Abstract
We surveyed the diversity, structural organization, and patterns of evolution of DNA transposons in rotifers of the class Bdelloidea, a group of basal triploblast animals that appears to have evolved for millions of years without sexual reproduction. Representatives of five superfamilies were identified: ITm (IS630/Tc/mariner), hAT, piggyBac, helitron, and foldback. Except for mariners, no fully intact copies were found. Mariners, both intact and decayed, are present in high copy number, and those described here may be grouped in several closely related lineages. Comparisons across lineages show strong evidence of purifying selection, whereas there is little or no evidence of such selection within lineages. This pattern could have resulted from repeated horizontal transfers from an exogenous source, followed by limited intragenomic proliferation, or, less plausibly, from within-host formation of new lineages under host- or element-based selection for function, in either case followed by eventual inactivation and decay. Unexpectedly, the flanking sequences surrounding the majority of mariners are very similar, indicating either insertion specificity or proliferation as part of larger DNA segments. Members of all superfamilies are present near chromosome ends, associated with the apparently domesticated retroelement Athena, in large clusters composed of diverse DNA transposons, often inserted into each other, whereas the examined gene-rich regions are nearly transposon-free.
Keywords: foldback, hAT, helitron, mariner, piggyBac
Rotifers of the class Bdelloidea constitute a particularly interesting system for investigating the dynamics and evolution of transposable elements (TEs) and their dependence on reproductive mode. These small freshwater invertebrates, members of the early-branching triploblast phylum Rotifera, are the taxonomic group for which evidence for long-term evolution without sexual reproduction is strongest. Classified in four families and 370 described species in which males and hermaphrodites have never been documented, bdelloids appear to have diverged from rotifers of their facultatively sexual sister class, the monogononts, tens of millions of years ago (1–3).
Two major categories of TEs inhabit eukaryotic genomes: retrotransposons move via an RNA intermediate copied back into DNA by an element-encoded reverse transcriptase, whereas DNA transposons move only as DNA, most of them using an element-encoded transposase (TPase) to perform cut-and-paste transposition (4, 5). The first category outnumbers the second in many species, presumably due in part to their intrinsically higher proliferative potential, although DNA TEs achieve high copy numbers in some species. TEs in both categories typically generate short target-site duplications (TSDs) in the host DNA upon insertion, the length of which is a characteristic of the particular TE family and is determined by the element-encoded enzymatic activity that makes staggered cuts in host DNA. Otherwise, however, they share few similarities beyond their ability to cause insertional mutations and ectopic exchange.
The proliferation of TEs within eukaryotic genomes is limited by a number of mechanisms, including selective mechanisms that depend on sexual reproduction. One of these is ectopic crossing-over, leading to inviable karyotypes. An additional possibility dependent on sexual reproduction is recombination with synergistic epistasis, and, at least in some species and perhaps generally, there are silencing mechanisms associated with meiosis. Therefore, we have suggested that the absence of these various mechanisms in asexuals may lead to the unchecked intragenomic proliferation of retrotransposons and may be a major factor responsible for the relatively early extinction of lineages that abandon sexual reproduction and, correspondingly, for the advantage of sex over asex (6, 7). In that case, ancient asexuals, if they exist, would be expected to lack intact transposable elements, except for those that are introduced by horizontal transmission or that serve some function advantageous to their host, and are subject to limiting mechanisms not dependent on sex.
Consistent with this expectation, nested PCR screens for reverse transcriptase genes of two major superfamilies of retrotransposons did not find them in any of five diverse bdelloid species tested, whereas they were readily found in all [long interspersed nuclear element (LINE)-like elements] or nearly all (gypsy-like elements) of 39 sexually reproducing species, representing 23 animal phyla, including monogonont rotifers (6). However, similar PCR screens for TPase genes revealed the presence in bdelloid genomes of mariner-like TPases, and the suggestion was made that, in contrast to retrotransposons, the propensity of mariners for horizontal transfer, as well as their capabilities for self-limitation and lower mutagenicity, might account for their presence (6, 7). Here we report on the diversity, properties, and patterns of evolution of several types of DNA transposons identified in bdelloid genomes by PCR screens, genome walks, and sequencing of genomic cosmids from telomeric and internal chromosome regions, and evaluate these findings in the context of genome structure and possible horizontal transfer.
Materials and Methods
Universal fast walking (UFW), a PCR-based procedure, was done according to Myrick and Gelbart (8). Primers for UFW and for amplification of full-length mariner and piggyBac elements are listed in Table 2, which is published as supporting information on the PNAS web site. PCR products were cloned with the aid of the Topo-TA kit (Invitrogen). Cosmids are from a Philodina roseola genomic library (9); cosmid inserts were purified on agarose gels, sonicated, and size-selected to yield ≈2-kb fragments, which were blunt-ended with T4 DNA polymerase, cloned into pBluescriptIISK+ (Stratagene), sequenced with Big-Dye Terminator version 3.1 and the standard T3/T7 primers on the ABI3730XL, and assembled with phrap/phred/consed (CodonCode, Dedham, MA). Divergence, Ks, and Ka values were calculated with the diverge program of Wisconsin Package version 10.3 (Accelrys). Phylogenetic analysis was performed with mega version 3.0 with default settings (Kimura two-parameter model) and mrbayes version 3.0 (four Markov chains, 106 generations, with each 10th tree sampled and the first 1,000 trees discarded as burn-in) (10, 11).
Results
IS630/Tc/Mariner (ITm) Superfamily. Members of the ITm superfamily, including IS630-like, Tc-like, and mariner-like elements, are probably the most numerous and certainly the most phylogenetically widespread among DNA transposons, occurring sporadically in nearly all major taxonomic groups from bacteria to humans (12–14). Mariners are among the smallest autonomous DNA transposons (≈1.3 kb), consisting only of a TPase gene and short (≈30 bp) terminal inverted repeats (TIRs), and are known for their propensity for horizontal transfer. No host factors other than basic DNA repair functions are required for their transposition, explaining the ability of insect mariners to transpose even in bacteria and protists (15, 16). The active center of ITm TPases is composed of the D,D(E/D) catalytic triad, which takes the form of D,D(34)D in the mariner family and D,D(35)E in the Tc family, where the number indicates the characteristic spacing between the two amino acids. More recently discovered members of the superfamily include the D,D(37)E, D,D(37)D, and D,D(39)D families (13).
Mariner Family. Short (≈120-bp) D(34)D TPase fragments from two mariner subfamilies, elegans and lineata, were previously amplified by nested PCR from genomic DNA of three of five bdelloid species tested, Adineta vaga, Macrotrachela quadricornifera, and Habrotrocha constricta, representing three different bdelloid families, and Southern blot analysis revealed that they are present in hundreds of copies (6). We designed primers matching the elegans subfamily TPase fragments as a starting point to obtain full-length mariners of this subfamily from A. vaga genomic DNA by a procedure involving three steps (Fig. 1A Top): UFW in both directions to obtain sequences adjacent to elements closely homologous to the known D(34)D TPase fragment; comparison of 5′ and 3′ arms to determine TIR sequences; and amplification with TIR primers. Analysis of UFW products also allowed us to examine divergence between 5′ (and 3′) TIRs among themselves, and to compare all 5′ TIRs with all 3′ TIRs. We sequenced 10 amplicons from each side of D(34)D-containing elements, obtained by two UFWs from the D(34)D fragment, and found a few variations among 5′ (and 3′) TIRs and between TIRs from one side or the other (Fig. 2A). The primers corresponding to every detected TIR variant were pooled for the TIR-PCR step to maximize the diversity of amplified full-length copies.
Fig. 1.
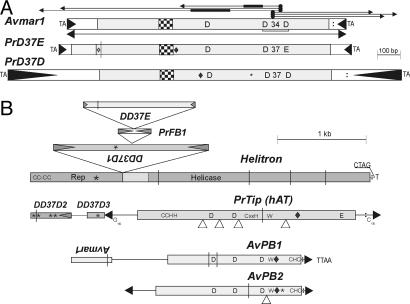
Structure and coding capacity of bdelloid DNA TEs. (A) IS630/Tc/mariner. (B) Helitron, hAT, foldback, and piggyBac superfamilies. TIRs are designated by filled triangles; frameshifts are designated by vertical lines; in-frame stop codons are designated by asterisks; introns are designated by open triangles; NLS are designated by diamonds; poly(A) signals are designated by colons; and helix-turn-helix motifs are designated by checkered boxes. Letters indicate the corresponding conserved amino acids and TSDs. For Avmar1, thin horizontal lines with arrows indicate PCR amplification strategy, and thick lines mark the position of internal deletions in two amplified copies.
Fig. 2.
Terminal sequences of bdelloid DNA transposons. (A) Sequences of Avmar1 TIRs from different 5′ and 3′ UFW clones, showing divergence in a sequence logo format (http://weblogo.berkeley.edu/logo.cgi). Positions differing between 5′ and 3′ TIRs are marked by asterisks. (B–E) TIRs of ITm (mariner, DD37D, DD37E), foldback, piggyBac, and hAT superfamily members from bdelloids (underlined) and other species, including C. elegans, D. melanogaster, Haematobia irritans, Homo sapiens, Anopheles albimanus, Aedes triseratus, C. briggsae, Bombyx mori, Chironomus thummi, Xenopus laevis, and Daphnia pulex (13, 17–20) in the BoxShade format. Numbers indicate TIR length. (F) Conserved 3′ termini of Helitrons from P. roseola, A. vaga, C. elegans, Oryza sativa, and A. thaliana, with underlined hairpins (21).
Amplification of A. vaga DNA with the pooled elegans subfamily TIR primers gave a prominent PCR product of ≈1.3 kb, which was subjected to cloning and sequencing, yielding 25 full-length sequences. All of these were distinct, indicating the presence of a large number of elements belonging to this subfamily. The consensus full-length element, named Avmar1 following the nomenclature in ref. 22, is a canonical mariner encoding a TPase with the D,D(34)D catalytic triad and the helix-turn-helix motif (Figs. 1 and 2B, Table 1, and Fig. 4, which is published as supporting information on the PNAS web site). Its short 5′ and 3′ noncoding regions comprise 133 and 74 bp, respectively. A TATA-like sequence, (TA)5, at position 86 is expanded to (TA)6–8 in some copies.
Table 1. Properties of members of five DNA transposon superfamilies from bdelloid rotifers.
| Superfamily/family | TE family | Identified in | TSD | TIR/mismatch | Length | ORF/intact | Top blastp hit (% identity/% similarity) |
|---|---|---|---|---|---|---|---|
| ITm/mariner | Avmar 1 | PCR, UFW | TA | 38/2 (44/5) | 1,242 bp | 345 aa/yes | Cemar1T (C. Telegans) (61/77) |
| ITm/DD37 (mori) | PrD37D1 | Cosmids T1, T2 | TA | 223/0 (235/3) | 1,753 bp | 341 aa/* | Cemar6 (C. elegans) (32/54) |
| ITm/DD37E | PrD37E | Cosmid T1 | TA | 32/0 | 1,329 bp | 351 aa/# | ITmD37E1 (A. gambiae) (31/51) |
| foldback | PrFT1 | Cosmid T1 | 9 bp | 153/5 | 382 bp | NA | NA |
| PrFT2 | Cosmid G1 | 9 bp | 83/5 | 521 bp | NA | NA | |
| hAT | PrTip1 | Cosmid T2 | – | 16/2 plus G15 | 3,483 bp | 833 aa/* | Tip100 (I. purpurea) (28/47) |
| piggyBac | AvPB1 | UFW | TTAA | 13 | 2,577 bp | 499 aa/## | Yabusame1 (B. mori) (33/52) |
| AvPB2 | TIR-PCR | TTAA | 13 | 2,340 bp | 559 aa/#** | Yabusame1 (B. mori) (36/54) | |
| Helitron | Pr,AvHeli | Cosmid T1, UFW | – | NA | 5.4 kb | 1,281 aa/4# | Helitron (A. gambiae) (36/48) |
NA, not applicable; * and # denote in-frame stop codons and frameshifts, respectively.
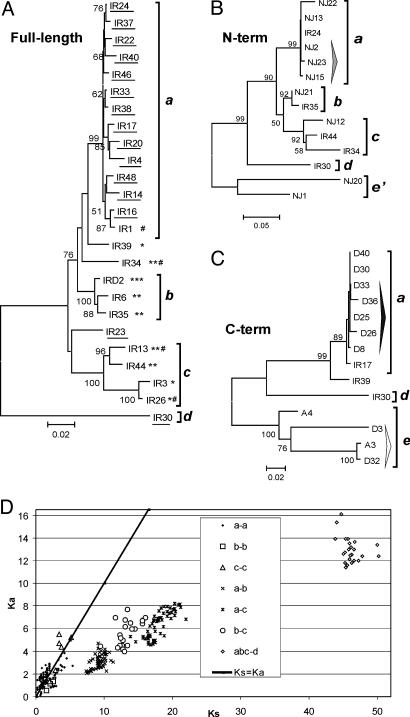
This set of 25 Avmar1 sequences obtained by TIR-PCR was subjected to phylogenetic analysis, revealing four different lineages, designated a–d (Fig. 3A). All but one of the 14 members of the a lineage are intact, and the average pairwise divergence between them is 2% (range 0.4–5.5%), consistent with recent intragenomic proliferation, certainly within the bdelloid radiation. The coding sequence of the 15 intact copies yields a set of 15 TPase variants. All seven copies from lineages b and c are defective and contain one to three in-frame stop codons, nearly all of them shared among members of a lineage (indicating trans-mobilization by an active TPase), and three of these copies also contain frameshifts (Fig. 3A). Inter-lineage divergence between copies belonging to lineages a–c ranges from 5% to 10%. The single amplified representative of lineage d contains no defects in the ORF and is nearly 20% divergent from the other three lineages. Three copies (IR23, IR34, and IR39) could not be assigned to a specific lineage and have sequences showing them to be recombinants between members of the a and c lineages, either PCR-mediated or, as described in ref. 23, occurring naturally.
Fig. 3.
Phylogeny and selection patterns in Avmar1. Neighbor-joining (NJ) trees of full-length Avmar1 copies (A) and 900-bp N-terminal (B) and 300-bp C-terminal (C) UFW fragments, excluding TIRs, are shown. Copies with intact ORFs are underlined; * and # denote in-frame stop codons and frameshifts, respectively. Lineages are designated a–d; three copies in A (IR23, IR34, and IR39) cannot be assigned to a specific lineage because of recombination (see text); two additional divergent lineages in B and C obtained by UFW but not by TIR-PCR are designated e and e′. For lineage assignment, appropriate segments of several IR copies from A are included in B and C phylogenies. Bootstrap support values >50% from 10,000 replications are indicated; Bayesian analysis gave the same tree topology (data not shown) and the clade credibility values for clades that had >50% support in NJ trees were all higher. Tall arrowheads in B and C denote copies sharing common flanking sequences. (D) Plot of Ka against Ks in all pairwise combinations within and between the Avmar1 lineages a–d.
The ratio of nonsynonymous to synonymous substitutions (Ka/Ks), which provides a measure of selection acting to maintain amino acid sequence, was compared within and between Avmar1 lineages to learn whether such purifying selection has acted on Avmar1 sequences, as suggested by our earlier inspection of 120-bp TPase fragments (6). Ka/Ks = 1 indicates neutral evolution, whereas Ka/Ks < 1 indicates purifying selection. Evidence of purifying selection between lineages is clearly seen in comparison of all interlineage Ka versus Ks values, whereas there is little or no indication of purifying selection within lineages (Fig. 3D). In a codon-based Z test, the hypothesis of neutral evolution is rejected for between-lineage, but not within-lineage, comparisons (Table 3, which is published as supporting information on the PNAS web site). In Caenorhabditis elegans, purifying selection was observed between, and not within, two divergent mariner lineages, and was attributed to horizontal transfer of active copies of each lineage, followed by their limited expansion within the recipient and eventual inactivation and decay (24).
Of the 45 cloned Avmar1 PCR products (25 full-length and 10 from each side), two carried internal deletions (85 and 173 bp, apparently mediated by 5- and 7-bp microhomologies, respectively) (Fig. 1A Top). Deletion derivatives of DNA transposons are frequently observed and are thought to occur during abortive gap repair at the excision site, by means of aborted synthesis-dependent strand annealing and subsequent nonhomologous end-joining via microhomologies within the element (25, 26).
In addition to the TE sequence, the UFW procedure yields the sequence of its adjacent flanking DNA. Comparison of 3′ and 5′ UFW products revealed an unusual pattern: of 15 clones extending into flanking DNA, only five had unique flanking sequences, whereas the majority could be subdivided into three groups, each having common flanking sequences that extend at least several hundred base pairs beyond the TIRs and the TSD (Fig. 3 B and C). Such similarity between flanks is clearly evident when the total UFW product is sequenced, yielding easily readable sequences of flanking DNA in addition to the predominant Avmar1 family sequence (Fig. 5 Top, which is published as supporting information on the PNAS web site). This finding may be interpreted as evidence of Avmar1 insertion preference for a specific target site in a multicopy sequence, which is unusual for DNA TEs, but not unprecedented (19, 27). Alternatively, a copy of Avmar1 may have been inserted into another TE with a higher proliferative potential, leading to preferential amplification of this copy within a larger transposable unit.
D,D(37)D and D,D(37)E Families. Members of the D,D(37)E family have been found only in mosquito species, whereas the D,D(37)D family has been found in other insects (Bombyx, Sarcophaga) and in nematodes (Caenorhabditis spp.) (13). We found representatives of both families (Figs. 1 A and 4, and Fig. 6A, which is published as supporting information on the PNAS web site) in the bdelloid P. roseola, which, although negative for mariner and Tc elements in PCR screens, does contain other members of the large ITm superfamily. They were identified in the course of sequencing two telomere-associated cosmids (T1 and T2), which were selected by probing with the apparently domesticated retroelement Athena and which hybridize to chromosome ends (ref. 28 and unpublished data). A full-length PrD(37)E element found on cosmid T1 is the top-most member of a pileup of four nested DNA TEs (Fig. 1). If the first or second ATG triplet is used for translation initiation, addition of an A to the A9 run at the beginning of the element would create a 381/351-aa ORF with a bipartite nuclear localization signal (NLS) and the other functional motifs depicted in Fig. 1, whereas use of the third ATG gives a 294-aa ORF that may or may not be functional. The PrD(37)D1 element, which served as a target for the PrD37E/PrFB1 insertion described above, carries an in-frame stop codon that interrupts the TPase ORF with a bipartite NLS. It belongs to the ITm D,D(37)D family (13), which occupies an intermediate position between the Tc and mariner families and is also known as the basal mori subfamily of the mariner family (14). Two other members of this family (D2 and D3) were found on cosmid T2. These incomplete copies have only 34% amino acid identity (51% similarity) to PrD(37)D1 on cosmid T1 and contain multiple defects in the TPase ORF, indicating that they became inactive long ago (Fig. 4 and Fig. 6A).
Foldback TEs. Foldback TEs are characterized by long inverted repeats, notable for their rapid reannealing during measurements of reassociation kinetics. It is believed that most foldback TEs are nonautonomous and that their mobility results from the action of a TPase encoded elsewhere, so that they may be reassigned to existing or new superfamilies when the associated TPase is identified, as has happened with miniature inverted-repeat transposable elements (MITEs) (29). A foldback TE we identified on cosmid T1 (Table 1 and Fig. 7A, which is published as supporting information on the PNAS web site) apparently belongs to type 3 (30): nonautonomous hairpins with no tandem subrepeats and a relatively short loop in the middle. This 382-bp near-perfect hairpin element, PrFT1, is inserted into PrD(37)D1, causing a 9-bp TSD, and itself was the target for insertion of PrD(37)E (Fig. 1B). A different foldback element, PrFT2, was identified as an inverted repeat, which also caused a 9-bp TSD upon insertion into the intergenic region of cosmid G1, a member of a pair of gene-rich P. roseola cosmids containing the hsp82 heat shock gene, which hybridizes to an internal region of a chromosome (ref. 9; D. Mark Welch and M.M., unpublished data). The right half of PrFT2 contains an imperfect tandem duplication, which may result in alternative secondary structures, as in foldback elements of type 2 (30) (Fig. 7B). A 20-bp terminal segment of the PrFT2 TIR, when used as a primer, yields a PCR product in another bdelloid, H. constricta, indicating that the element is present in members of two different bdelloid families (D. Mark Welch, personal communication).
The only DNA TE superfamily known to cause a 9-bp TSD, and to have large TIRs, is Mutator/IS256, found mainly in plants and fungi (31, 32), and it is possible that a Mutator-like TPase is responsible for PrFT mobility. Although the TIRs of PrFT1 and PrFT2 are not similar to the ends of Mutator-like elements, the TIRs of this superfamily exhibit little conservation (32). However, limited similarity may be noted between TIRs of PrFT, the FB elements of Drosophila melanogaster, and TFB1 from Chironomus thummi for which the corresponding TPases have not been identified (Fig. 2C) (17, 18). The absence of identifiable coding sequences complicates determination of the current mobility status of foldback TEs in bdelloids.
hAT Superfamily. The hAT superfamily, which is also phylogenetically widespread, got its name from hobo, Ac, and Tam3 transposons from fruit flies, maize, and snapdragons, respectively (14, 33). The best-studied active representatives are McClintock's Ac elements and their nonautonomous Ds derivatives from maize, hobo from D. melanogaster, and Hermes from Musca domestica (33–35). The hAT elements encode relatively large TPases (500–800 aa) and are usually found in lower copy numbers than members of the ITm superfamily (14). The hAT element identified on cosmid T2 (Fig. 1B) has a frameshift and an in-frame stop codon interrupting the TPase ORF, in which four introns can be deduced from alignment with other hAT elements, but the TIRs are too short to enable amplification of related copies. A curious feature of the TIRs is the internal asymmetrically distributed oligo(G)15–16 stretch (Figs. 1B and 2E). Originally, this hAT could have been inserted into PrD(37)D3 with a subsequent rearrangement, as neither the expected 8-bp TSD nor the other half of PrD(37)D3 ORF at its 3′ flank can be identified. Phylogenetically, it belongs to the poorly explored hAT clade that includes Tip100 from Ipomoea purpurea and other plants (14, 36) (Fig. 6B). The deduced ORF contains an N-terminal C2H2 BED Zn finger domain, a highly conserved C-terminal hAT dimerization domain, identifiable DDE catalytic residues, the CxxC motif, a bipartite NLS, and the conserved tryptophan (W) implicated in hairpin formation and processing (35) (Fig. 1B).
piggyBac Superfamily. Formerly known as the TTAA family because of this characteristic TSD, these low-copy-number DNA TEs are now considered to constitute a superfamily, being found in insects, fungi, crustaceans, sea urchins, and many vertebrates (37). Although less studied mechanistically, they probably belong to the transposase/integrase megafamily and share the RNaseH-like fold and the DDD catalytic residues, not yet confirmed experimentally, with hAT TPases and retroviral integrases (35). Unlike other DNA TEs, they seem capable of precise excision (38, 39).
Only defective piggyBac copies could be detected in bdelloids. The element AvPB1, which contains two frameshifts and only one TIR, was identifed in A. vaga in the course of UFW from Avmar1. Identification of TTAA in the target allowed us to deduce a putative TIR, which begins with a characteristic CCC, as do many other piggyBac TIRs (19) (Fig. 2D). However, the use of this TIR sequence, together with the TTAA target, as a PCR primer yielded only a weak and diffuse band, and sequencing of cloned amplicons revealed two defective (one frameshift and two stop codons in each) copies of a related (60% amino acid identity) family, designated AvPB2, which are 97% identical and share an intron in the TPase ORF. A conserved Cys5–His–Cys2 motif at the C terminus of the TPase and the preceding bipartite NLS (19, 37) are detectable in AvPB1 and AvPB2, as is the putative DDD catalytic triad, which can be described as DE (77)DN(97)D (2)D. Moreover, a highly conserved tryptophan with adjacent basic residues, a motif essential for base flipping during hairpin formation and resolution in Tn5 and possibly in hAT and in RAG recombinase (35, 40), is found between the catalytic domain and the Cys-rich motif (Fig. 1B), indicating that a hairpin intermediate may be formed during piggyBac transposition.
Helitrons. Helitrons are the only eukaryotic DNA TEs that are not expected to transpose by a cut-and-paste mechanism: they are thought to move by rolling-circle replication, similar to prokaryotic IS91-like transposons. These large elements (up to 15 kb in length) insert into the AT dinucleotide without causing a TSD, and carry TC··CTRR sequences at their ends (21). Their distribution is patchy, albeit phylogenetically widespread: they have been identified in C. elegans, rice, Arabidopsis, maize, mosquitoes, white rot fungus, sea urchin, Ciona, and fish (41, 42).
A helitron was found in a P. roseola Athena-containing telomeric cosmid T1, and another was encountered during UFW from an A. vaga Athena element. These helitrons display 71% nucleotide sequence identity in the coding region (60% in the 3′ UTR) and 76/91% ORF amino acid identity/similarity. The characteristic CTAG at the 3′ terminus can be identified in both elements, although the expected hairpins in the 3′ UTR are not readily recognizable (Figs. 1B and 2E). The N-terminal Zn finger, the Rep (rolling circle replication initiation protein) domain, and all of the conserved helicase motifs can be discerned within the helitron ORF, which in P. roseola was a target for a nested set of three DNA TE insertions associated with a deletion in Rep (Fig. 1B). Consistent with its bottom location in this pileup of TEs, its reading frame contains four frameshifts and an in-frame stop codon. Overall, the sequences cloned in the telomeric cosmids T1 and T2 are reminiscent of heterochromatic regions in Drosophila and Arabidopsis in that they are composed of multiple TE copies inserted into each other (43–45), with the important difference that we find no commonly occurring retroelements, only a putative chromovirus on cosmid T1 and the apparently domesticated Athena.
Discussion
This study provides a detailed characterization of DNA transposon content and distribution in the genomes of bdelloid rotifers, the taxonomic group for which evidence of ancient asexuality is strongest, and also represents a survey of such elements in the poorly explored assemblage of basal triploblasts to which rotifers belong. We used two approaches: characterization of full-length copies and surroundings of DNA TEs obtained by various PCR procedures using TE-specific primers, and searches within genomic DNA cloned in cosmids derived from telomeric regions and from a more proximal gene-rich region. Bdelloids were found to possess a diversified portfolio of DNA transposons that includes members of at least five superfamilies. Even representatives of superfamilies with extremely patchy phylogenetic distribution, such as piggyBac and helitrons, were easily found (Table 1). The fact that representatives of all five superfamilies identified in this survey are present on cosmids that yield fluorescent in situ hybridization signals at chromosome ends and, for a foldback TE, at another specific chromosomal site, shows that these elements are bona fide components of bdelloid genomes.
Examination of nearly a megabase of P. roseola genomic DNA, cloned in cosmids, revealed two distinct categories: gene-rich and TE-rich, hybridizing to internal and telomeric chromosome regions, respectively, the latter selected for hybridization to Athena probes (ref. 28 and unpublished data) and containing little else than Athenas and DNA TEs. In a total of 10 gene-rich cosmids (ref. 9 and D. Mark Welch and J. L. Mark Welch, personal communication), representing ≈0.5 Mb, the only TE identified was a 521-bp foldback element. For comparison, euchromatin in D. melanogaster (3.86% total TE content, which is relatively low among animals) has an average density of 13.5 TE/Mb, and, even excluding TE-rich proximal euchromatin, contains 7.7–12.3 retrotransposons and 2.3–3.6 DNA transposons per Mb (46). In C. elegans, the average density of retrotransposons in both gene-rich and gene-poor regions is ≈7 per Mb, whereas that of DNA TEs is 19 per Mb in gene-rich and 55 per Mb in gene-poor regions (47). It may be that the distinct compartmentalization of TEs into telomeric versus gene-rich regions results from regional insertion preferences, from selective advantage of heterochromatin-forming sequences in such nongenic regions as telomeres and centromeres (48, 49), or selection against TE insertions in gene-rich regions.
The hAT, piggyBac, and Helitron superfamilies, each present at low copy number, were found only as defective copies. However, the abundant ITm superfamily is represented by decayed as well as intact copies, evidence of both ancient and more recent activity. Repeated horizontal input of these elements, perhaps from some closely associated source, such as a parasite or a symbiont, is a definite possibility, and this scenario is commonly invoked for evolution of mariner/Tc transposons (12, 14, 50). Indeed, horizontal escape of a Tc-like transposon has been reported from a lepidopteran genome into a closely associated granulovirus (51), and a Tc-like transposon has been detected in the genome of the prokaryotic Wolbachia endosymbiont of Drosophila ananassae (gi accession no. 58698412) and in the assembled genomic contigs of its host (http://genome.ucsc.edu/cgi-bin/hgBlat?db=droAna1). In this connection, it may be noted that prokaryotes cannot serve as hosts for intron-containing elements, such as PrTip or AvPB2 (Fig. 1B) and TEs that, unlike mariners, require eukaryote-specific host cofactors or modifications.
Among bdelloid DNA transposons, mariners exhibit evidence of the most recent activity, are present in high copy numbers, do not contain introns, and are able to transpose even in prokaryotes (16), making them particularly likely candidates for repeated horizontal transmission. We sequenced and compared numerous full-length Avmar1 copies to characterize patterns of their evolution. In particular, we sought to verify indications of purifying selection from our earlier analysis of short Avmar and Hcmar TPase fragments (6), and to compare the degree of divergence within a cloned set of copies to detect evidence of recent activity. We conclude from Ks/Ka ratios that there is purifying selection between lineages, and little or no evidence of such selection within lineages. This pattern agrees well with repeated episodes of the commonly accepted cycle of mariner evolution: each lineage takes its origin from a horizontally transmitted founder copy, undergoes an increase in copy number, but eventually becomes inactive and decays (12, 14, 52).
Mechanisms of self-limitation of DNA transposons may include competitive inhibition by binding of TPase to the TIRs of defective copies, dominant-negative complementation caused by defective subunits within TPase dimers, overproduction inhibition, and RNA-based silencing. Such self-limiting effects have been observed in diverse superfamilies of TPase-encoding elements, including mariner (12, 53), P-element (54), and Ac (55, 56). If such effects are common to all TPase-based systems, and if, as in mariners, they eventually result in the inactivation of lineages, the presence of intact or nearly intact copies, along with decaying copies, as we have seen for members of the ITm superfamily, would be consistent with horizontal entry of all of these elements into bdelloid genomes. Although this is the most plausible scenario and horizontal transmission of mariners is well known in other species (12, 14, 50, 52), the evolution of new lineages within the host cannot be ruled out. Such within-host origin of new lineages would require either host benefit or cycles of reactivation of previously defective copies (57). Among possible host-related functions, one may entertain those in which active copies give rise to nested TE clusters, possibly with dsRNA-forming potential such as the one shown in Fig. 1B, which may be important for heterochromatin formation (45, 49). The only other eukaryotic systems in which host benefit has been suggested to account for observed purifying selection acting on TPase sequences are the ciliated protozoa, in connection with macronuclear development. However, the basis of TPase purifying selection in ciliates remains uncertain (58, 59).
In summary, we find that bdelloid genomes contain a wide variety of DNA TEs, especially in telomeric regions where they form nested clusters that are often associated with the apparently domesticated retroelement Athena, whereas such elements appear to be far rarer in gene-rich, proximal regions. The only intact DNA TEs we find are mariners, and the observed pattern of their sequence divergence is most easily explained as the result of successive horizontal invasions, each followed by expansion, subsequent inactivation via a trans-acting mechanism of self-limitation, and gradual decay. Such cycles of reinfection would not be expected for retrotransposons, owing to their much lower or, for long interspersed nuclear element (LINE)-like elements, even nonexistent propensity for horizontal transmission and their preference for cis-action. Notably, the density of TEs in the gene-rich regions of the model animal species D. melanogaster and C. elegans is much greater than that in the gene-rich bdelloid cosmids we examined. A further difference is the near absence in bdelloids of retrotransposons other than Athena, even within telomeric transposon clusters, consistent with the failure to detect LINE- and gypsy-like retrotransposons in PCR screens of genomic DNA (6). However, it remains to be determined how bdelloids became largely free of deleterious retrotransposons, elements that are found in virtually all other eukaryotes (7).
Supplementary Material
Acknowledgments
We thank D. Mark Welch for critical comments and the U.S. National Science Foundation for continued support.
Author contributions: I.R.A. and M.M. designed research; I.R.A. performed research; I.R.A. analyzed data; and I.R.A. and M.M. wrote the paper.
Abbreviations: TE, transposable element; TSD, target-site duplication; UFW, universal fast walking; TIR, terminal inverted repeat; NLS, nuclear localization signal.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY179351 and DQ138240–DQ138289).
References
- 1.Mayr, E. (1963) Animal Species and Evolution (Harvard Univ. Press, Cambridge, MA).
- 2.Mark Welch, D. & Meselson, M. (2000) Science 288, 1211-1215. [DOI] [PubMed] [Google Scholar]
- 3.Normark, B. B., Judson, O. & Moran, N. (2003) Biol. J. Linn. Soc. 79, 69-84. [Google Scholar]
- 4.Curcio, M. J. & Derbyshire, K. M. (2003) Nat. Rev. Mol. Cell. Biol. 4, 865-877. [DOI] [PubMed] [Google Scholar]
- 5.Kazazian, H. H., Jr. (2004) Science 303, 1626-1632. [DOI] [PubMed] [Google Scholar]
- 6.Arkhipova, I. & Meselson, M. (2000) Proc. Natl. Acad. Sci. USA 97, 14473-14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkhipova, I. & Meselson, M. (2005) BioEssays 27, 76-85. [DOI] [PubMed] [Google Scholar]
- 8.Myrick, K. V. & Gelbart, W. M. (2002) Gene 284, 125-131. [DOI] [PubMed] [Google Scholar]
- 9.Mark Welch, J. L., Mark Welch, D. B. & Meselson, M. (2004) Proc. Natl. Acad. Sci. USA 101, 1618-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar, S., Tamura, K. & Nei, M. (2004) Briefings Bioinformatics 5, 150-163. [DOI] [PubMed] [Google Scholar]
- 11.Ronquist, F. & Huelsenbeck, J. P. (2003) Bioinformatics 19, 1572-1574. [DOI] [PubMed] [Google Scholar]
- 12.Hartl, D. L., Lohe, A. R. & Lozovskaya, E. R. (1997) Annu. Rev. Genet. 31, 337-358. [DOI] [PubMed] [Google Scholar]
- 13.Shao, H. & Tu, Z. (2001) Genetics 159, 1103-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson, H. M. (2002) in Mobile DNA II, eds. Craig, N. L, Craigie R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 1093-1110.
- 15.Gueiros-Filho, F. J. & Beverley, S. M. (1997) Science 276, 1716-1719. [DOI] [PubMed] [Google Scholar]
- 16.Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter, S. S. (1982) Nature 297, 201-204. [DOI] [PubMed] [Google Scholar]
- 18.Hankeln, T. & Schmidt, E. R. (1990) J. Mol. Biol. 215, 477-482. [DOI] [PubMed] [Google Scholar]
- 19.Penton, E. H., Sullender, B. W. & Crease, T. J. (2002) J. Mol. Evol. 55, 664-673. [DOI] [PubMed] [Google Scholar]
- 20.Rubin, E., Lithwick, G. & Levy, A. A. (2001) Genetics 158, 949-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapitonov, V. V. & Jurka, J. (2001) Proc. Natl. Acad. Sci. USA 98, 8714-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson, H. M. & Asplund, M. L. (1996) Insect. Biochem. Mol. Biol. 26, 945-954. [DOI] [PubMed] [Google Scholar]
- 23.Fischer, S. E., Wienholds, E. & Plasterk, R. H. (2003) Genetics 164, 127-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witherspoon, D. J. & Robertson, H. M. (2003) J. Mol. Evol. 56, 751-769. [DOI] [PubMed] [Google Scholar]
- 25.Gloor, G. B., Nassif, N. A., Johnson-Schlitz, D. M., Preston, C. R. & Engels, W. R. (1991) Science 253, 1110-1117. [DOI] [PubMed] [Google Scholar]
- 26.Rubin, E. & Levy, A. A. (1997) Mol. Cell. Biol. 17, 6294-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bigot, Y., Hamelin, M. H., Capy, P. & Periquet, G. (1994) Proc. Natl. Acad. Sci. USA 91, 3408-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arkhipova, I. R., Pyatkov, K. I., Meselson, M. & Evgen'ev, M. B. (2003) Nat. Genet. 33, 123-124. [DOI] [PubMed] [Google Scholar]
- 29.Feschotte, C., Jiang, N. & Wessler, S. R. (2002) Nat. Rev. Genet. 3, 329-341. [DOI] [PubMed] [Google Scholar]
- 30.Rebatchouk, D. & Narita, J. O. (1997) Plant Mol. Biol. 34, 831-835. [DOI] [PubMed] [Google Scholar]
- 31.Eisen, J. A., Benito, M. I. & Walbot, V. (1994) Nucleic Acids Res. 22, 2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walbot, V. & Rudenko, G. N. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington DC), pp. 533-560.
- 33.Calvi, B. R., Hong, T. J., Findley, S. D. & Gelbart, W. M. (1991) Cell 66, 465-471. [DOI] [PubMed] [Google Scholar]
- 34.Kunze, R. & Weil, C. F. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 565-612.
- 35.Zhou, L., Mitra, R., Atkinson, P. W., Hickman, A. B., Dyda, F. & Craig, N. L. (2004) Nature 432, 995-1001. [DOI] [PubMed] [Google Scholar]
- 36.Habu, Y., Hisatomi, Y. & Iida, S. (1998) Plant J. 16, 371-376. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar, A., Sim, C., Hong, Y. S., Hogan, J. R., Fraser, M. J., Robertson, H. M. & Collins, F. H. (2003) Mol. Genet. Genomics 270, 173-180. [DOI] [PubMed] [Google Scholar]
- 38.Fraser, M. J., Ciszczon, T., Elick, T. & Bauser, C. (1996) Insect Mol. Biol. 5, 141-151. [DOI] [PubMed] [Google Scholar]
- 39.Thibault, S. T., Luu, H. T., Vann, N. & Miller, T. A. (1999) Insect Mol. Biol. 8, 119-123. [DOI] [PubMed] [Google Scholar]
- 40.Ason, B. & Reznikoff, W. S. (2002) J. Biol. Chem. 277, 11284-11291. [DOI] [PubMed] [Google Scholar]
- 41.Lal, S. K., Giroux, M. J., Brendel, V., Vallejos, C. E. & Hannah, L. C. (2003) Plant Cell 15, 381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulter, R. T, Goodwin, T. J. & Butler, M. I. (2003) Gene 313, 201-212. [DOI] [PubMed] [Google Scholar]
- 43.Vaury, C., Bucheton, A. & Pelisson, A. (1989) Chromosoma 98, 215-224. [DOI] [PubMed] [Google Scholar]
- 44.Nurminsky, D. I, Shevelyov, Y. Y., Nuzhdin, S. V. & Gvozdev, V. A. (1994) Chromosoma 103, 277-285. [DOI] [PubMed] [Google Scholar]
- 45.Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., McCombie, W. R., Lavine, K., Mittal, V., May, B., Kasschau, K. D., et al. (2004) Nature 430, 471-476. [DOI] [PubMed] [Google Scholar]
- 46.Kaminker, J. S., Bergman, C. M., Kronmiller, B., Carlson, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D. A., Lewis, S. E., Rubin, G. M., et al. (2002) Genome Biol. 3, RESEARCH0084. [DOI] [PMC free article] [PubMed]
- 47.Duret, L., Marais, G. & Biemont, C. (2000) Genetics 156, 1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall, I. M., Noma, K. & Grewal, S. I. (2003) Proc. Natl. Acad. Sci. USA 100, 193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, F. L., Haynes, K., Simpson, C. L., Lee, S. D., Collins, L., Wuller, J., Eissenberg, J. C. & Elgin, S. C. (2004) Mol. Cell. Biol. 24, 8210-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva, J. C., Loreto, E. L. & Clark, J. B. (2004) Curr. Issues Mol. Biol. 6, 57-71. [PubMed] [Google Scholar]
- 51.Jehle, J. A., Nickel, A., Vlak, J. M. & Backhaus, H. (1998) J. Mol. Evol. 46, 215-224. [DOI] [PubMed] [Google Scholar]
- 52.Lampe, D. J., Witherspoon, D. J., Soto-Adames, F. N. & Robertson, H. M. (2003) Mol. Biol. Evol. 20, 554-562. [DOI] [PubMed] [Google Scholar]
- 53.De Aguiar, D. & Hartl, D. L. (1999) Genetica 107, 79-85. [PubMed] [Google Scholar]
- 54.Witherspoon, D. J. (1999) Mol. Biol. Evol. 16, 472-478. [DOI] [PubMed] [Google Scholar]
- 55.Scofield, S. R., English, J. J. & Jones, J. D. (1993) Cell 75, 507-517. [DOI] [PubMed] [Google Scholar]
- 56.Kunze, R., Behrens, U., Courage-Franzkowiak, U., Feldmar, S., Kuhn, S. & Lutticke, R. (1993) Proc. Natl. Acad. Sci. USA 90, 7094-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lampe, D. J., Walden, K. K. & Robertson, H. M. (2001) Mol. Biol. Evol. 18, 954-961. [DOI] [PubMed] [Google Scholar]
- 58.Witherspoon, D. J., Doak, T. G., Williams, K. R., Seegmiller, A., Seger, J. & Herrick, G. (1997) Mol. Biol. Evol. 14, 696-706. [DOI] [PubMed] [Google Scholar]
- 59.Doak, T. G., Witherspoon, D. J., Jahn, C. L. & Herrick, G. (2003) Eukaryot. Cell 2, 95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.