Abstract
To create high-affinity antibodies, B cells target a high rate of somatic hypermutation (SHM) to the Ig variable-region genes that encode the antigen-binding site. This mutational process requires transcription and is triggered by activation-induced cytidine deaminase (AID), which converts deoxycytidine to deoxyuridine. Mistargeting of AID to non-Ig genes is thought to result in the malignant transformation of B cells, but the mechanism responsible for targeting SHM to certain DNA regions and not to others is largely unknown. Cis-acting elements have been proposed to play a role in directing the hypermutation machinery, but the motifs required for targeting SHM have been difficult to identify because many of the candidate elements, such as promoters or enhancers, are also required for transcription of Ig genes. Here we describe a system in cultured hybridoma cells in which transcription of the endogenous heavy-chain Ig gene continues in the absence of the core intronic enhancer (Eμ) and its flanking matrix attachment regions (MARs). When AID is expressed in these cells, SHM occurred at the WT frequency even when Eμ and the MARs were absent together. Interestingly, SHM occurred at less than the WT frequency when Eμ or the MARs were individually absent. Our results suggest that these intronic regulatory elements can exert a complex influence on SHM that is separable from their role in regulating transcription.
Keywords: cis-acting elements, enhancer, matrix attachment region
During the adaptive immune response, the variable regions (V regions) of Ig genes undergo somatic hypermutation (SHM) to generate the high-affinity antibodies required to protect against pathogenic organisms. SHM depends on the targeting of activation-induced cytidine deaminase (AID) to the V regions of heavy and light chain genes, whereas the constant regions (C regions) of the Ig genes are protected from high rates of SHM. AID is expressed primarily in centroblast B cells in the germinal centers of secondary lymphoid organs; thus, its restricted expression spares other cells from its mutagenic effects (reviewed in ref. 1). In centroblast B cells, some highly expressed non-Ig genes do not undergo SHM, indicating that a high rate of transcription is not sufficient to make a gene accessible to AID (2). AID can also cause mutations in some highly expressed non-Ig genes, including many protooncogenes, especially in tumor cells (3–5), whereas ubiquitous expression of AID in mice resulted in tumorigenesis (6). It is therefore important to understand the mechanisms that are responsible for the preferential targeting of AID and its associated proteins to the V regions, not only because SHM is required for affinity maturation during the antibody response but also because the mistargeting of AID to non-Ig genes is thought to be associated with malignant transformation.
SHM is dependent on and roughly proportional to the rate of transcription of the targeted genes in vivo and in cultured cells (7–9). Furthermore, SHM begins just downstream from the transcription start site and ends ≈1.5 kb downstream from the promoter (10, 11). These findings have led to the suggestion that cis-acting elements, such as promoters and enhancers that direct and regulate transcription, also target the hypermutation machinery to particular DNA regions (12, 13). Although there is some evidence for this suggestion, it has been especially difficult to determine whether there are specific cis-acting motifs required for targeting SHM because many of the obvious candidates are located in the promoters and enhancers and are also required for transcription of the Ig genes (12). Mutation or deletion of such motifs usually reduces the rate of transcription, and this alone results in a lower rate of mutation.
In this paper, we have investigated the role in SHM of the IgH intronic elements in their native chromosomal location in a tissue culture system that does not depend on these elements for heavy chain transcription. This system is based on a series of hybridoma cells in which deletion of core intronic enhancer (Eμ) and/or its associated matrix attachment regions (MARs) from the endogenous IgH locus leads to bimodal or variegated expression of IgH, in which some cells express IgH at WT levels and other cells do not express IgH at all (14). We have used these highly expressing cells to test the role of these cis-acting elements in targeting SHM and found that, together, Eμ and its associated MARs are dispensable for high levels of SHM in the IgH locus. However, the presence of the MARs in the absence of Eμ results in a complete loss of SHM. The presence of Eμ in the absence of the MARs results in a decrease in SHM compared with WT cells.
Materials and Methods
Cell Lines. Sp6 is a WT hybridoma cell line that produces IgM specific for the hapten 2,4,6-trinitrophenyl (15). All recombinants were derived by homologous recombination from a derivative of Sp6, igm692 (16). The WT recombinant has the WT complement of intronic regulatory elements (17). In Eμ, only the core Eμ enhancer is present in the J–C intron (18). The MARs recombinant, in which the MARs are the only intronic regulatory elements in the J–C intron, was used to generate the MARs-μ+ and MARs-μ- subclones, which are active and silent for IgH expression, respectively (18, 19). Δ, a recombinant with a complete deletion of the intronic elements, was used to generate the subclones Δ-μ+ and Δ-μ-, in which IgH is active or silent, respectively (14, 20).
Generation of Stably AID-Expressing Transfectants. Hybridoma cells were transfected with a vector encoding human AID (hAID) or a control vector pCEP4 (pCEP), as described in ref. 21. Both vectors conferred resistance to puromycin. Cells (1 × 107 to 2 × 107) were electroporated with 10 μg of plasmid DNA that had been digested with EcoRV and NruI. Cells were plated immediately after electroporation in 96-well plates at a density of 103 cells per 100 μl per well. Medium (100 μl) containing 12 μg/ul puromycin was added to wells 24 h afterward. Colonies were selected ≈10 days after electroporation, expanded, and AID-expressing, and control subclones were maintained in medium containing 6 μg/ml puromycin.
Real-Time RT-PCR. Total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Genomic DNA was digested with DNaseI (Worthington) for 15 min at room temperature. Total RNA (1 μg) was reverse-transcribed by using iScript reverse transcriptase (BioRad). The resulting cDNA was analyzed by quantitative PCR in a final volume of 15 μl in a 96-well format using SYBR green (Applied Biosystems) and DNA Engine Opticon 2 (MJ research, Cambridge, MA) with the following primers: QhAID forward, 5′-CTTCGCAATAAGAACGGCTG-3′; QhAID reverse, 5′-GAGGTGAACCAGGTGACGC-3′; Sp6V forward, 5′-GCAGACAAATCCTCCAGCACAG-3′; Sp6V reverse, 5′-CCCCAGTAAGCAAGCCCGTAGC-3′; QRTmGAPDH forward, 5′-GAGGCCGGTGCTGAGTATGTCGTG-3′; and QRTm-GAPDH reverse, 5′-TCGGCAGAAGGGGCGGAGAT-3′.
Analysis of Mutation Rates and Targeting. Genomic DNA was prepared with the DNeasy kit (Qiagen, Valencia, CA). The variable and constant Ig regions and a region encompassing the J–C intron and the human AID transgene were amplified by PCR using Pfu-Turbo (Stratagene) and the following primer pairs: V region, 5′-TTACCTGGGTCTATGGCAGT-3′ (5′ primer) and 5′-TGAAGGCTCAGAATCCCCC-3′ (3′ primer); J–C intron, 5′-TTGTGATTAACTATGCTATGGACTACTGG-3′ (5′ primer) and 5′-CTGCTATTTCCTTGTTGCTACTC-3′ (3′ primer); Cμ region, 5′-CCCCTCCTTTGCCGACATCTTCC-3′ (5′ primer) and 5′-TTCCATTCCTCCTCGTCACAGTC-3′ (3′ primer); and hAID, 5′-GAGGCAAGAAGACACTCTGG-3′ (5′ primer) and 5′-GTGACATTCCTGGAAGTTGC-3′ (3′ primer). PCR products were cloned with the Zero Blunt TOPO PCR cloning kit (Invitrogen) according to the manufacturer's specifications. The presence of the insert was verified by digestion with EcoRI. Plasmid DNA was sequenced at the Albert Einstein College of Medicine Cancer Center DNA Sequencing Facility and at GENEWIZ (North Brunswick, NJ) and SeqWright (Houston, TX). DNA sequences were aligned with the seqmanii program of DNASTAR (Madison, WI). For statistical analysis, the unpaired Student's t test was used.
Results
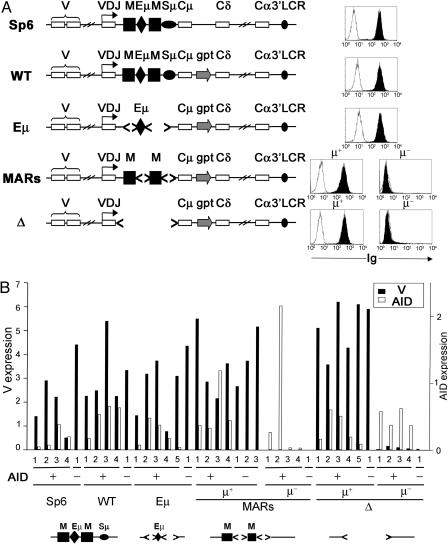
A System to Separate the Role of Cis-Acting Elements in Transcription and SHM in the Endogenous IgH Locus. We have previously described a hybridoma cell system in which deletion of the intronic cis-acting elements from the endogenous IgH locus resulted in bimodal IgH expression, in which some cells expressed IgM and the μ heavy-chain mRNA at WT levels and others did not express the μ heavy chain at all (14). Thus, virtually all cells in the parental population of Sp6 hybridoma cells produce high levels of IgM (Fig. 1A, WT). The WT recombinant was generated by inserting the guanyl phophorybosyl transferase (gpt) between the Cμ region and Cδ region (Fig. 1 A) (17), resulting in insulation from the influence of the 3′ locus control region on the expression of the μ heavy chain (20). In WT, all of the cells still expressed WT levels of IgM (14). A similar high level of expression was retained in the Eμ recombinant in which the Eμ without the MARs and μ switch region was introduced into the J–C intron (Fig. 1 A). Cells with a J–C intron bearing only the MARs or a deletion of all of the intronic elements (Δ) generated IgM-producing (MARs-μ+ and Δ-μ+) and IgM-nonproducing (MARs-μ- and Δ-μ-) subclones. Because μ transcription in subclones from these latter two recombinants varied in their degree of stability (19), only subclones that stably retained their epiphenotype (μ+ or μ-) for more than 2 months in culture were used in this study.
Fig. 1.
A hybridoma cell system to study the role of the intronic elements in SHM. (A) In the IgH locus of the hybridoma cells used in this study, exons are shown as white rectangles, and cis-acting elements are shown as black shapes. M, MAR; Sμ, μ switch region, 3′LCR, 3′ α regulatory elements. The gpt selectable marker is shown as an arrow. < > indicate deletions. The WT, Eμ, MARs, and Δ recombinants are derived by homologous recombination from a mutant of Sp6, as described in the text. Flow cytometry profiles of hybridomas for IgM (black histograms) and IgG (white histograms) are shown to the right. Note that MARs and Δ recombinants generate μ+ or μ- subclones. (B) Quantitative RT-PCR results of V region and hAID mRNA measurements. Expression levels relative to GADPH are shown on the y axis as indicated. Several transfectants of each recombinant, indicated by numbers, were analyzed.
Like most hybridomas, Sp6 and the recombinants described here can only undergo SHM in the presence of exogenously expressed hAID (21). Thus, the retention of equivalent μ heavy-chain expression in the various recombinant clones shown in Fig. 1 made it possible to examine the effect of AID expression on V- and C-region mutation in the endogenous locus in the absence of different combinations of the intronic regulatory elements. To perform these experiments, we transfected the subclones shown in Fig. 1 A with a vector expressing hAID or a control vector (pCEP). AID-expressing and control clones were then propagated in culture for 2 months. Only transfectants that retained hAID mRNA expression at the end of the 2-month period were analyzed further. RNA from these cultures was analyzed by real-time RT-PCR, as shown in Fig. 1B. As expected, transfectants derived from Sp6, WT, Eμ, MARs-μ+, and Δ-μ+ produced high levels of μ mRNA, whereas transfectants of MARs-μ- and Δ-μ- cells did not. Cultures varied in the amount of AID expression, as indicated in Fig. 1B, with subclones that had been transfected with pCEP vector not showing hAID expression.
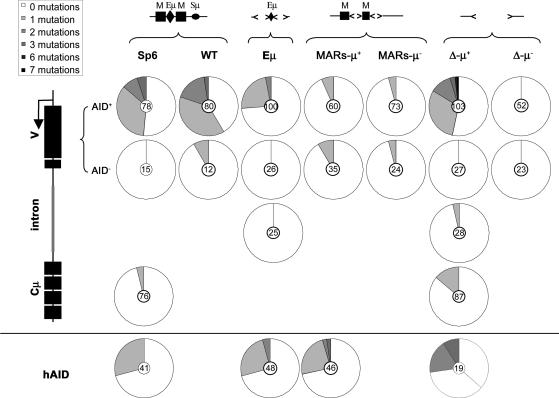
The Intronic Cis-Acting Elements Are Dispensable for SHM, but MARs and, to a Lesser Degree, Eμ by Themselves Do Not Support or May Inhibit SHM. The rate of SHM was measured in AID-expressing and control transfectants. As shown in Fig. 2 and Table 1, for the functional V region, subclones derived from Sp6 mutated at a frequency of 1.1 (±0.3) × 10-3 per base, a value comparable with that shown previously for other hybridomas (21). Similarly, the WT recombinant and Δ-μ+, a μ-expressing subclone lacking the intronic regulatory elements, showed high frequencies of mutation in the V region that were not significantly different from Sp6. Control subclones transfected with the empty vector had low frequencies of mutation of ≈9.1 × 10-5 per base in all subclones, a frequency that approached that of the PCR error rate. It therefore appears that the intronic elements are dispensable for high levels of SHM in this system. As expected, clones derived from MARs-μ- and Δ-μ-, which did not express μ (Fig. 1), did not show mutation frequencies above background (Fig. 2), confirming that transcription is needed for hypermutation.
Fig. 2.
The extent of SHM in hybridoma transfectants. Mutations were assessed in the regions indicated in AID transfectants (V-AID+, Intron, Cμ, and hAID) and pCEP-transfected controls (AID-). The structure of the J–C intron of the hybridomas analyzed is shown above the pie charts. Pie charts indicate the fraction of DNA molecules analyzed bearing 0–7 mutations. The numbers in the middle of the pie charts indicate the total number of DNA molecules analyzed.
Table 1. Average of mutation frequencies (±SD) ×10–4 in hybridoma transfectants.
| V
|
|||||
|---|---|---|---|---|---|
| Cell lines | AID+ | AID– | Intron | Cμ | hAID |
| Sp6 | 10.9 ± 3.3 (45/40,914) | <1.6 (0/6,368) | ND | 0.5 (2/32,046) | 5.3 ± 0.1 (13/24,395) |
| WT | 15.5 ± 2.0 (70/44,676) | 1.5 (1/6,801) | ND | ND | ND |
| Eμ | 2.0 ± 1.9 (29/49,889) | <0.7 (0/15,357) | <0.8 (0/13,010) | ND | 4.4 ± 1.3 (13/29,199) |
| MARs-μ+ | 1.0 ± 0.7 (4/36,835) | 0.9 ± 1.6 (2/20,407) | ND | ND | 6.1 (± 6.5) (13/11,437) |
| MAR-μ– | 1.1 ± 1.0 (4/35,357) | 0.8 ± 1.6 (1/12,974) | ND | ND | ND |
| Δ-μ+ | 13.0 ± 4.8 (80/60,161) | <0.6 (0/15,928) | 1.3 ± 1.9 (1/6,127) | 2.2 ± 1.9 (12/50,068) | 11.4 ± 6.1 (13/11,437) |
| Δ-μ– | <1.0 (0/28,789) | <0.8 (0/12,613) | ND | ND | ND |
Averages of mutation frequency are of several hAID (V AID+, Intron, Cμ, and hAID) or pCEP (V AID–) transfectants. Values in parentheses indicate mutations/bases sequenced. ND, not determined.
Because it appeared that SHM could occur in the absence of both Eμ and the MARs, it was surprising that the mutation frequencies in the Eμ and MARs-μ+-derived subclones that retained the core Eμ or the MARs, respectively, were lower than Sp6 and WT (Fig. 2 and Table 1). Although mutation frequencies in Eμ were significantly lower compared with Sp6 (P = 0.019), they were also significantly higher than the background (P = 0.0014). MARs-μ+ subclones are significantly lower than Sp6 (P = 0.0011) and similar to the background (P = 0.83). To confirm that these cell lines were able to undergo SHM, we measured SHM in the hAID transgene that had been shown previously to mutate in a variety of cell lines (21). We found that hAID mutated at similar frequencies of 4.4 × 10-4, 6.1 × 10-4 and 11.4 × 10-4, respectively, in the Eμ, MARs-μ+ and Δ-μ+ recombinants, which indicates that the SHM machinery was equally effective when assayed on the ectopically integrated non-Ig AID transgene in these cell lines, even though the V region was much more highly targeted in the Δ-μ+ clones. Thus, although the Eμ and the MARs (and μ switch regions) together are dispensable for SHM, the core Eμ alone inhibits or does not facilitate the targeting of SHM to the V region, whereas the presence of MARs by themselves reduces SHM of the V region to background levels.
Targeting of SHM to the V Region in the Presence and Absence of the Intronic Elements. In B cells undergoing SHM, the V region is mutated at much higher levels than the IgH C region, even in AID-overexpressing hybridoma cells (21). It has been suggested that lack of mutation in IgH C regions is due to their distance from the promoter (22–24). We compared the extent of SHM in the Cμ regions of Sp6 and Δ-μ+ subclones, in which the C region is 7.7 kb closer to the promoter than in Sp6 (Fig. 2 and Table 1). There was an increase in the frequency of mutation in the C region in the Δ-μ+ cells compared with Sp6 cells, but this difference was not statistically significant (P = 0.138). Low levels of mutation were also seen in the intron of Δ-μ+ and Eμ cells (Fig. 2).
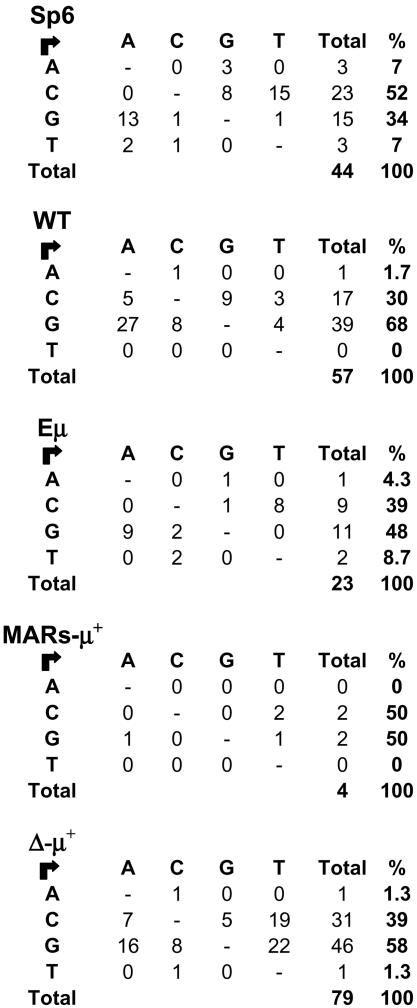
AID is targeted to G/C residues that are part of the hotspot motifs DGWY and WRCH (targeting the italic bases in the two sequences, where D is A, G, or T; W is A or T; Y is C or T; R is A or G; and H is T, C, or A). In vivo there is an equal distribution of mutations in G/C and A/T bases, and the latter are attributed to the second mismatch repair-dependent phase of SHM (21, 25). Moreover, in culture cells, for reasons that are not completely clear, ≈80% of mutations are found in G/C. Here we tested whether the intronic cis-acting elements might affect targeting of SHM to these hotspot motifs. As shown in Fig. 3 and Table 2, the vast majority of mutations in the Sp6 and all of the recombinants occurred at C or G residues, and most of these mutations were transitions. Moreover, most mutations at C or G residues occurred at hotspot motifs (Table 2). Furthermore, a naive assessment of the motifs targeted by SHM in which the hotspots were determined on the basis of the frequency of neighboring bases around the mutated residues revealed that these consensus sequences are indeed subsets of the DGWY/WRCH motifs (data not shown). This profile is consistent with that seen in other cell lines and suggests that the repair machinery, and not just AID alone, operates in these cells. In all mutating lines with the exception of the WT recombinant, there were equivalent levels of C→T and G→A mutations, suggesting that there was no strand bias (Fig. 3).
Fig. 3.
Characteristics of mutations in hybridoma cells. The diagram shows the nature of the mutations in various hAID-transfected hybridomas. Arrows point from the nonmutated base to the mutated one. Recombinants are as indicated in Fig. 1.
Table 2. Characteristics of mutations in the IgH V regions of AID-transfected hybridoma recombinants.
| Cell lines | Unique mutations | CG (%) | Transitions (%) | Hotspots (% of C/G mutations) |
|---|---|---|---|---|
| Sp6 | 45 | 37 (82) | 26 (58) | 20 (54) |
| WT | 41 | 38 (93) | 15 (37) | 28 (74) |
| Eμ | 23 | 20 (87) | 17 (74) | 16 (80) |
| Δ (μ+) | 61 | 59 (97) | 31 (49) | 34 (58) |
Discussion
AID deaminates deoxycytidine residues in ssDNA to produce deoxyuridine, which is then processed by DNA repair enzymes, resulting in mutations (reviewed in ref. 1). Although transcription has been postulated as a prerequisite for SHM because the transcription bubble could create the ssDNA substrate needed for AID deamination (ref. 26; see also ref. 27 for a review), transcription is not sufficient for SHM because some highly transcribed genes in centroblasts do not undergo SHM (2). Moreover, within the Ig transcriptome, only the V (not the C regions) undergo SHM (28). It has been suggested that cis-acting elements, such as the promoters and enhancers that flank the V region and direct and regulate transcription, could also target the hypermutation machinery to V and not to C. In fact, when the V region promoter is placed just 5′ to the C region, mutation in that C region occurs (24). Furthermore, the Eμ and its flanking MARs in the J–C intron have been shown to contribute to high levels of SHM in transgenes (29–32). The 3′ regulatory regions, which are hundreds of kilobases downstream from the Ig heavy-chain V region, contribute to the regulation of IgH expression and class switch recombination, but they are not required for SHM in the heavy chain (30, 33, 34). In contrast, the κ light-chain 3′ enhancer, which is only a few kilobases downstream from the light-chain V region, does appear to be required for SHM of the light-chain V region (35–38). In one set of ectopically integrated transgenes, E boxes that bind E47 were shown to up-regulate SHM without affecting transcription (39), but that motif is present in the IgH C, as well as V regions, and has not been shown to play a role in the mutation of the endogenous heavy-chain gene. Although these and other studies point to a requirement for cis-acting elements for SHM (40, 41), other studies on randomly integrated transgenes argue that hypermutation is independent of integration site and does not require Ig promoters or enhancers and, thus, is not targeted by particular cis-acting elements (42, 43). It has been especially difficult to determine whether there are specific cis-acting motifs in the promoter or enhancer that are required for targeting SHM because mutation or deletion of individual motifs within these regulatory elements will reduce the rate of transcription, and this alone will result in a lower rate of mutation.
To overcome this problem, we used a set of hybridoma cell lines derived from the Sp6 hybridoma cell line in which the Eμ and MARs have been deleted from the endogenous locus in various combinations. All of the recombinants contained a gpt gene between the Cμ and Cδ regions, which insulates the μ gene from the 3′α activating elements (Fig. 1 A) (ref. 20 and M.J.S., unpublished observation). Loss of Eμ or of both Eμ and the MARs resulted in cells in which expression was variegated, such that two types of stable subclones could be isolated: those that continued to make WT levels of the μ heavy chain and those that had lost the ability to express the μ heavy chain. By transfecting AID into these subclones, it became possible to compare V region SHM in the presence and absence of Eμ and the MARs in cells that were all transcribing the μ heavy-chain gene at similar rates.
A number of independent AID transfectants with different levels of AID mRNA were examined for each recombinant. There was no significant correlation between the steady-state levels of AID mRNA and SHM frequency (Fig. 1B and Table 3, which is published as supporting information on the PNAS web site). Moreover, the levels of AID after propagation of cells in culture for 2 months did not correlate with SHM frequency. We have previously reported that increasing AID mRNA above the endogenous levels in the Ramos cells did not increase the rate of mutation (21), and AID expression in the hybridoma cells studied here might also have reached a threshold beyond which it is not possible to increase the rate of V region mutation. Deletions and insertions in the V regions were found in Sp6 and Δ(μ+) transfectants (Table 3).
Our analysis of these recombinant hybridomas yielded several very interesting results. First, the μ-expressing recombinants that lacked Eμ and MARs underwent SHM at the same rate as the recombinants that retained both these elements. Thus, after 2 months of exposure to AID, the frequency of mutated V regions and the number of mutations per V region were similar in the parental Sp6 cell line, the WT cells, and the Δ-μ+ recombinant that lack both Eμ and the MARs. These mutation frequencies were also similar to those for constitutively mutating human Ramos Burkitt's lymphoma cells (21, 44, 45). The loss of Eμ and the MARs did not affect the targeting of SHM to G/C bases in hot spots in the V region. Furthermore, no significant differences in mutation of the C region were found between Δ-μ+ and Sp6 recombinants (Fig. 2). These results indicate that all aspects of hypermutation that we measured with this system were normal in the combined absence of Eμ and MARs. In contrast, the μ-nonexpressing subclones did not mutate above background (Fig. 2), confirming that transcription is needed for hypermutation. The μ-nonexpressing subclones contain all of the soluble factors necessary for transcription (19, 46), demonstrating that epigenetic silencing of the IgH locus in cis is enough to ablate SHM. Another interesting result was our finding that the MARs recombinant (that retained the MARs but lacked Eμ) did not mutate the V region. This recombinant was competent to carry out AID-induced mutation, because the AID-expressing transfectants still targeted the AID transgene itself for mutation (Fig. 2). A lesser but still marked deficiency in SHM was also evident in the recombinant that retained Eμ but lacked the MARs. The low frequency of mutation in these recombinants suggests the existence of a cis-acting mechanism that can prevent hypermutation. It is possible that core Eμ and the MARs by themselves are inhibitory for SHM. Alternatively, new sequences created at junctions in the Eμ and MARs recombinants might have had negative regulatory effects on SHM. Although it is unclear whether these or other unknown mechanisms are responsible for the results observed in these particular recombinant hybridoma clones, these results suggest that cis-acting sequences can play an inhibitory and activating role in the targeting of SHM.
Supplementary Material
Acknowledgments
We thank Drs. Barbara Birshtein and Alberto Martin and members of the M.D.S. Laboratory for critical reading of the manuscript and members of the A. Melnick Laboratory for help with real-time PCR. This work was supported by National Institutes of Health Grants CA72649 and AI53362. M.D.S. is supported by the Harry Eagle Chair provided by the National Women's Division of the Albert Einstein College of Medicine. D.R. is supported by a Cancer Research Institute Postdoctoral Fellowship and the Harry Eagle Fellowship. M.D.I.-U. was a fellow of the Ministerio de Educacion, Cultura, y Deporte of Spain and is currently supported by Northeast Biodefense Center Fellowship AI57158.
Author contributions: D.R. and M.D.S. designed research; D.R., M.D.I.-U., and M.F. performed research; D.R. and M.J.S. contributed new reagents/analytic tools; D.R. and M.D.I.-U. analyzed data; and D.R., M.J.S., and M.D.S. wrote the paper.
Abreviations: AID, activation-induced cytidine deaminase; hAID, human AID; C region, constant region; gpt, guanyl phophorybosyl transferase; MAR, matrix attachment region; SHM, somatic hypermutation; V region, variable region.
References
- 1.Li, Z., Woo, C. J., Iglesias-Ussel, M. D., Ronai, D. & Scharff, M. D. (2004) Genes Dev. 18, 1-11. [DOI] [PubMed] [Google Scholar]
- 2.Shen, H. M., Michael, N., Kim, N. & Storb, U. (2000) Int. Immunol. 12, 1085-1093. [DOI] [PubMed] [Google Scholar]
- 3.Kuppers, R. & Dalla-Favera, R. (2001) Oncogene 20, 5580-5594. [DOI] [PubMed] [Google Scholar]
- 4.Pasqualucci, L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature 412, 341-346. [DOI] [PubMed] [Google Scholar]
- 5.Shen, H. M., Peters, A., Baron, B., Zhu, X. & Storb, U. (1998) Science 280, 1750-1752. [DOI] [PubMed] [Google Scholar]
- 6.Okazaki, I. M., Hiai, H., Kakazu, N., Yamada, S., Muramatsu, M., Kinoshita, K. & Honjo, T. (2003) J. Exp. Med. 197, 1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachl, J., Carlson, C., Gray-Schopfer, V., Dessing, M. & Olsson, C. (2001) J. Immunol. 166, 5051-5057. [DOI] [PubMed] [Google Scholar]
- 8.Fukita, Y., Jacobs, H. & Rajewsky, K. (1998) Immunity 9, 105-114. [DOI] [PubMed] [Google Scholar]
- 9.Maizels, N. (1995) Cell 83, 9-12. [DOI] [PubMed] [Google Scholar]
- 10.Rada, C. & Milstein, C. (2001) EMBO J. 20, 4570-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebecque, S. G. & Gearhart, P. J. (1990) J. Exp. Med. 172, 1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storb, U., Peters, A., Klotz, E., Kim, N., Shen, H. M., Hackett, J., Rogerson, B. & Martin, T. E. (1998) Immunol. Rev. 162, 153-160. [DOI] [PubMed] [Google Scholar]
- 13.Wagner, S. D. & Neuberger, M. S. (1996) Annu. Rev. Immunol. 14, 441-457. [DOI] [PubMed] [Google Scholar]
- 14.Ronai, D., Berru, M. & Shulman, M. J. (1999) Mol. Cell. Biol. 19, 7031-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler, G. & Milstein, C. (1976) Eur. J. Immunol. 6, 511-519. [DOI] [PubMed] [Google Scholar]
- 16.Baar, J. & Shulman, M. J. (1995) J. Immunol. 155, 1911-1920.7636242 [Google Scholar]
- 17.Oancea, A. E. & Shulman, M. J. (1994) Int. Immunol. 6, 1161-1168. [DOI] [PubMed] [Google Scholar]
- 18.Wiersma, E. J., Ronai, D., Berru, M., Tsui, F. W. & Shulman, M. J. (1999) J. Biol. Chem. 274, 4858-4862. [DOI] [PubMed] [Google Scholar]
- 19.Ronai, D., Berru, M. & Shulman, M. J. (2004) Genetics 167, 411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oancea, A. E., Berru, M. & Shulman, M. J. (1997) Mol. Cell. Biol. 17, 2658-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J. & Scharff, M. D. (2002) Nature 415, 802-806. [DOI] [PubMed] [Google Scholar]
- 22.Weber, J. S., Berry, J., Litwin, S. & Claflin, J. L. (1991) J. Immunol. 146, 3218-3226. [PubMed] [Google Scholar]
- 23.Rada, C., Yelamos, J., Dean, W. & Milstein, C. (1997) Eur. J. Immunol. 27, 3115-3120. [DOI] [PubMed] [Google Scholar]
- 24.Peters, A. & Storb, U. (1996) Immunity 4, 57-65. [DOI] [PubMed] [Google Scholar]
- 25.Phung, Q. H., Winter, D. B., Cranston, A., Tarone, R. E., Bohr, V. A., Fishel, R. & Gearhart, P. J. (1998) J. Exp. Med. 187, 1745-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bransteitter, R., Pham, P., Scharff, M. D. & Goodman, M. F. (2003) Proc. Natl. Acad. Sci. USA 100, 4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barreto, V. M., Ramiro, A. R. & Nussenzweig, M. C. (2005) Trends Immunol. 26, 90-96. [DOI] [PubMed] [Google Scholar]
- 28.O'Brien, R. L., Brinster, R. L. & Storb, U. (1987) Nature 326, 405-409. [DOI] [PubMed] [Google Scholar]
- 29.Bachl, J. & Wabl, M. (1996) Immunogenetics 45, 59-64. [DOI] [PubMed] [Google Scholar]
- 30.Lin, M. M., Green, N. S., Zhang, W. & Scharff, M. D. (1998) Int. Immunol. 10, 1121-1129. [DOI] [PubMed] [Google Scholar]
- 31.Giusti, A. M. & Manser, T. (1993) J. Exp. Med. 177, 797-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackett, J., Stebbins, C., Rogerson, B., Davis, M. M. & Storb, U. (1992) J. Exp. Med. 176, 225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morvan, C. L., Pinaud, E., Decourt, C., Cuvillier, A. & Cogne, M. (2003) Blood 102, 1421-1427. [DOI] [PubMed] [Google Scholar]
- 34.Tumas-Brundage, K. M., Vora, K. A. & Manser, T. (1997) Mol. Immunol. 34, 367-378. [DOI] [PubMed] [Google Scholar]
- 35.Betz, A. G., Milstein, C., Gonzalez-Fernandez, A., Pannell, R., Larson, T. & Neuberger, M. S. (1994) Cell 77, 239-248. [DOI] [PubMed] [Google Scholar]
- 36.Goyenechea, B., Klix, N., Yélamos, J., Williams, G. T., Riddell, A., Neuberger, M. S. & Milstein, C. (1997) EMBO J. 16, 3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi, M., Wu, P., Trevorrow, K. W., Claflin, L. & Garrard, W. T. (1999) J. Immunol. 162, 6029-6039. [PubMed] [Google Scholar]
- 38.Kodama, M., Hayashi, R., Nishizumi, H., Nagawa, F., Takemori, T. & Sakano, H. (2001) Int. Immunol. 13, 1415-1422. [DOI] [PubMed] [Google Scholar]
- 39.Michael, N., Shen, H. M., Longerich, S., Kim, N., Longacre, A. & Storb, U. (2003) Immunity 19, 235-242. [DOI] [PubMed] [Google Scholar]
- 40.Green, N. S., Lin, M. M. & Scharff, M. D. (1998) Immunol. Rev. 162, 77-87. [DOI] [PubMed] [Google Scholar]
- 41.Neuberger, M. S. & Milstein, C. (1995) Curr. Opin. Immunol. 7, 248-254. [DOI] [PubMed] [Google Scholar]
- 42.Wang, C. L., Harper, R. A. & Wabl, M. (2004) Proc. Natl. Acad. Sci. USA 101, 7352-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, A. & Scharff, M. D. (2002) Nat. Rev. Immunol. 2, 605-614. [DOI] [PubMed] [Google Scholar]
- 44.Martin, A. & Scharff, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 12304-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, W., Bardwell, P. D., Woo, C. J., Poltoratsky, V., Scharff, M. D. & Martin, A. (2001) Int. Immunol. 13, 1175-1184. [DOI] [PubMed] [Google Scholar]
- 46.Ronai, D., Berru, M. & Shulman, M. J. (2002) J. Immunol. 169, 6919-6927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.