Abstract
Severe acute respiratory syndrome (SARS) is caused by an emergent coronavirus (SARS-CoV), for which there is currently no effective treatment. SARS-CoV mediates receptor binding and entry by its spike (S) glycoprotein, and infection is sensitive to lysosomotropic agents that perturb endosomal pH. We demonstrate here that the lysosomotropic-agent-mediated block to SARS-CoV infection is overcome by protease treatment of target-cell-associated virus. In addition, SARS-CoV infection was blocked by specific inhibitors of the pH-sensitive endosomal protease cathepsin L. A cell-free membrane-fusion system demonstrates that engagement of receptor followed by proteolysis is required for SARS-CoV membrane fusion and indicates that cathepsin L is sufficient to activate membrane fusion by SARS-CoV S. These results suggest that SARS-CoV infection results from a unique, three-step process: receptor binding and induced conformational changes in S glycoprotein followed by cathepsin L proteolysis within endosomes. The requirement for cathepsin L proteolysis identifies a previously uncharacterized class of inhibitor for SARS-CoV infection.
Keywords: SARS, viral entry, proteolysis, membrane fusion, viral envelope
Severe acute respiratory syndrome (SARS) is an acute respiratory illness caused by a newly described coronavirus (SARS-CoV) (1), the result of a zoonosis of a highly related animal coronavirus (2). There continues to be potential for further zoonotic transmission events, leading to the reintroduction of SARS-CoV into the human population. No effective antiviral treatments have been described for SARS, and, although several promising studies are ongoing, there is currently no licensed protective vaccine.
SARS-CoV entry into target cells is initiated by engagement of its cellular receptor, angiotensin-converting enzyme 2 (ACE2) by spike (S) glycoprotein (3). Subsequent infection is sensitive to inhibitors of endosomal acidification such as ammonium chloride (4–6), suggesting that SARS-CoV requires a low-pH milieu for infection. On the other hand, S protein can mediate cell–cell fusion at neutral pH (3, 4), indicating that S protein-mediated fusion does not include an absolute requirement for an acidic environment. Given these discordant findings, we hypothesized that cellular factors sensitive to ammonium chloride, such as pH-dependent endosomal proteins, may play a role in mediating SARS-CoV entry. In this study, the requirements for proteases in the activation of viral infectivity and the effect of protease inhibitors on SARS-CoV infection are examined. Our results are consistent with a model in which SARS-CoV employs a unique three-step method for membrane fusion, involving receptor-binding and induced conformational changes in S glycoprotein followed by cathepsin L (CTSL) proteolysis and activation of membrane fusion within endosomes.
Methods
Cell Lines and Plasmids. Human ACE2 was amplified by PCR from a cDNA library and cloned into pcDNA3.1. pCAGGS SARS-CoV S, as described in ref. 4. pCB6 vesicular stomatitis virus (VSV)-G, amphotropic murine leukemia virus (MLV-A) envelope, and avian sarcoma and leukosis virus (ASLV-A) envelope are described in refs. 4 and 7.
Cells were maintained in DMEM10 (DMEM supplemented with 10% FBS). A HeLa/Tva cell line was produced by using pcDNA6-Tva and selection with blasticidin. The 293T cells were transiently transfected with human ACE2 (293T/ACE2), by using standard calcium phosphate transfection techniques and challenged 48 h posttransfection.
Pseudotype Preparation. Pseudotypes were produced, essentially as described in ref. 4, by using 10 μg of luciferase of GFP vector (pNL-luc or pNL-gfp) (8) and 30 μg of plasmid-encoding viral envelope or ACE2. Dual-envelope-expressing virions were transfected with 10 μg of pNL-GFP, 15 μg of pCB6 ASLV-A envelope, and 20 μg of pCAGGS SARS-CoV S. If required, virions were concentrated by ultracentrifuge concentration at 40,000 rpm in a SW41 rotor (Beckman) through a 20% sucrose cushion for 1 h at 4°C. The pellets were resuspended in PBS overnight at 4°C.
Trypsin Pretreatment. Concentrated pseudovirions were exposed to l-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma) for 10 min at 25°C. DMEM10 supplemented with 75 μg/ml soybean trypsin inhibitor (STI) was then added. Samples were used to spin-infect 293T/ACE2 cells at 1,200 × g for 2 h at 4°C. After incubation for 5 h at 37°C, the medium was changed, and the cells were incubated for an additional 40 h. The cells were analyzed for luciferase activity by using a commercial assay (Promega).
Trypsin Bypass. Preincubation of 293T/ACE2 cells took place at 37°C for 45 min with DMEM10 in the presence or absence of ammonium chloride (20 mM). The medium was replaced with cold DMEM10 in the presence or absence of ammonium chloride (40 mM) and incubated for an additional 15 min at 4°C. An equal volume of diluted cold virus was added [a 1-in-10 dilution of HIV-luc(SARS S) or a 1-in-100 dilution of HIV-luc(VSV-G)], and the cells were spin-infected at 4°C to allow virus-binding to cells. The medium was replaced with warm serum-free DMEM in the presence or absence of ammonium chloride (20 mM) and incubated at 37°C for 15 min. The medium was removed, and fresh DMEM in the presence or absence of TPCK-trypsin (15 μg/ml) was added for 10 min at 25°C. The trypsin was removed, and DMEM10 supplemented with STI (75 μg/ml) in the presence or absence of ammonium chloride (20 mM) was added. The medium was replaced with fresh DMEM10 12 h later. Cells were analyzed for luciferase activity 36 h later.
Replication-Competent SARS-CoV Assays. SARS-CoV (strain Tor2) was handled under biosafety-level 3 conditions and grown and titered on Vero E6 cells. For trypsin-bypass experiments, Vero E6 cells were incubated on ice for 1 h with DMEM2.5 (in the presence or absence of 25 mM ammonium chloride or 500 μg/ml leupeptin). SARS-CoV, at a multiplicity of infection of ≈0.5, was then added, and the cells were spin-infected at 4°C for 1 h at 1,200 × g. The virus was removed, and the cells were incubated for 10 min with serum-free DMEM at 37°C. The medium was then replaced with DMEM in the presence or absence of TPCK-trypsin (15 μg/ml), and the cells were incubated at room temperature for 10 min. The trypsin was removed and replaced with DMEM2.5 containing STI (75 μg/ml) in the presence or absence of ammonium chloride (25 mM) or leupeptin (500 μg/ml). The cells were incubated at 37°C for 4 h, the medium was replaced with DMEM2.5 without inhibitors, and the cells were incubated for an additional 40 h. The cells were fixed for 10 min in cold methanol/acetone, washed in PBS, and incubated for 2 h at 65°C. The cells were immunostained with anti-S protein antibodies IMG-557 and IMG-5010 (Imgenex, San Diego), at 0.5 μg/ml, followed by a mixture of anti-rabbit and anti-mouse FITC conjugates.
For leupeptin sensitivity assays, 293T/ACE2 cells were pretreated for 1 h with DMEM2.5 in the presence or absence of leupeptin and challenged with an equal volume of virus at a multiplicity of infection of ≈5. After 3 h, the cells were washed twice and incubated with DMEM2.5 in the presence or absence of leupeptin for an additional 4 h. The medium was then replaced with DMEM2.5, and the cells were incubated for 72 h. The supernatant was harvested, centrifuged to remove cell debris, and incubated at 65°C for 1 h in 1% Empigen (Calbiochem). Samples were analyzed for SARS-CoV nucleocapsid by using a commercial ELISA kit (Imgenex).
Intervirion Fusion. HIV-luc(ACE2) (500 ng of p24) was mixed with 1,000 ng of p24 of HIV-gfp particles incorporating ASLV-A envelope, SARS-CoV S protein, or both envelopes in PBS at 4°C for 30 min to allow binding. Samples were raised to 37°C for 15 min to allow for conformational rearrangements. Virions were adjusted to the desired pH with 0.1 M citric acid. PBS, TPCK-trypsin (final concentration 10 μg/ml), CTSL, cathepsin B (CTSB) (final concentrations 2 μg/ml) or CTSL buffer alone was then added. Recombinant CTSL (R & D Systems) was preactivated by incubation for 15 min at 10 μg/ml in 50 mM Mes, pH 6.0, on ice. Recombinant CTSB (R & D Systems) was preactivated in 25 mM Mes, 5 mM DTT, pH 5.0, for 30 min at 25°C. After a 10-min incubation at 25°C, proteolysis was halted by the addition of 300 μl of DMEM10 containing leupeptin (25 μg/ml) and STI (75 μg/ml). Virions were then incubated at 37°C for 30 min to allow membrane fusion. 100 μl of the virion mixture was added in quadruplicate to HeLa-Tva cells pretreated for 1 h with leupeptin (20 μg/ml). The cells were spin-infected and incubated at 37°C for 5 h. The medium was replaced with fresh DMEM10 and the cells were assayed for luciferase activity 40 h later.
Temperature-Sensitivity Intervirion-Fusion Assay. Intervirion-fusion assays were performed as above, except that binding was performed wholly at 4°C for 50 min for some samples, whereas others were allowed to bind at 4°C for 30 min, followed by 15 min at 37°C. The samples incubated at 37°C were returned to 4°C for 5 min, and cold TPCK-trypsin (final concentration of 10 μg/ml) was added. After a 15-min incubation at 4°C, proteolysis was halted by the addition of DMEM10 with STI (75 μg/ml) and leupeptin (25 μg/ml). Virions were then incubated at 37°C for 30 min to allow membrane fusion to occur, and the assay was completed as described above.
Protease Inhibitors. Vero E6 cells or 293T cells were pretreated for 1 h with leupeptin (Roche Molecular Biochemicals), CA-074, E64c, aprotinin, Z-leu-leu-leu-f luoromethyl ketone (Z-lll-FMK), or MDL28170 (Sigma). Inhibitors were removed and replaced with the same inhibitors at double the final concentration. An equal volume of pseudotypes was then added, and cells were spin-infected as described above. After spin-infection, the cells were incubated for 5 h, and the medium was replaced with fresh DMEM10 without drug. Cells were assayed for luciferase activity after 40 h.
Chemical-Library Screening for Cathepsin L. A library of 1,000 pharmacologically active compounds in DMSO was diluted to 100 μM in 50% glycerol and printed in triplicate on polysine-coated glass, as described in ref. 9. The library was screened for inhibitors of human CTSL at 1 μM in 400 mM NaCl, 20 mM malonate buffer, and 1 mM EDTA, pH 5.5, with fluorogenic substrate Z-Phe-Arg-AMC (Bachem) at 1 mM for detection. Leupeptin and blank spots with no compounds were used as controls. After the addition of enzyme and substrate, the reactions were incubated for 4 h before imaging the slide, as described in ref. 9.
IC50 Determination Protease Inhibitor MDL28170. IC50 determination was carried out by mixing 20 μl of 50 nM CTSL with 60 μl of buffer (400 mM NaOAc/4 mM EDTA, pH 5.5) containing MDL28170, at a final concentration ranging from 10 μM to 100 pM. The reaction was activated by the addition of 20 μl of 10 μM Z-Phe-Arg-7-amino-4-methylcoumarin (AMC). Fluorescence (Ex, 355 nM; Em, 460 nM) from cleaved AMC was detected in a kinetic mode by using an Ascent Fluoroskan FL plate reader (Thermo Electron LabSystems, San Jose, CA), with eight replicates on the same plate. The kinetic data were plotted, and the IC50 curve was determined by using software from GraphPad (San Diego).
Results
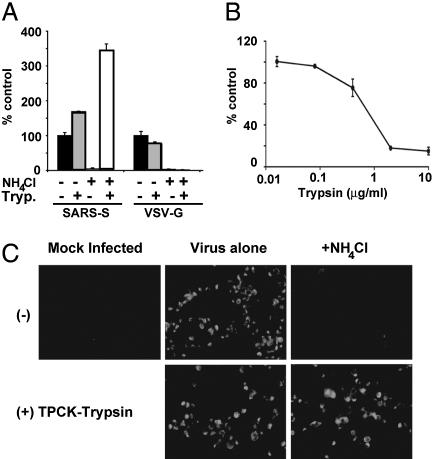
Proteolysis Activates SARS-CoV S Protein's Membrane-Fusion Potential. Fusion between Vero and 293T cells expressing SARS-CoV S protein occurs at neutral pH and is greatly enhanced by trypsin activation; yet, lysosomotropic agents block SARS-CoV infection (4). To reconcile the observed effects of pH and proteolysis on SARS-CoV membrane fusion, we posited that exogenous trypsin cleavage mimics the action of a pH-dependent endosomal protease (4). This hypothesis predicts that protease treatment of cell-associated virus should overcome the block to viral entry mediated by lysosomotropic agents like ammonium chloride. As demonstrated in ref. 4, pretreatment of cells with ammonium chloride dramatically reduced infection mediated by SARS-CoV S glycoprotein (Fig. 1A) and the pH-dependent viral glycoprotein VSV-G incorporated into HIV virions. However, when cell-bound HIV(SARS S) pseudovirions were exposed to trypsin, infection occurred in the presence or absence of ammonium chloride (Fig. 1 A). In fact, the combination of trypsin proteolysis and ammonium chloride increased viral infectivity by 3-fold. Proteolysis of replication-competent SARS-CoV (Tor2 strain) bound to Vero E6 cells also overcame the block to viral infection otherwise mediated by ammonium chloride (Fig. 1C). Thus, proteolysis of SARS-CoV bypasses the requirement for acid pH during the viral entry process.
Fig. 1.
Effect of trypsin on SARS-CoV infection. (A) Trypsin treatment bypasses ammonium chloride inhibition. HIV-luc(SARS S) or HIV-luc(VSV-G) were bound to mock (black and gray bars) or ammonium chloride-treated (third set of bars and white bars) 293T/ACE2 cells. The cells were incubated with either PBS (black bars and third set of bars) or TPCK-trypsin (gray and white bars). The results are presented as a percentage of no-ammonium-chloride (NH4Cl), no-trypsin (Tryp.) controls (≈4,000 and 10,000 RLU for SARS S and VSV-G, respectively) and represent the means of samples run in triplicate (±SD). Similar results were seen in two subsequent assays. (B) Trypsin pretreatment of S protein inactivates infectivity. HIV-luc(SARS S) infection of 293T/ACE2 cells was assessed as luciferase activity, presented as a percentage of no-trypsin control (≈40,000 RLU). The results represent the means of samples run in triplicate (±SD). (C) Trypsin treatment bypasses ammonium chloride inhibition of SARS-CoV. Mock- (Center) or 25 mM ammonium chloride-pretreated (Right) Vero E6 cells were spin-infected with replication-competent SARS-CoV at a multiplicity of infection of 0.5 and incubated with either DMEM (Upper) or DMEM containing TPCK-trypsin (Lower). After 48 h, the cells were immunostained for S protein.
In marked contrast to the studies in which SARS-CoV or HIV(SARS S) virions were bound to cells before trypsin treatment, proteolysis of free HIV(SARS S) pseudovirions dramatically diminished infectivity (Fig. 1B). Trypsin concentrations 10-fold lower than those used to activate fusion of cell-associated HIV(SARS S) were able to effectively inhibit infection by free virus. Similarly, in cell–cell fusion assays, proteolysis after mixing SARS-CoV S-expressing cells with target cells also resulted in more robust membrane fusion, compared with pretreatment with trypsin (data not shown). In addition, trypsin was unable to bypass the requirement for ACE2 on receptor-null cell lines, such as QT6 cells, even upon stable expression of the attachment factors DC-SIGN or DC-SIGNR (data not shown), suggesting a requirement for receptor engagement. The finding that, in solution, proteolysis leads to SARS-CoV S inactivation, whereas proteolysis leads to activation when the virus is bound to receptor-expressing membranes, demonstrates that the context in which proteolysis occurs is an important determinant of SARS-CoV infectivity.
Sensitivity of SARS-CoV S Protein-Mediated Entry to Protease Inhibitors. The ability of trypsin cleavage to overcome inhibition of endosomal acidification suggested a requirement for endosomal protease activity. To test this hypothesis, the infection of 293T cells with HIV(SARS S) was examined in the presence of leupeptin, an inhibitor of endosomal trypsin-like serine and cysteine proteases (Fig. 2A). Similar results were seen with 293T/ACE2 and Vero E6 cells (data not shown). Entry mediated by SARS-CoV S protein was efficiently blocked by leupeptin, with >95% inhibition observed at 10 μg/ml. Infection mediated by VSV-G, a pH-dependent viral membrane-fusion protein, and the pH-independent envelope from amphotropic MLV was not inhibited by leupeptin (Fig. 2 A).
Fig. 2.
Protease-inhibitor sensitivity. (A) Leupeptin inhibits S protein-mediated infection. The 293T cells were preincubated with leupeptin and challenged with HIV-luc SARS S (solid line, ♦), VSV-G (dashed line, ▪), or MLV-Ampho (dotted line, ▴). The results are presented as a percentage of infection of untreated cells (≈3,000 RLU) for each envelope) and represent the means of samples run in triplicate (±SD). Similar results were seen in two subsequent assays. (B) Leupeptin inhibits replication-competent SARS-CoV infection. Cells were either preincubated with leupeptin for 1 h and then exposed to virus for 3 h in the continued presence of leupeptin (solid line) or exposed to virus for 3 h and incubated for an additional 4 h with leupeptin (dashed line). At 3 days postexposure, the supernatant was analyzed for nucleoprotein by ELISA. The results are expressed as OD and represent the means of samples run in triplicate (±SD). Similar results were seen in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium cytoxicity assay. (C) Trypsin treatment bypasses leupeptin inhibition of live SARS-CoV. Mock- (Center) or 500 μg/ml leupeptin-pretreated (Right) Vero E6 cells were spin-infected with replication-competent SARS-CoV at a multiplicity of infection of 0.5 and incubated with either DMEM (Upper) or DMEM containing TPCK-trypsin (Lower). After 48 h, the cells were immunostained for S protein. (D) E64c blocks SARS-CoV S protein-mediated entry. The 293T cells were preincubated with E64c (solid lines) or aprotinin (dashed lines) and challenged with HIV-luc SARS S (black lines) or VSV-G (gray lines). The results are presented as a percentage of infection of untreated cells (≈1,500 RLU for VSV-G and 6,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen in two additional experiments. (E) Z-lll-FMK inhibits S protein-mediated infection. Vero E6 cells were preincubated with Z-lll-FMK (solid lines) or CA-074 (dashed lines) and then challenged with HIV-luc SARS S (black lines) or VSV-G (gray lines). The results are presented as a percentage of infection of untreated cells (≈15,000 RLU for VSV-G and 20,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen on 293T and 293T/ACE2 cells.
Infection by replication-competent SARS-CoV was also inhibited by leupeptin (Fig. 2B). Efficient inhibition was observed only if leupeptin was present 1 h before and during the 3-h exposure to the virus. When leupeptin was added to cells after exposure to SARS-CoV and then removed 4 h later, there was little or no effect on SARS-CoV replication, even at a concentration of 250 μg/ml. Thus, it is unlikely that the concentrations of leupeptin required to efficiently inhibit a spreading SARS-CoV infection are inhibiting postentry steps of replication or are merely toxic to the cells. Rather, leupeptin appears to inhibit an early step in viral entry. In a manner similar to inhibition by ammonium chloride (Fig. 1C), the leupeptin-mediated block to SARS-CoV infection of Vero E6 cells could be bypassed by proteolysis of virus bound to the cell surface (Fig. 2C). These findings are consistent with exogenous trypsin treatment compensating for cleavage normally mediated by leupeptin-sensitive endosomal proteases.
To more precisely define the protease(s) involved in SARS-CoV infection, a series of inhibitors were analyzed. E64c, an inhibitor of cysteine proteases, specifically inhibited infection by HIV(SARS S) pseudovirions, whereas aprotinin, an inhibitor of serine-type proteases, had no effect (Fig. 2D). Inhibitors of other classes of proteases, such as pepstatin, an aspartate protease inhibitor, also had no effect on either S protein- or VSV-G-mediated infection (data not shown). CA-074, a selective inhibitor of CTSB (10) did not dramatically affect infection by either HIV(SARS-CoV S) or HIV(VSV-G) (Fig. 2E; and see Table 1, which is published as supporting information on the PNAS web site). In contrast, Z-lll-FMK, an inhibitor of both CTSB and CTSL (11), efficiently inhibited infection by HIV(SARS S), but not by HIV(VSV-G) (Fig. 2E). In addition, a panel of four commercially available CTSL inhibitors specifically inhibited HIV(SARS S) infection (Table 1). Overall, these inhibitor results suggest that a pH-dependent cysteine protease, perhaps CTSL, is important for SARS-CoV infection.
Screen for Pharmacologically Active Inhibitors. To identify potential lead candidates for therapeutic inhibition of CTSL, a high-throughput screening of a library of pharmacologically active compounds was performed (see Methods). MDL28170 was identified as an efficient inhibitor of CTSL-mediated substrate cleavage, with an IC50 of 2.5 nM (Fig. 3A). MDL28170 (also known as calpain inhibitor III, or Z-Val-Phe-CHO) is an inhibitor of cytosolic calpains (12, 13). Inhibition of CTSB has also been noted (12). Interestingly, related calpain inhibitors have already been described as inhibitors of SARS-CoV replication (14), although it was assumed the action was through inhibition of viral proteases. Similarly, we found efficient inhibition of SARS-CoV replication using MDL28170 (data not shown). In addition, MDL28170 efficiently inhibited infection by HIV(SARS S), but not by HIV(VSV-G) pseudovirions (Fig. 3B and Table 1). Given that the pseudotype infection assay is a direct measure of S protein-mediated viral entry, these results suggest that MDL28170's action is due to inhibition of endosomal protease activity during viral entry. Thus, these experiments identify MDL28170 as a strong initial candidate for antiviral inhibitors of SARS-CoV viral entry.
Fig. 3.
Cathepsin-L-specific inhibitor blocks infection. (A) MDL28170 inhibits CTSL activity with an IC50 of 2.5 nM. A 1,000-compound library was screened for inhibitors of CTSL activity (Inset, bottom left). MDL28170 (Inset, top right) was found to be a potent inhibitor. The compound library was screened against several other cathepsins, including CTSB, with no hits. The activity of MDL28170 was confirmed in an in vitro CTSL-cleavage assay (inhibition curve). (B) MDL28170 inhibits S protein-mediated infection. The 293T cells were preincubated with MDL28170 and challenged with HIV-luc SARS S (solid line) or VSV-G (dashed line). The results are presented as a percentage of infection of untreated cells (≈100,000 RLU for VSV-G and 20,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen on Vero E6 and 293T/ACE2 cells.
Protease-Mediated Activation of Membrane Fusion. To further study the relative contributions of acid pH and specific proteases on SARS-CoV infection, we developed a cell-free, virus–virus membrane-fusion assay employing virions that carry either S glycoprotein or the SARS-CoV cellular receptor ACE2 (3). The HIV(ACE 2) pseudotypes encode luciferase, whereas S glycoprotein particles encode GFP and have on their surface not only SARS-CoV S but also the envelope glycoprotein from subgroup A ASLV-A envelope. Membrane fusion between the virions carrying SARS-CoV S and those with ACE2 is indicated by transfer of the genome encoding luciferase to HeLa/tva cells expressing the cellular receptor for ASLV-A but not SARS-CoV. A similar cell-free membrane-fusion assay has been used to analyze HIV and MLV-envelope-mediated membrane fusion and, in both instances, has been shown to accurately reflect normal virus infection requirements (15, 16).
Characterization of the peseudovirions demonstrated the efficient production of HIV particles containing ACE2 in their lipid coats, as determined by Western analysis of purified virions (data not shown). These HIV-luc(ACE2) particles were able to efficiently and specifically infect 293T cells expressing SARS-CoV S protein, as demonstrated by high levels of luciferase activity in the target cells (see Fig. 5, which is published as supporting information on the PNAS web site). HIV particles encoding GFP and incorporating both SARS-CoV S and ASLV-A envelope [referred to as HIV-gfp(SARS S/ASLV-A)] were also efficiently produced and infectious on cell lines expressing either ACE2 or the ASLV-A receptor Tva (data not shown).
The ability of SARS-CoV S and ACE2, on the surface of their respective virions, to mediate intervirion membrane fusion was assessed by coincubating the pseudotypes before infection of HeLa/Tva cells. In contrast to the results seen when individual pseudovirions were used, a mixture of HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) resulted in expression of the luciferase-encoding genome in HeLa/Tva cells (Fig. 4A). Luciferase activity was not observed when a pseudotype that did not carry ACE2 [termed HIV-luc(bald)] was mixed with HIV-gfp(SARS S/ASLV-A) or when HIV-gfp particles expressing ASLV-A env alone were mixed with HIV-luc(ACE2) (Fig. 4A). Thus, luciferase activity appears to be a measure of SARS-CoV S-mediated intervirion membrane fusion.
Fig. 4.
S protein-mediated intervirion fusion. (A) Intervirion fusion requires ACE2 and S protein. Bald or ACE2 particles encoding luciferase (x axis) were incubated with particles encoding GFP (SARS S and ASLV-A envelope, gray bars; SARS S alone, black bars; or ASLV-A envelope alone, white bars). Virions were mixed and used to infect HeLa/Tva cells that had been pretreated with medium in the presence and absence of leupeptin (Leu) (20 μg/ml). Intervirion fusion was measured as luciferase activity 48 h postinfection. Results represent the means of samples run in triplicate (±SD). (B) Trypsin cleavage promotes fusion mediated by S protein. Intervirion fusion between HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) treated with TPCK-trypsin (10 μg/ml) for 10 min at 25°C or pulsed at pH 5.0 was quantified by luciferase activity 48 h postinfection of HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in triplicate (±SD). Mixtures of HIV-gfp(SARS S), HIV-gfp(ASLV-A), and HIV-luc(ACE2) could not be activated by trypsin cleavage, suggesting that S and ASLV-A envelope are required to be incorporated into the same particle in order for transduction of target cells by fused particles. (C) Receptor interactions at elevated temperature are required before trypsin cleavage. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and incubated at 4°C to allow binding. Samples were then incubated at the noted temperatures. TPCK-trypsin digestion was carried out at 4°C for 15 min. The results represent the means of samples run in quadruplicate (±SD). Similar results were observed in two additional experiments. Temp., temperature. (D) CTSL enhances intervirion fusion. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and incubated for 10 min at 25°C with preactivated CTSB (at pH 5.0), CTSL (at pH 6.0), CTSL buffer alone (at pH 6.0), or TPCK-trypsin (at pH 7.0). The mixed virus was used to infect HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in quadruplicate (±SD). Similar results were observed in two subsequent experiments. (E) Acidic conditions are required for CTSL-mediated S protein activation. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and adjusted to various pHs and CTSL was added. After neutralization of acid conditions, the mixed virus was used to infect HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in quadruplicate (±SD). Tryp, trypsin. Similar results were observed in an additional experiment.
We used this virus–virus membrane-fusion assay to examine the effects of pH and proteolysis on SARS-CoV-mediated membrane fusion. Pretreatment of the HeLa/Tva cells with leupeptin before the addition of mixed virions abrogated S protein-mediated intervirion fusion, as demonstrated by the background levels of luciferase activity observed (Fig. 4A). As a control, leupeptin was found to have no effect on ASLV-A envelope-mediated infection of HeLa/Tva cells (data not shown). These results suggest that, for virus–virus membrane fusion to occur, the particles must be coendocytosed into endosomes, where proteases sensitive to leupeptin are able to alleviate a block to fusion between the virus particles. Thus, in all subsequent assays, target cells were pretreated with leupeptin to determine the effect of the addition of exogenous protease on virus–virus fusion before plating on target cells.
To more directly assess the requirement for proteolytic activation of S protein, we incubated the two pseudovirion populations to allow S protein and ACE2-mediated virus–virus binding. Trypsin proteolysis of the bound virus particles dramatically increased luciferase expression in target HeLa/Tva cells, despite endosomal proteolysis inhibition by leupeptin (Fig. 4B). In contrast to trypsin, a brief low-pH pulse did not facilitate virus–virus membrane fusion, as assayed by luciferase gene transfer in the leupeptin-treated target cells (Fig. 4B). The higher levels of luciferase activity seen in these experiments compared with Fig. 4A may reflect a more efficient membrane fusion reaction, because this assay does not rely on traffic of the bound virions to the endosome for intervirion fusion. These results confirm that a low-pH environment does not appear to act as a direct trigger for SARS-CoV entry. In agreement with the studies above, showing proteolytic bypass of lysosomotropic-agent-mediated inhibition, these membrane-fusion data are most consistent with a model in which the low-pH environment of the endosome is needed for proteolytic activation of membrane-fusion activity.
Temperature-Dependence of Protease Activation. The fact that S protein needs to bind ACE2 in order for trypsin treatment to have an effect on membrane fusion (Fig. 1) suggested that conformational changes induced by SARS-CoV S protein–receptor interaction may be required before proteolysis. Conformational changes are generally slowed or arrested at low temperatures. Thus, we examined whether incubation of mixtures of HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) virions at 4°C, compared with 37°C before treatment with protease, affected subsequent membrane-fusion activity, possibly by preventing conformational changes in S protein induced by ACE2 binding. Only a small increase in intervirion fusion was seen with HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) virus particles maintained at 4°C, despite trypsin treatment (Fig. 4C). When the mixture of HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) particles was preincubated at 37°C for 15 min, however, before being returned to 4°C for trypsin treatment, efficient intervirion fusion was observed (Fig. 4C). These results indicate that a receptor and temperature-dependent step occurs before proteolysis of SARS-CoV S protein, possibly involving receptor-induced conformational changes within S to either expose a protease cleavage site or to undergo some of the steps leading up to membrane fusion.
Cathepsin L Activates SARS-CoV Membrane Fusion. The ability of specific inhibitors to block SARS-CoV entry and the requirement for proteolysis for S-mediated intervirion membrane fusion suggested that CTSL may play a role in directly modulating the fusion activity of SARS-CoV S. To test this hypothesis, recombinant cathepsins common to cellular endosomes, such as CTSB and CTSL, were used in the virus–virus membrane-fusion assay. Treatment of mixed HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) particles with CTSL at pH 6.0 mediated intervirion fusion as efficiently as did trypsin (Fig. 4D). In contrast, CTSB treatment did not produce a reproducible increase in intervirion fusion. Additionally, CTSL buffer alone at pH 6.0 had no effect. The sensitivity of SARS-CoV S protein-mediated entry to lysosomotropic agents is likely explained by the fact that endosomal proteases, such as CTSL, cleave more efficiently and are more stable at acidic pH. To address this question, CTSL-mediated activation of SARS-CoV S membrane fusion was performed at different pHs. With HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) particles and CTSL, a gradual reduction in levels of fusion was observed with increasing pH, and incubation at pH 7.1 resulted in no intervirion fusion (Fig. 4E).
Discussion
Distinct spikes of trimeric glycoproteins mediate the attachment, fusion, and entry of enveloped RNA viruses such as the orthomyxo-, paramyxo-, filo-, retro-, and coronaviruses. A hallmark of these class I viral membrane-fusion proteins is that they undergo a series of structural rearrangements that cause fusion between the viral and cellular membranes. The glycoproteins in the virion spikes are in an energetically unfavorable conformation, and an activating trigger is required to allow metastable protein complexes to refold into a more stable final form. For many viruses, binding to specific receptors can induce the conformational rearrangements within envelope proteins required for membrane fusion by binding to a single receptor, as is the case for Amphotropic MLV, or consecutive binding to a receptor and coreceptor, as is seen in HIV entry. Alternatively, viruses such as influenza require only an acidic milieu to be triggered (17). More recently, a fourth means of achieving glycoprotein triggering has been described for the avian retrovirus ASLV-A, whereby both binding to a specific receptor and low pH are required in order for membrane fusion to be completed (18). We describe here a potential fifth model for glycoprotein triggering that requires the involvement of endosomal protease activity subsequent to receptor interactions.
A number of possibilities exist for the role of CTSL in SARS-CoV entry, including the cleavage of S protein, ACE2, or another cellular protein that aids in membrane fusion. One explanation is that, as in influenza, cleavage is required to expose the hydrophobic fusion peptide. Indeed, protease activation of influenza hemagglutinin can occur during entry in certain cell types (19). However, in the case of SARS-CoV, it appears that interaction with receptor is required before such cleavage. Although a fusion peptide has not been established for SARS-CoV S protein by mutagenesis mapping, prediction models place it immediately amino-terminal of the membrane-distal leucine/isoleucine heptad repeat (HR1) (20). Another likely scenario is that S protein is physically constrained from undergoing the necessary conformational changes required for fusion peptide insertion. Cleavage at sites exposed by receptor-binding then either relieves these constraints or even actively induces the conformational rearrangements leading to fusion peptide insertion. In this model, one can view proteolytic cleavage of S as the fusion-activating trigger comparable to pH for influenza HA or coreceptor-binding for HIV envelope. Analogous to the conformations of the influenza and HIV proteins induced by pH or coreceptor binding, it seems likely that the CTSL-cleaved SARS-CoV S may be a transient intermediate in the membrane-fusion process. It is, perhaps, this transient nature or the rather nonspecific character of cathepsin L that has made identification of the cleavage sites in S difficult. However, preliminary mutational analysis of the residues near the S1–S2 boundary of SARS S suggest that trypsin activation does not require cleavage at this location (G.S., A.J.R., and P.B., unpublished data).
An alternative model is that receptor-binding mediates the early conformational changes in the S protein, including fusion peptide insertion into the target membrane but that uncleaved S protein is constrained in such a way that the later steps in membrane fusion, such as stable six-helix bundle formation or fusion pore formation, cannot occur. The act of cleavage then releases this constraint. In support of this model, the ASLV envelope protein is thought to use receptor binding to activate the early steps of membrane fusion, including fusion peptide insertion and at least partial refolding into a six-helix bundle, but needs a low-pH step to complete membrane fusion (21–23). It may be that SARS-CoV S uses protease in an analogous manner to pH for ASLV as a second trigger acting late in the membrane-fusion process. The role, if any, that extracellular proteases commonly found in sites of SARS-CoV replication (such as the airways and the gut) may play in this model for viral entry is unclear. It is even possible that extracellular cleavage after receptor engagement would negate the requirement for endocytosis, as seen in the trypsin-bypass experiments.
Overall, these experiments suggest a previously undescribed paradigm for viral entry into target cells. Namely, that for SARS-CoV S protein, receptor-mediated conformational changes induce exposure of cryptic cleavage sites within viral envelope glycoprotein. Proteolysis by cellular proteases is then necessary to fully activate the viral glycoprotein's membrane-fusion potential. Further characterization of this phenomenon is likely to highlight steps in the activation of S protein that may yield targets for specific inhibitors of entry. Indeed, the finding that CTSL is an important activating protease for SARS-CoV infection suggests CTSL as a target for therapeutic intervention. MDL28170 represents an attractive starting point for specific inhibitors of CTSL as antiviral therapeutics targeting SARS-CoV entry.
The entry process described here for SARS-CoV S protein and the inhibitors of this process also raise the question whether other classically defined pH-dependent viruses display this dependence because of a requirement for acidic protease involvement and not pH-induced structural rearrangements, as is commonly assumed. Indeed, it has recently been suggested that Ebola glycoprotein undergoes similar processing by endosomal proteases (see ref. 24; G.S., A.J.R., and P.B., unpublished observations). Future investigation will reveal whether SARS-CoV and Ebola represent initial members of a previously uncharacterized category of viral fusion proteins.
Supplementary Material
Acknowledgments
We thank Jim Wilson and Gary Kobinger (University of Pennsylvania) for Tor-2 virus, Arwen Vermeulen for selecting the HeLa-Tva cells, Stefan Pöhlmann for comments, and Ben Doranz for advice. This work was funded by National Institutes of Health (NIH) Mid-Atlantic Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Grant U54 AI057168 and NIH Grants R01 AI43455, R21 AI059172, and R21 AI 058701. J.D.R. is supported by American Foundation for AIDS Research Fellowship 106437-34-RFGN.
Author contributions: G.S., J.D.R., S.L.D., and P.B. designed research; G.S., D.N.G., A.J.R., and J.D.R. performed research; D.N.G. and S.L.D. contributed new reagents/analytic tools; G.S., D.N.G., A.J.R., J.D.R., and P.B. analyzed data; and G.S. and P.B. wrote the paper.
Abbreviations: ACE2, angiotensin-converting enzyme 2; ASLV, avian sarcoma and leukosis virus; CTSB, cathepsin B; CTSL, cathepsin L; MLV, murine leukemia virus; TPCK, L-1-tosylamido-2-phenylethyl chloromethyl ketone; RLU, relative light units; S, spike (glycoprotein); SARS, severe acute respiratory syndrome; SARS-CoV, SARS-associated coronavirus; STI, soybean trypsin inhibitor; VSV, vesicular stomatitis virus; Z-lll-FMK, Z-leu-leu-leu-fluoromethyl ketone.
References
- 1.Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. [DOI] [PubMed] [Google Scholar]
- 2.Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., Luo, S. W., Li, P. H., Zhang, L. J., Guan, Y. J., et al. (2003) Science 302, 276-278. [DOI] [PubMed] [Google Scholar]
- 3.Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., Somasundaran, M., Sullivan, J. L., Luzuriaga, K., Greenough, T. C., et al. (2003) Nature 426, 450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons, G., Reeves, J. D., Rennekamp, A. J., Amberg, S. M., Piefer, A. J. & Bates, P. (2004) Proc. Natl. Acad. Sci. USA 101, 4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang, Z. Y., Huang, Y., Ganesh, L., Leung, K., Kong, W. P., Schwartz, O., Subbarao, K. & Nabel, G. J. (2004) J. Virol. 78, 5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofmann, H., Hattermann, K., Marzi, A., Gramberg, T., Geier, M., Krumbiegel, M., Kuate, S., Uberla, K., Niedrig, M. & Pohlmann, S. (2004) J. Virol. 78, 6134-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilbert, J. M., Bates, P., Varmus, H. E. & White, J. M. (1994) J. Virol. 68, 5623-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor, R. I., Chen, B. K., Choe, S. & Landau, N. R. (1995) Virology 206, 935-944. [DOI] [PubMed] [Google Scholar]
- 9.Gosalia, D. N. & Diamond, S. L. (2003) Proc. Natl. Acad. Sci. USA 100, 8721-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montaser, M., Lalmanach, G. & Mach, L. (2002) Biol. Chem. 383, 1305-1308. [DOI] [PubMed] [Google Scholar]
- 11.Hanisch, F. G., Schwientek, T., Von Bergwelt-Baildon, M. S., Schultze, J. L. & Finn, O. (2003) Eur. J. Immunol. 33, 3242-3254. [DOI] [PubMed] [Google Scholar]
- 12.Brana, C., Benham, C. D. & Sundstrom, L. E. (1999) Eur. J. Neurosci. 11, 2375-2384. [DOI] [PubMed] [Google Scholar]
- 13.Lubisch, W. & Moller, A. (2002) Bioorg. Med. Chem. Lett. 12, 1335-1338. [DOI] [PubMed] [Google Scholar]
- 14.Barnard, D. L., Hubbard, V. D., Burton, J., Smee, D. F., Morrey, J. D., Otto, M. J. & Sidwell, R. W. (2004) Antivir. Chem. Chemother. 15, 15-22. [DOI] [PubMed] [Google Scholar]
- 15.Sparacio, S., Zeilfelder, U., Pfeiffer, T., Henzler, T. & Bosch, V. (2000) Virology 271, 248-252. [DOI] [PubMed] [Google Scholar]
- 16.Zhou, J. & Aiken, C. (2001) J. Virol. 75, 5851-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531-569. [DOI] [PubMed] [Google Scholar]
- 18.Mothes, W., Boerger, A. L., Narayan, S., Cunningham, J. M. & Young, J. A. (2000) Cell 103, 679-689. [DOI] [PubMed] [Google Scholar]
- 19.Boycott, R., Klenk, H. D. & Ohuchi, M. (1994) Virology 203, 313-319. [DOI] [PubMed] [Google Scholar]
- 20.Bosch, B. J., Martina, B. E., Van Der Zee, R., Lepault, J., Haijema, B. J., Versluis, C., Heck, A. J., De Groot, R., Osterhaus, A. D. & Rottier, P. J. (2004) Proc. Natl. Acad. Sci. USA 101, 8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melikyan, G. B., Barnard, R. J., Markosyan, R. M., Young, J. A. & Cohen, F. S. (2004) J. Virol. 78, 3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Earp, L. J., Delos, S. E., Netter, R. C., Bates, P. & White, J. M. (2003) J. Virol. 77, 3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Netter, R. C., Amberg, S. M., Balliet, J. W., Biscone, M. J., Vermeulen, A., Earp, L. J., White, J. M. & Bates, P. (2004) J. Virol. 78, 13430-13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandran, K., Sullivan, N. J., Felbor, U., Whelan, S. P. & Cunningham, J. M. (2005) Science 308, 1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.