscanus sequence. Primer-walking methods were used to fill in many of the sequence gaps and to obtain additional sequence. Draft S. franciscanus genomic sequence obtained by these means has been submitted to GenBank (see Table 3, which is published as supporting information on the PNAS web site).
Results and Discussion
Theory and Predictions. Several possible mechanisms could account for the relative preservation of genomic sequence since divergence from a common ancestor. The most obvious and potent of these mechanisms is selection against deleterious effects of sequence change, such as commonly operates within protein coding regions. Cis-regulatory modules are defined experimentally as DNA fragments that, as a whole, faithfully recreate given developmental patterns of expression in gene transfer experiments. They consist of the target sites for the transcription factors to which they respond, plus the sequence intervening between these sites. Although interspecific sequence comparisons indeed reveal cis-regulatory modules as long contiguous patches of sequence that is relatively well conserved with respect to the external sequence, it is not obvious why there would be deleterious effects of sequence change outside the specific base pairs that participate directly in chemical interactions with transcription factor amino acid side chains. In three-dimensional analyses of DNA–transcription factor complexes, detailed mutational studies, and “selex” assays, only a few base pairs per interaction are seen to be partially or wholly constrained, and these elements are commonly confined to short sequences typically ≈6–8 bp in length. Furthermore, for well studied examples, there is direct evidence that the actual transcription factor target sites often occupy less than half of the module length. This evidence is of several kinds, including (i) oligonucleotide mapping of all specific sites of DNA–protein interaction (e.g., ref. 38), (ii) numerous reconstruction and mutation studies in which modular sequences are altered without discernable effects on function except when constrained nucleotides within target sites are changed (e.g., refs. 14–16 and 39), (iii) studies on regulatory modules of which the transacting factors are known and the sites of their interaction can be recognized in the sequence (for reviews, see refs. 13 and 14), and (iv) comparative studies on orthologous cis-regulatory modules from animals that are so distant from one another that only the transcription factor target sites are unchanged (e.g., refs. 40–46). Here, it can be seen directly that the target sites themselves are spaced by intervening sequences that have undergone a great deal of change during evolution. The evidence combines to exclude the idea that the observed patterns of cis-regulatory module conservation are due to functional nucleotide-by-nucleotide selection across the whole length of the module.
A mechanism that might account for what is observed is as follows. In the evolution of cis-regulatory modules, the occurrence of indels that are large enough to be likely to affect adjacent target sites might be selectively disfavored, whereas the occurrence (fixation) of single-nucleotide substitutions and small indels between transcription factor target sites is not constrained, although change within the sites themselves is, of course, constrained. We now know for several cases that the rate of indel accumulation in unselected sequence is sufficiently high to account for a large fraction of the total sequence change during divergence (47–49). Given this fact, the relative suppression within cis-regulatory modules of large indels but not of small indels or single-nucleotide changes gives the following predictions: (i) Comparison of two genomes just sufficiently distant so that nonselected sequence cannot usually be aligned will indeed reveal cis-regulatory modules as internally aligned, and thus apparently conserved patches of sequence, because the occurrence of large indels rapidly generates sequence that cannot easily be aligned, whereas, until it approaches saturation, the occurrence of single-nucleotide substitutions or small indels does not. (ii) Within these patches, the rate of occurrence of single-nucleotide substitutions and of small (one or a few base pairs long) indels will be similar to the rate outside them after correcting for the fraction of the module included in the actually constrained target site sequence. (iii) At greater evolutionary distance, as small changes accumulate, the apparent conservation of the module as a whole will disappear, because similarities of the unconstrained portions of the intramodular sequence will be lost, and only the transcription factor target sites themselves will be retained as conserved sequence elements.
That cis-regulatory modules are in fact effectively identified by detection of patchy interspecific sequence conservation (2–12), consistent with prediction (i), is our starting point. Also consistent [with prediction (iii)] is the common observation that at great evolutionary distance, patchy sequence conservation of cis-regulatory modules can no longer be seen, even where gene transfer experiments reveal conserved target site function (e.g., refs. 40–46). However, to test this proposition directly, the requirements are (i) to ascertain sequence divergence within cis-regulatory modules that are already known experimentally to be functional, so that the comparison of sequences within and outside its boundaries is meaningful and (ii) that a species pair be used that is sufficiently close so that the genomic sequence can be unequivocally aligned both inside and outside selectively conserved features.
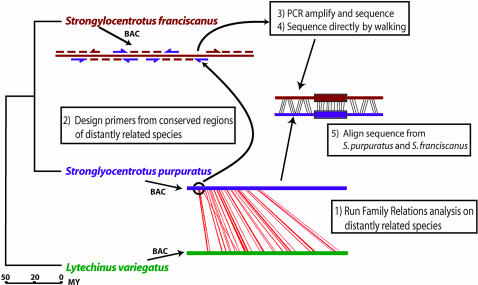
Sea Urchin Species. Extensive comparisons of genomic S. purpuratus vs. L. variegatus sequence around all genes included in this study had earlier revealed the conserved cis-regulatory modules to be flanked by sequence that is too divergent to be recognized (citations are given in Materials and Methods). The family Toxopneustidae, to which Lytechinus belongs, is believed to have diverged from the Strongylotrotidae ≈50 million years ago (50, 51). To be able to align and compare not only the orthologous cis-regulatory modules but also the flanking, freely diverging sequence, we turned to a member of the genus Strongylocentrotus known from earlier work, S. franciscanus (19, 52). The North Pacific radiation of the Strongylocentrotidae represented by S. franciscanus and S. purpuratus, which are today sympatric, is dated to ≈18 million years ago (51–54). The adult forms of these two species are in all respects very similar, except for the brick-red pigmentation and the much larger size of S. franciscanus. The phylogenetic relation of all three species is summarized in the diagram of Fig. 1.
Fig. 1.
The sea urchin evolutionary distances and the sequencing method. The phylogenetic tree derived from several sources (see text under the heading “Sea Urchin Species”) is depicted on the left side. The scale of divergence times in millions of years appears below the tree. To the right, the sequencing strategy is shown as a cartoon (“1”). A family relations comparison made between the BAC sequences of the more distantly related species, S. purpuratus (purple) and L. variegatus (green), is displayed as red lines (“2”). The conserved patches thus revealed are then used to design primers. An example of a conserved region thus used is circled, and an arrow points to the assortment of these primers used on the S. franciscanus BAC sequence (red) (“3” and “4”). Both standard PCR followed by sequencing and direct sequencing from the S. franciscanus BAC template were used with these primers (“5”). The resulting S. franciscanus sequence was aligned with the S. purpuratus sequence, and the number of gaps and substitutions was tallied.
Five genes were chosen, of which cis-regulatory modules had been discovered and characterized in other studies (see references in Materials and Methods). Although we had available whole BAC sequences covering the respective gene regions of L. variegatus and S. purpuratus (see Table 2), it was necessary to obtain the desired S. franciscanus sequence de novo. The starting point was to screen an S. franciscanus BAC library (55) so that we would have directly accessible genomic sequence in and around the test genes. As summarized in Fig. 1 and detailed in Materials and Methods, the S. franciscanus sequence desired for the present comparisons was obtained by two different approaches. Where the sequence similarity between L. variegatus and S. purpuratus genomes was very high (that is, in particularly conserved exons and in known and putative cis-regulatory modules), we included elements of these sequences in pairs of PCR primers that would be expected also to recognize the orthologous S. franciscanus sequence. The intervening DNA was thereby amplified from the S. franciscanus BAC and could be sequenced directly. Otherwise, the S. franciscanus sequence was obtained by “walking” directly on the BAC DNA, beginning with a conserved primer site. Maps of the S. purpuratus and S. franciscanus cis-regulatory and flanking sequences with respect to the exonic structure of each of the five genes are shown in Fig. 2.
Fig. 2.
The active and flanking region coverage from the S. franciscanus BAC sequence is shown for the five genes mapped onto the S. purpuratus BAC clones with the S. purpuratus exon positions for reference. The genomic sequence in the S. franciscanus BAC is depicted in light gray for flanking regions and in dark gray for active regions. The coordinates of the S. purpuratus BAC are indicated in kb at the end of the black line representing the sequence. The orientation of the sequence with respect to the direction of transcription is indicated outboard of the number (5′ and 3′).
Divergence Processes Within Cis-Regulatory Modules and in the Flanking Sequence. The intrageneric sequence comparisons that we obtained for the five cis-regulatory modules and their respective nearby external sequences are shown in Table 1 and Fig. 3 (sequence comparisons are available in Figs. 5–10, which are published as supporting information on the PNAS web site). Balhoff and Wray (21) carried out a comparable analysis of sequence divergence in the cis-regulatory domains of the endo16 gene within the species S. purpuratus, and we have recalculated their data in the same manner as that used for the S. franciscanus–S. purpuratus sequence comparisons obtained in this study. These results are also included in both Fig. 3 and Table 1. The endo16 gene resides in a rapidly evolving region of the genome; for example, unlike the case for all of the other genes in this study, none of the endo16 cis-regulatory modules that were identified experimentally (56, 57) display patchy sequence conservation between S. purpuratus and L. variegatus except for the proximal module A (58), whereas module B is partially conserved (C. H. Yuh and E.H.D., unpublished data). Here, we have taken modules A and B, for which every target site has been studied functionally in S. purpuratus (16), and considered them as bona fide cis-regulatory modules; the upstream regions sequenced by Balhoff and Wray (21), which contain the repressive modules F and E, part of D, and the distal booster module G (56, 57), are taken as the flanking sequence because it is entirely nonconserved to L. variegatus. This maneuver is a conservative one, for there could indeed be some conservation in these regions relative to true flanking sequence. Note, however, that the intraspecific divergence of these flanking regions among the 11 different individual genomes included in this analysis is equivalent in magnitude to the interspecific sequence divergence for the other five genes (Table 3). This divergence was compared with that among three different alleles of modules A and B (see alignments in the supporting information for details).
Table 1. The distribution of sequence features in the active and flanking regions of six genes.
| Gene | SNPs | Indels 1–5 | Indels 6–10 | Indels 11–15 | Indels 16–20 | Indels 21+ |
|---|---|---|---|---|---|---|
| brachyury | ||||||
| Active | 550.0 | 87.5 | 25.0 | 0.0 | 0.0 | 0.0 |
| Flanking | 1,498.6 | 162.9 | 32.2 | 8.5 | 3.4 | 13.6 |
| Ratio | 0.4 | 0.5 | 0.8 | 0.0 | 0.0 | 0.0 |
| delta | ||||||
| Active | 636.9 | 67.0 | 13.4 | 10.1 | 3.4 | 3.4 |
| Flanking | 880.3 | 111.6 | 18.6 | 15.1 | 1.2 | 12.8 |
| Ratio | 0.7 | 0.6 | 0.7 | 0.7 | 2.9 | 0.3 |
| gatae | ||||||
| Active | 657.0 | 33.4 | 22.3 | 0.0 | 0.0 | 0.0 |
| Flanking | 1,077.5 | 135.2 | 26.8 | 11.3 | 5.6 | 9.9 |
| Ratio | 0.6 | 0.2 | 0.8 | 0.0 | 0.0 | 0.0 |
| otx | ||||||
| Active | 287.3 | 62.8 | 9.0 | 0.0 | 0.0 | 0.0 |
| Flanking | 1,183.4 | 118.5 | 21.3 | 9.1 | 4.6 | 9.1 |
| Ratio | 0.2 | 0.5 | 0.4 | 0.0 | 0.0 | 0.0 |
| wnt8 | ||||||
| Active | 837.6 | 110.2 | 7.3 | 7.3 | 0.0 | 0.0 |
| Flanking | 2,249.1 | 165.4 | 40.7 | 21.6 | 10.2 | 17.8 |
| Ratio | 0.4 | 0.7 | 0.2 | 0.3 | 0.0 | 0.0 |
| endo16 | ||||||
| Active | 261.2 | 72.6 | 14.5 | 0.0 | 0.0 | 0.0 |
| Flanking | 927.6 | 117.7 | 23.2 | 13.9 | 11.1 | 22.2 |
| Ratio | 0.3 | 0.6 | 0.6 | 0.0 | 0.0 | 0.0 |
| Total | ||||||
| Active | 556.6 | 72.7 | 14.1 | 4.7 | 1.2 | 1.2 |
| Flanking | 1,271.3 | 133.0 | 26.7 | 13.7 | 6.4 | 14.9 |
| Ratio | 0.4 | 0.5 | 0.5 | 0.3 | 0.2 | 0.1 |
The data are arranged vertically to allow comparison of the active and flanking region values. The number in each category is normalized to the length of sequence examined. The third row for each gene is the number of features in the active region divided by the number of features in the flanking region.
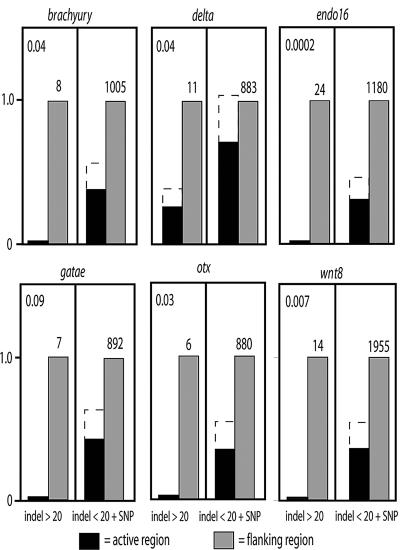
Fig. 3.
The distribution of indels and SNPs for active cis-regulatory sequence (black) compared with adjacent inactive sequence (gray). For each gene, the indels >20 bp are compared on the left side of the graph, and the indels <20 bp plus the SNPs are compared on the right side. The values are first normalized to the length of sequence measured and then divided by the value of the inactive fraction. The number of features in the inactive sequence is shown above the bar. The probability that the obtained results could occur randomly was tested by the relationship P(0) = e** - (ΣGF·LA/LF), where ΣGF is the sum of gaps in flanking sequence larger than the largest gap in the active sequence, LA is total length of active sequences analyzed for the given gene, and LF is total length of flanking sequence analyzed for the given gene.
Comparison within vs. outside the cis-regulatory regions consistently yielded two revealing statistics. First, single base pair changes and small indels indeed occur frequently within the cis-regulatory module sequences (shown on the right side of each graph in Fig. 3). In contrast, as shown on the left side of the graphs, larger indels are almost totally suppressed inside the regulatory modules with respect to their rate of occurrence in the flanking sequence. A simple Poisson metric shows that in five of the six cases (i.e., except for the gatae module), long indel suppression is highly improbable (P < 0.05) on random expectation, using the rate of occurrence of the large indel class in the flanking sequence as the model expectation. Larger indels are lacking within the gatae regulatory module as well but are also sufficiently rare in the flanking sequence to obscure the inside/outside difference. Details are given for each gene in the legend of Fig. 3. These comparisons indicate that the patchy sequence conservation relative to flanking regions of the genome that is so useful for identification of cis-regulatory modules has two separate causes. An important qualitative difference is the near absence of large indels within conserved modules; in addition, there is typically an ≈30–50% decrease in the frequency of small changes within, which could be due to restriction in change inside and immediately adjacent to target sites. Outside the modules, the much greater change in nearby sequence is due to not only accumulation of single-base changes and small indels but also the occurrence of large indels. Another gene not included in the sequence study presented here in which evolution is proceeding at a particularly rapid rate is the gcm gene (29). Here, as illustrated in Fig. 4, there is a remarkable incidence of large indels, which distinguish two alleles recovered from different S. purpuratus genomes. However, these large indels again occur exclusively outside, not inside, the known cis-regulatory modules.
Significance and Implications. Comparisons carried out at greater evolutionary distances have clearly illustrated the relative constraint on sequence change within target sites for transcription factors that are known to be important for function of cis-regulatory modules. For example, interspecific sequence comparison of enhancers that control pair rule gene expression in Drosophila [i.e., eve stripe 2 (44) and hairy (59) enhancers], hox gene enhancers in vertebrates (40, 41), and many others (27, 60) all show that target sites within these modules are generally spared the numerous single-base changes, insertions, and deletions that characterize much of the remaining sequence of the “minimum essential” regulatory module. In all of the cited examples, however, the evolutionary separation of the species compared is too great to allow an assessment of the amount of change in the less constrained intramodular sequence relative to the amount in the flanking extramodular sequence. The comparisons shown between S. franciscanus and S. purpuratus in Table 1 are illuminating in this respect: The ratio of single-base changes to small indels is generally similar gene to gene. Note, however, that the absolute rate of single base pair change varies ≈2-fold in the various flanking regions of the genome sampled but that the rate of small indel occurrence is quite remarkably invariant in these regions. For all cases, the ratio of the rates for single-base alterations and small indels, inside to outside the modules, lies between ≈0.3 and 0.8. The fact that we know from much other work (40–46) that the target sites themselves are relatively inflexible suggests that, depending on the module, from ≈30% to ≈80% of the length of the overall conserved module sequence patches are as unconstrained as are the sequence-independent flanking sequences. Thus, in the test set, as expected, for the cases where the important target sites are known [that is, for otx (27), wnt8 (T. Minokawa and E.H.D., unpublished data), and endo16 modules A and B (16)], almost none of the single-base changes and small indels occur within these known sites (see the supporting information). It is therefore not the case that patchy sequence conservation extending all across regulatory modules implies that we are missing a large fraction of sequence-specific intramodular interactions or that when viewed microscopically at this relatively close evolutionary distance, the whole of the module is constrained. Instead, it is correct to assume that the alterations in sequence that do occur have taken place in the fraction of the respective modules that is between transcription factor target sites, substantiating the above calculation.
Because a considerable fraction of the intersite distance within the modules is free to be altered by single-base changes and small indel events, probably at the unconstrained rate, it is to be expected that with time and divergence, the patchy conservation detectable by interspecific comparison algorithms will eventually disappear and only the sites themselves will survive, as is indeed seen (40–46). The mutual spacing of these sites, as well as their sequence environment, will be changed by the continuing superimposition of insertions as well as small deletions, except where spacing is a functional aspect of transcription factor interaction. At closer evolutionary distances, the selective constraint on large indels that this study reveals acts as a force that maintains the ancestral alignment of the module sequence longer than in flanking regions, where occurrence of these indels is less constrained. The argument that this constraint is a selective one that reflects the likelihood that large deletions will impinge on adjacent target sites is reasonable, given the average spacing of such sites: e.g., in the well studied modules A and B of the endo16 gene of S. purpuratus, there are 13 sites in a total domain of ≈500 bp, which includes an empty intermodular space, but also several regions where the sites are present in some proximity to one another (15, 16). In the cyIIIa gene (39), the sites are of similar density. In both regulatory systems, randomly positioned deletions >20 bp in length (Fig. 3) would frequently be dangerous. In addition, there could be a selective constraint on large insertions, given the fundamental and definitive requirement for transcription factor clustering in cis-regulatory modules (14, 61): The factors that interact with a given module must interact with one another to generate a combinatorial output (62). In essence, both larger insertions and larger deletions will tend to degrade the site cluster morphology of the regulatory module, and that is the general explanation of their suppression.
In summary, the sequence conservation that allows our computers to identify cis-regulatory modules at the right interspecific distance is the result of the more orderly process of single base pair and small indel fixation in free regions of the module sequence than occurs in external regions, where larger changes that destroy alignment relatively rapidly are permitted to take place. It is not caused, except indirectly, by conservation of transcription factor target sites per se, which persists at evolutionary distances after patchy conservation across the whole modular sequence has disappeared. Thus, an interesting note on the history of this field: cis-regulatory sequence is indeed conserved (for a while) but not exactly for the reason originally thought (1).
A practical consequence may follow from this view of near-term cis-regulatory sequence evolution. By comparing two genomes that are closely enough related so that they can be aligned everywhere (so that most indels can be detected), it should be possible to formulate a library of putative cis-regulatory sequence modules around any given gene through a computational search for domains of large indel suppression. This approach might provide an additional tool for cis-regulatory module prediction to add to those now available. It would depend on one specific kind of genomic change among many, and it could lessen the requirement for use of different interspecific species choices for detection of meaningful conservation in different regions of the genome (e.g., ref. 63).
Supplementary Material
Acknowledgments
We thank Applied Biosystems for their support. This work was also supported by National Science Foundation Grant IOB-0212869 and the California Institute of Technology Beckman Institute.
Author contributions: R.A.C. and E.H.D. designed research; R.A.C., S.H.C., K.B., T.-Y.C., Q.-A.Y., A.K., and A.H. performed research; A.R. contributed new reagents/analytic tools; R.A.C., E.H.D., and K. B. analyzed data; and R.A.C. and E.H.D. wrote the paper.
Abbreviation: BAC, bacterial artificial chromosome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ088382–DQ088386).
References
- 1.Britten, R. J. & Davidson, E. H. (1971) Q. Rev. Biol. 46, 111-138. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, M. A., Charchar, F. J., Kinston, S., Bird, C. P., Grafham, D., Rogers, J., Grutzner, F., Marshall, J. A., Green, A. R. & Göttgens, B. (2003) Genomics 81, 249-259. [DOI] [PubMed] [Google Scholar]
- 3.Göttgens, B., Barton, L. M., Gilbert, J. G. R., Bench, A. J., Sanchez, M.-J., Bahn, S., Mistry, S., Grafham, D., McMurray, A., Vaudin, M., et al. (2000) Nat. Biotechnol. 18, 181-186. [DOI] [PubMed] [Google Scholar]
- 4.Göttgens, B., Gilbert, J. G. R., Barton, L. M., Graham, D., Rogers, J., Bentley, D. R. & Green, A. R. (2001) Genome Res. 11, 87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin, C., Kleinjan, D. A., Doe, B. & van Heyningen, V. (2002) Mech. Dev. 112, 89-100. [DOI] [PubMed] [Google Scholar]
- 6.Hardison, R., Slightom, J. L., Gumucio, D. L., Goodman, M., Stojanovic, N. & Miller, W. (1997) Gene 205, 73-94. [DOI] [PubMed] [Google Scholar]
- 7.Kammandel, B., Chowdhury, K., Stoykova, A., Aparicio, S., Brenner, S. & Gruss, P. (1999) Dev. Biol. 205, 79-97. [DOI] [PubMed] [Google Scholar]
- 8.Kimura-Yoshida, C., Kitajima, K., Oda-Ishii, I., Tian, E., Suzuki, M., Yamamoto, M., Suzuki, T., Kobayashi, M., Aizawa, S. & Matsuo, I. (2003) Development (Cambridge, U.K.) 131, 57-71. [DOI] [PubMed] [Google Scholar]
- 9.Kleinjan, D. A., Seawright, A., Childs, A. J. & van Heyningen, V. (2004) Dev. Biol. 265, 462-477. [DOI] [PubMed] [Google Scholar]
- 10.Kurokawa, D., Takasaki, N., Kiyonari, H., Nakayama, R., Kimura-Yosida, C., Matsuo, I. & Aizawa, S. (2004) Development (Cambridge, U.K.) 131, 3307-3317. [DOI] [PubMed] [Google Scholar]
- 11.Margulies, E. H., NISC Comparative Sequencing Program & Green, E. D. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 255-263. [DOI] [PubMed] [Google Scholar]
- 12.Pennacchio, L. A., Baroukh, N. & Rubin, E. M. (2003) Cold Spring Harbor Symp. Quant. Biol. 68, 303-309. [DOI] [PubMed] [Google Scholar]
- 13.Arnone, M. & Davidson, E. H. (1997) Development (Cambridge, U.K.) 124, 1851-1864. [DOI] [PubMed] [Google Scholar]
- 14.Davidson, E. H. (2001) Genomic Regulatory Systems: Development and Evolution (Academic, San Diego).
- 15.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (1998) Science 279, 1896-1902. [DOI] [PubMed] [Google Scholar]
- 16.Yuh, C.-H., Bolouri, H. & Davidson, E. H. (2001) Development (Cambridge, U.K.) 128, 617-628. [DOI] [PubMed] [Google Scholar]
- 17.Oliveri, P. & Davidson, E. H. (2004) Curr. Opin. Genet. Dev. 14, 351-360. [DOI] [PubMed] [Google Scholar]
- 18.Levine, M. & Davidson, E. H. (2005) Proc. Natl. Acad. Sci. USA 102, 4936-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, T. J., Grula, J. W., Davidson, E. H. & Britten, R. J. (1980) J. Mol. Evol. 16, 95-110. [DOI] [PubMed] [Google Scholar]
- 20.Brown, C. T., Xie, Y., Davidson, E. H. & Cameron, R. A. (2005) BMC Bioinformatics 6, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balhoff, J. P. & Wray, G. A. (2005) Proc. Natl. Acad. Sci. USA 102, 8591-8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, P. Y. & Davidson, E. H. (2005) Gene Expression Patterns 5, 161-165. [DOI] [PubMed] [Google Scholar]
- 23.Li, X. T., Wikramanayake, A. H. & Klein, W. H. (1999) Dev. Biol. 212, 425-439. [DOI] [PubMed] [Google Scholar]
- 24.Sweet, H. C., Gehring, M. & Ettensohn, C.A. (2002) Development (Cambridge, U.K.) 129, 1945-1955. [DOI] [PubMed] [Google Scholar]
- 25.Wikramanayake, A., Peterson, R., Chen, J., Huang, L., Bince, J., McClay, D. & Klein, W. (2004) Genesis 39, 194-205. [DOI] [PubMed] [Google Scholar]
- 26.Soltysik-Espaqola, M., Klinzing, D. C., Pfarr, K., Burke, R. D.& Ernst, S. G. (1994) Dev. Biol. 165, 73-85. [DOI] [PubMed] [Google Scholar]
- 27.Yuh, C.-H., Dorman, E. R., Howard, M. L. & Davidson, E. H. (2004) Dev. Biol. 269, 536-551. [DOI] [PubMed] [Google Scholar]
- 28.Revilla-i-Domingo, R., Minokawa, T. & Davidson, E. H. (2004) Dev. Biol. 274, 438-451. [DOI] [PubMed] [Google Scholar]
- 29.Ransick, A., Rast, J. P., Minokawa, T., Calestani, C. & Davidson, E. H. (2002) Dev. Biol. 246, 132-147. [DOI] [PubMed] [Google Scholar]
- 30.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Science 295, 1669-1678. [DOI] [PubMed] [Google Scholar]
- 31.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C.-H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Dev. Biol. 246, 162-190. [DOI] [PubMed] [Google Scholar]
- 32.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 33.Rozen, S. & Skaletsky, H. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds. Krawetz, S. & Misener, S. (Humana, Totowa, NJ), pp. 365-386.
- 34.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 35.Ewing, B., Hillier, L., Wendl, M. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 36.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 24, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuh, C.-H., Ransick, A., Martinez, P., Britten, R. J. & Davidson, E. H. (1994) Mech. Dev. 47, 165-186. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhamer, C. V. & Davidson, E. H. (1996) Development (Cambridge, U.K.) 122, 333-348. [DOI] [PubMed] [Google Scholar]
- 40.Tümpel, S., Maconochie, M., Wiedemann, L. M. & Krumlauf, R. (2002) Dev. Biol. 246, 45-56. [DOI] [PubMed] [Google Scholar]
- 41.Shashikant, C. S., Kim, C. B., Borbely, M. A., Wang, W. C. H. & Ruddle, F. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15446-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, C.-B., Amemiya, C., Bailey, W., Kawasaki, K., Mezey, J., Miller, W., Minoshima, S. Shimizu, N., Wagner, G. & Ruddle, F. (2000) Proc. Natl. Acad. Sci. USA 97, 1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oda-Ishii, I., Bertrand, V., Matsuo, I., Lemaire, P. & Saiga, H. (2005) Development (Cambridge, U.K.) 132, 1663-1674. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig, M. Z., Patel, N. H. & Kreitman, M. (1998) Development (Cambridge, U.K.) 125, 949-958. [DOI] [PubMed] [Google Scholar]
- 45.Langeland, J. A. & Carroll, S. B. (1993) Development (Cambridge, U.K.) 117, 585-596. [DOI] [PubMed] [Google Scholar]
- 46.Williams, J. A., Paddock, S. W., Vorwerk, K. & Carroll, S. B. (1994) Nature 368, 299-305. [DOI] [PubMed] [Google Scholar]
- 47.Britten, R. J., Rowen, L., Williams, J. & Cameron, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 4661-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Britten, R. (2002) Proc. Natl. Acad. Sci. USA 99, 13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T. D., Itoh, T., Tsai, S.-F., Park, H.-S., Yaspo, M.-L., Lehrach, H., Chen, Z., et al. (2002) Science 295, 131-134. [DOI] [PubMed] [Google Scholar]
- 50.Littlewood, D. T. J. & Smith, A. B. (1995) Philos. Trans. R. Soc. London B 347, 213-234. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez, P. & Lessios, H. A. (1999) Mol. Biol. Evol. 16, 938-952. [DOI] [PubMed] [Google Scholar]
- 52.Springer, M. S., Tusneem, N. A., Davidson, E. H. & Britten, R. J. (1995) Mol. Biol. Evol. 12, 219-230. [DOI] [PubMed] [Google Scholar]
- 53.Lee, Y.-H. (2003) Mol. Biol. Evol. 20, 1211-1221. [DOI] [PubMed] [Google Scholar]
- 54.Biermann, C. H., Kessing, B. D. & Palumbi, S. R. (2003) Evol. Dev. 5, 360-371. [DOI] [PubMed] [Google Scholar]
- 55.Cameron, R. A., Mahairas, G., Rast, J. P., Martinez, P., Biondi, T. R., Swartzell, S., Wallace, J. C., Poustka, A. J., Livingston, B. T., Wray, G. A., et al. (2002) Proc. Natl. Acad. Sci. USA 97, 9514-9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuh, C.-H., Moore, J. G. & Davidson, E. H. (1996) Development (Cambridge, U.K.) 122, 4045-4056. [DOI] [PubMed] [Google Scholar]
- 57.Yuh, C.-H. & Davidson, E. H. (1996) Development (Cambridge, U.K.) 122, 1069-1082. [DOI] [PubMed] [Google Scholar]
- 58.Kirchhamer, C. V., Yuh, C.-H. & Davidson, E. H. (1996) Proc. Natl. Acad. Sci. USA 93, 9322-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim, J. (2001) J. Exp. Zool. 291, 175-185. [DOI] [PubMed] [Google Scholar]
- 60.Erives, A. & Levine, M. (2004) Proc. Natl. Acad. Sci. USA 101, 3851-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romano, L. A. & Wray, G. A. (2003) Development (Cambridge, U.K.) 130, 4187-4199. [DOI] [PubMed] [Google Scholar]
- 62.Istrail, S. & Davidson, E. H. (2005) Proc. Natl. Acad. Sci. USA 102, 4954-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uchikawa, M., Ishida, Y., Takemoto, T., Kamachi, Y. & Kondoh, H. (2003) Dev. Cell 4, 509-519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.