Abstract
Nematode mitochondria expresses two types of extremely truncated tRNAs that are specifically recognized by two distinct elongation factor Tu (EF-Tu) species named EF-Tu1 and EF-Tu2. This is unlike the canonical EF-Tu molecule that participates in the standard protein biosynthesis systems, which basically recognizes all elongator tRNAs. EF-Tu2 specifically recognizes Ser-tRNASer that lacks a D arm but has a short T arm. Our previous study led us to speculate the lack of the D arm may be essential for the tRNA recognition of EF-Tu2. However, here, we showed that the EF-Tu2 can bind to D arm-bearing Ser-tRNAs, in which the D–T arm interaction was weakened by the mutations. The ethylnitrosourea-modification interference assay showed that EF-Tu2 is unique, in that it interacts with the phosphate groups on the T stem on the side that is opposite to where canonical EF-Tu binds. The hydrolysis protection assay using several EF-Tu2 mutants then strongly suggests that seven C-terminal amino acid residues of EF-Tu2 are essential for its aminoacyl-tRNA-binding activity. Our results indicate that the formation of the nematode mitochondrial (mt) EF-Tu2/GTP/aminoacyl-tRNA ternary complex is probably supported by a unique interaction between the C-terminal extension of EF-Tu2 and the tRNA.
INTRODUCTION
In the elongation cycle of protein biosynthesis in bacteria and eukaryotic organelles, one of the most essential steps is the formation of an active ternary complex between elongation factor Tu (EF-Tu), aminoacyl-tRNA (aa-tRNA) and GTP, after which the aa-tRNA is delivered to the ribosomal A-site (1,2). The canonical EF-Tu molecule associates with all kinds of elongator aa-tRNAs, except for selenocysteinyl-tRNA, by recognizing the acceptor and T stems (3–5). Canonical EF-Tu consists of three domains. The crystal structure of the ternary complex showed that GTP binds to domain 1, that the aminoacyl-group and the acceptor stem of aa-tRNA are sandwiched between domains 1 and 2, and that the T stem of aa-tRNA interacts with domain 3 (3,4). This aa-tRNA-recognition mechanism is highly conserved among bacterial EF-Tu. However, there are a few exceptions. Elongation factor SelB, which binds to selenocysteinyl-tRNASec and delivers it to the ribosome (6), possesses not only the three N-terminal domains homologous to EF-Tu but also a C-terminal domain 4 which recognizes a specific mRNA structure (7). The crystal structure of SelB/GppNHp suggested that it may recognize the extra long acceptor stem of tRNASec (8), although SelB binds to the acceptor and T stems of tRNASec, like EF-Tu. Other exceptions have been found in nematode mitochondria as described below.
It is known that a variety of mitochondrial (mt) tRNAs encoded by metazoan mtDNA lack some parts, although almost all tRNAs in prokaryotes and the eukaryotic cytoplasm have a conserved cloverleaf secondary structure (9). One of the most extreme cases of these variant tRNAs occurs in nematode mitochondria. The nematodes Caenorhabditis elegans, Ascaris suum and Onchocerca volvulus have two structurally distinct types of tRNA, one that lacks a T arm (20 tRNA species), and the other that lacks a D arm but bears a short T arm (two tRNASers) (10–12).
Previously, we found that these two types of truncated tRNAs in C.elegans are specifically recognized by two distinct EF-Tu species named EF-Tu1 and EF-Tu2 (13,14). EF-Tu1 specifically recognizes the T-armless aa-tRNAs, probably via its 57 amino acid C-terminal extension, which is not seen in canonical EF-Tu molecules (13). EF-Tu2 also possesses a C-terminal extension, albeit a shorter one consisting of 17 amino acids, but its role is unknown. EF-Tu2 has two unique properties in aa-tRNA recognition: (i) it exclusively recognizes the seryl-moiety of Ser-tRNA, unlike the canonical EF-Tu, which can bind to all 20 amino acids in the aa-tRNAs, and (ii) it is specific for a particular tRNA moiety since it bound only to D-armless Ser-tRNAs among four Ser-tRNAs used in our previous study (14). In other words, we found EF-Tu2 did not recognize bovine mitochondrial (mt) that possesses a D arm, despite its non-canonical secondary structure (15,16), which led us to speculate that EF-Tu2 may recognize ‘the lack of the D arm’.
Like the C.elegans EF-Tu molecules, mammalian mt EF-Tu also contains a C-terminal extension, albeit one that is only 11 residues long (17). Andersen et al. determined the crystal structure of the mammalian mt EF-Tu/GDP complex (18) and found this C-terminal extension adopts a helical structure. If its position would be maintained in the EF-Tu/aa-tRNA/GTP complex, it is likely that this helical C-terminal extension would interact with the tRNA (18). If this speculation is correct, it suggests that the somewhat longer C-terminal extension of EF-Tu2 in C.elegans may also interact with its tRNA species.
Given these observations, we sought to investigate the mechanism by which EF-Tu2 recognizes the tRNA moiety of aa-tRNA. Two specific questions were addressed: (i) What role does the C-terminal extension of EF-Tu2 play in the aa-tRNA recognition by EF-Tu2? and (ii) Is the lack of the D arm in the aa-tRNA essential for this recognition event?
MATERIALS AND METHODS
Preparation of tRNA transcripts
All tRNAs used in this study except for native Escherichia coli were prepared by in vitro transcription. To generate DNA templates for transcription, primer extension reactions were performed using two primers designed to complement each other at their 3′ regions (∼20 nt). In the DNA templates, the promoter sequence for T7 RNA polymerase is directly connected to the upstream region of the tRNA sequence. The transcription reaction was performed at 37°C for 4 h in a reaction mixture including 40 mM Tris–HCl (pH 8.0), 6 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM spermine, 0.01% Triton X-100, 50 μg/ml bovine serum albumin (BSA), 20 mM GMP, 1 mM each of ATP, GTP, CTP and UTP, 90 μg/ml T7 RNA polymerase and 10 μg/ml template double-stranded DNA. The products were purified by 10% denaturing polyacrylamide gel electrophoresis (PAGE).
Preparation of aa-tRNAs
All tRNAs used in our study were charged with serine using bovine mt SerRS (19). The reaction was performed at 37°C for 30 min in a reaction mixture that contained 100 mM HEPES–KOH (pH 7.8), 15 mM MgCl2, 60 mM KCl, 2 mM ATP, 5 mM DTT, 40 μM [3H] L-serine (74 GBq/mmol), 320 μg/ml bovine mt SerRS and 0.2–0.5 A260 U of tRNA. The serylated tRNA was purified as described (13) and finally dissolved with 6 mM KOAc (pH 5) at a concentration of 2 μM. The concentration of Ser-tRNA was estimated from the labeled amino acids incorporated into the tRNA.
Hydrolysis protection assay
The assay was basically performed according to (13) and (20). The deacylation reaction mixture contained 75 mM Tris–HCl (pH 7.5), 75 mM NH4Cl, 15 mM MgCl2, 7.5 mM DTT, 60 μg/ml BSA, 0.1 mM GTP, 2.375 mM phosphoenolpyruvate, 2.5 U/ml pyruvate kinase, 1.2 μM EF-Tu and 0.2 μM Ser-tRNA. The reaction mixture was preincubated at 30°C for 10 min without Ser-tRNA, after which Ser-tRNA was added. The deacylation reaction was performed at 30°C.
Modification interference assay using ethylnitrosourea (ENU)
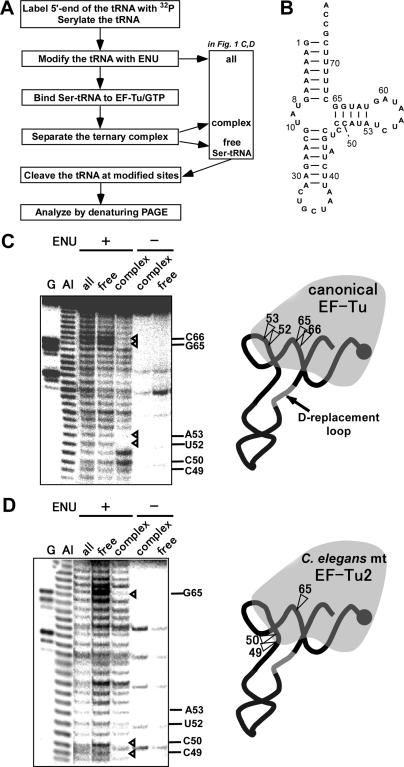
The 5′ 32P-labeled tRNAs were prepared as described (21) and purified by 10% denaturing gel electrophoresis. The phosphate backbone of the tRNA was ethylated with ENU according to (19,22,23) and the modification interference assay using ENU was performed according to (24,25) with slight modifications. The overall scheme of this assay is shown in Figure 1A. Ser-tRNA was ethylated at 37°C for 30 min in a 25 μl reaction mixture that contained 30 mM sodium cacodylate (pH 7.4), 0.2 mM EDTA, 80 μg/ml 5′-labeled Ser-tRNA and 0.2 vol of saturated ENU (in ice-cold ethanol solution). In the control experiment without ethylation, ethanol substituted for the ENU solution. The alkylated Ser-tRNAs were mixed with EF-Tu on ice for 10 min in a reaction mixture containing 50 mM Tris–HCl (pH 7.6), 65 mM NH4OAc, 10 mM Mg(OAc)2, 15 μM EF-Tu/GTP and 80 μg/ml ethylated or unethylated Ser-tRNA. The mixture was loaded onto a 5% polyacrylamide gel containing 50 mM Tris–HCl (pH 7.6), 65 mM NH4OAc, 10 mM Mg(OAc)2, 1 mM EDTA, 1 mM DTT and 10 μM GTP, and PAGE was performed at 4°C for 1.5 h (50 mA) to separate the EF-Tu/GTP/Ser-tRNA ternary complex from free Ser-tRNA. The ternary complex and free Ser-tRNAs were then eluted from the gel. Each tRNA was extracted from the eluate with phenol/chloroform/isoamylalcohol (25:24:1) and collected by ethanol-precipitation. Each tRNA was further purified to remove deacyl tRNA by subjecting it to electrophoresis on a 7.5% denaturing polyacrylamide gel containing 0.1 M NaOAc (pH 5.0). Each tRNA was cleaved at its ethylated position at 65°C for 8 min in a reaction buffer containing 100 mM Tris–HCl (pH 8) and 7 M urea and then analyzed by 15% denaturing PAGE.
Figure 1.
Identification of the EF-Tu-binding sites on bovine mt by the ENU modification interference assay. (A) overall scheme of the ENU-modification interference assay using EF-Tu and Ser-tRNA. (B), the secondary structure of the bovine mt transcript. Residues on the tRNA are numbered according to the numbering rule as described (30). (C) and (D), autoradiographs of the ENU-modification interference assay using bovine mt EF-Tu (C) and C.elegans mt EF-Tu2 (D) (left). Lanes: G, RNase T1 ladder; Al, alkaline ladder; all, free and complex, ENU-modified Ser-tRNA, the Ser-tRNA free from EF-Tu, and the Ser-tRNA in the ternary complex, respectively, as shown in Figure 1A; ENU + or − the Ser-tRNA with or without ENU modification. (C) and (D), the binding sites on bovine mt of each EF-Tu are shown (right). The tRNA is portrayed as a simplified backbone with the aminoacyl moiety depicted by a filled circle. The phosphate positions recognized by each EF-Tu are indicated by arrowheads in the left and right panels.
Preparation of C.elegans mt EF-Tu2 mutants
Vectors expressing the EF-Tu2 mutants were prepared using the pET-15b (Novagen)-derived expression vector encoding N-terminal His-tagged C.elegans mt EF-Tu2 (14). The mutations were introduced by employing the Stratagene QuikChange site-directed mutagenesis protocol and the primers as shown in Table 1. The sequences of the mutated plasmids were confirmed by using an ABI prism 310 Genetic Analyzer. Rosetta (DE3) E.coli cells were transformed with the plasmids and the expressed recombinant proteins were purified as described (13).
Table 1.
Oligodeoxynucleotides primers used to mutate EF-Tu2
| Name | Sequence (5′–3′) |
|---|---|
| (-)7aa-5′ | GGCTGTAGAGAAACATAATCTTTAGAAGTCCGCTGAAAAGATGTAG |
| (-)7aa-3′ | CTACATCTTTTCAGCGGACTTCTAAAGATTATGTTTCTCTACAGCC |
| (-)3aa-5′ | AACATAATCTTAAAAAGTCCGCTTAGAAGATGTAGGGATCCGGCTG |
| (-)3aa-3′ | CAGCCGGATCCCTACATCTTCTAAGCGGACTTTTTAAGATTATGTT |
| K429A-5′ | AGGATCATGTGGCTGTAGAGGCTCATAATCTTAAAAAGTCCGCTG |
| K429A-3′ | CAGCGGACTTTTTAAGATTATGAGCCTCTACAGCCACATGATCCT |
| K433A-5′ | GGCTGTAGAGAAACATAATCTTGCTAAGTCCGCTGAAAAGATGTAG |
| K433A-3′ | CTACATCTTTTCAGCGGACTTAGCAAGATTATGTTTCTCTACAGCC |
| K434A-5′ | CTGTAGAGAAACATAATCTTAAAGCTTCCGCTGAAAAGATGTAAG |
| K434A-3′ | CCTACATCTTTTCAGCGGAAGCTTTAAGATTATGTTTCTCTACAG |
| K438A-5′ | ATAATCTTAAAAAGTCCGCTGAAGCTATGTAGGGATCCGGCTGCT |
| K438A-3′ | AGCAGCCGGATCCCTACATAGCTTCAGCGGACTTTTTAAGATTAT |
The mutated positions are shown by bold letters.
RESULTS
EF-Tu2-binding sites on D-armless tRNA
To delineate where on the D-armless Ser-tRNA the C.elegans mt EF-Tu2 binds, the phosphate groups on the Ser-tRNA backbone were randomly ethylated by ENU and the ENU-modification interference assay was performed (Figure 1A). The basis of this experiment is that EF-Tu cannot bind to Ser-tRNA if it bears ethyl modifications at phosphodiester bonds at site(s) necessary for EF-Tu binding. In this experiment, D-armless bovine mt (Figure 1B) was used as a substitute for the native nematode mitochondrial (mt) tRNASer molecule because the ternary complex formed by EF-Tu2, GTP, and the native nematode mt Ser-tRNASer cannot be clearly observed on a gel (although EF-Tu2 does bind to nematode mt Ser-tRNASer, as shown in Figure 4). Acid PAGE analysis showed that ∼38% of the aminoacyl-tRNA was deacylated during the ethylation reaction. To compare the ethylated positions of Ser-tRNA bound to EF-Tu to those of free Ser-tRNA without interference from deacyl-tRNA, deacyl-tRNA was removed from the free Ser-tRNA fraction by acid PAGE as described in Materials and Methods. Control experiments using bovine mt EF-Tu and Thermus thermophilus EF-Tu were also performed. Figure 1C shows that ethylation of the 3′-phosphates of U52, A53, G65 and C66 on the tRNA interfered with the binding of the bovine mt EF-Tu. Quite similar results were obtained for T.thermophilus EF-Tu (Supplementary Figure 1). These positions are the binding sites of canonical EF-Tu molecules, as has been shown previously by co-crystallographic analysis of Thermus aquaticus EF-Tu and aa-tRNA (3,4). With regard to C.elegans mt EF-Tu2, however, while the 3′-phosphate of G65 was important for its binding to D-armless bovine mt , the 3′-phosphates of C49 and C50, with which normal EF-Tu cannot interact, were also important (Figure 1D). These results indicate that C.elegans mt EF-Tu2 possesses a unique aa-tRNA recognition mechanism that differs from that employed by canonical EF-Tu molecules.
Figure 4.
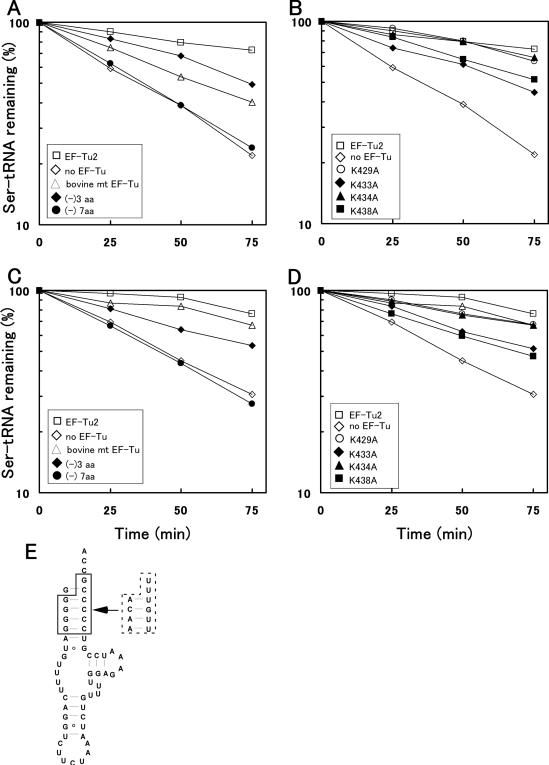
Ability of EF-Tu2 mutants to bind nematode mt or bovine mt . The deacylation protection assay was performed in with various EF-Tu mutants using nematode mt (A, B) or bovine mt (C, D). The experiments were performed with wild-type EF-Tu2 (open square), its 3-amino acid deletion mutant (filled diamond), its 7-amino acid-deletion mutant (filled circle), bovine mt EF-Tu (open triangle), or no EF-Tu (open diamond) (A, C), or with wild-type EF-Tu2 (open square), its K429A mutant (open circle), its K433A mutant (filled diamond), its K434A mutant (filled triangle), its K438A mutant (filled square), or no EF-Tu (open diamond) (B, D). (E) the secondary structure of A.suum mt . The residues within the broken line were changed into the sequence enclosed by the solid line to serylate the tRNA more efficiently.
Structural features of tRNA that are essential for recognition by EF-Tu2
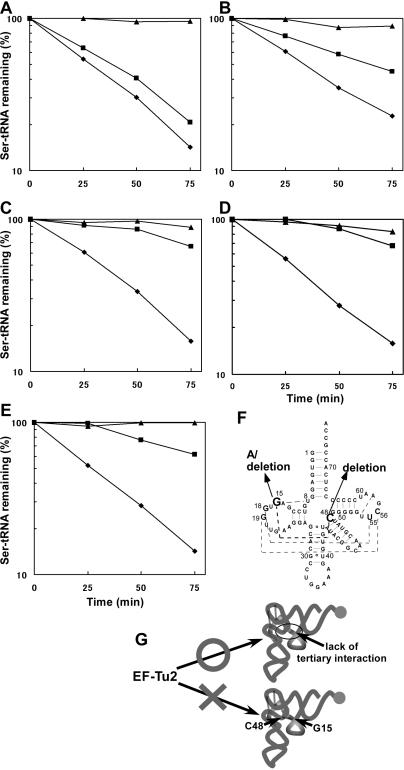
Our previous studies showed that EF-Tu2 bound to D-armless Ser-tRNAs but not to bovine mt possessing a D arm (14). Thus, we speculated that either ‘the lack of the D arm’ or ‘the region normally hidden by the D arm’ may be an essential structural feature of the tRNA that is recognized by EF-Tu2. (Also possibly contributing to the inability of EF-Tu2 to recognize bovine mt is its unusual secondary structure (15,16)). In this study, we confirmed that EF-Tu2 cannot bind to D arm-bearing Ser-tRNA by using native E.coli as a standard cloverleaf-structured tRNA (Figure 2A). However, unmodified E.coli (Figure 2F) was weakly recognized by EF-Tu2 (Figure 2B), implying that EF-Tu2 does not recognize ‘the lack of the D arm’ (i.e. the D arm-replacement loop). Therefore, ‘the region normally hidden by the D arm’ of tRNA is most likely a unique recognition site for EF-Tu2. To test this, we prepared E.coli mutants lacking the tertiary interaction between positions 15 and 48, which is near the unique binding positions of EF-Tu2 (positions C49 and C50, as shown in Figure 1). This tertiary interaction is known to be conserved among most tRNAs. The G15- and C48-deletion mutants of the tRNA bound strongly to EF-Tu2 (Figure 2C and D, respectively) despite possessing a D arm. The E.coli mutant whose G15 residue was replaced with A was also strongly recognized by EF-Tu2 (Figure 2E). These results support the notion that EF-Tu2 does not recognize ‘the lack of the D arm’. Rather, since EF-Tu2 binds to tRNA mutants lacking the 15–48 interaction, it seems to recognize a region that is normally hidden by this interaction. We also tested the G17C and G18A mutants of and found that they were also recognized by EF-Tu2 (data not shown). Each of these mutations destroys one of the conserved tertiary interactions between the D and T arms, suggesting EF-Tu2 can interact with the unique region around positions 49 and 50 when the interaction between the D and T arms (or the T arm-neighbouring nucleoside [position 48]) is weakened.
Figure 2.
Binding of EF-Tu2 to E.coli and its mutants. (A–E), deacylation-protection assay using wild-type tRNASer (A), transcribed tRNASer (B), the G15-deletion mutant (C), the C48-deletion mutant (D), and the G15A mutant (E). These assays were performed in the presence of C.elegans mt EF-Tu2 (square) or E.coli EF-Tu (triangle) and in the absence of either EF-Tu (diamond). In this assay, the protection of Ser-tRNA from hydrolysis reflects its EF-Tu-binding ability (20). (F), the secondary structure of E.coli . Arrows indicate the substitutions and deletions employed in this study. Broken lines indicate tertiary interactions. (G), schematic representation of the tertiary structures of the Ser-tRNAs that are and are not recognized by EF-Tu2. Broken lines indicate tertiary interactions. Filled circles show the aminoacyl-moieties of the Ser-tRNAs.
Role played by the C-terminal extension of EF-Tu2 in tRNA recognition
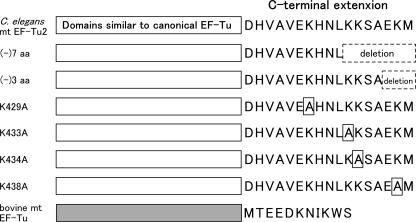
The unique aa-tRNA recognition features of EF-Tu2 shown in Figure 1 and 2 could be attributed to the C-terminal extension of EF-Tu2. If this is the case, any deletion of this C-terminal extension would impair the aa-tRNA-binding activity of EF-Tu2. To test this, we performed the hydrolysis protection assay to assess the aa-tRNA-binding activities of EF-Tu2 deletion mutants (Figure 3) using A.suum mt or bovine mt (Figure 4A, C). The acceptor stem of A.suum mt was mutated to facilitate its serylation (Figure 4E). This mutation did not affect interaction with EF-Tu2 because wild-type EF-Tu2 bound to the mutated Ser-tRNA as well as the Ser-tRNA with the original sequence as described in (14). EF-Tu2 lacking seven amino acids[(-)7 aa] did not bind to either Ser-tRNA, unlike the mutant lacking three amino acids [(-)3 aa]. This indicates that the seven amino acids of the C-terminus are important for the binding of EF-Tu2 to Ser-tRNA.
Figure 3.
Schematic depiction of the EF-Tu2 mutants. The C.elegans mt EF-Tu2 and bovine mt EF-Tu domains are depicted by white and gray boxes, respectively. The amino acid sequence of the C-terminal extensions is shown. Deletions and mutations are indicated by broken and solid squares, respectively.
To identify the residues in the C-terminal extension of EF-Tu2 that are important for its aa-tRNA-binding activity, we examined the aa-tRNA-binding activities of the K429A, K433A, K434A and K438A mutants of EF-Tu2 (Figure 3). In these mutants, each of the four lysine residues in the C-terminal extension was mutated to alanine. This is because it seems that basic residues are the most important RNA-binding sites. The K429A and K434A mutants bound to the Ser-tRNA with similar efficiency as wild-type EF-Tu2, but the K433A and K438A mutants bound more weakly (Figure 4B and D). Thus, K433 and K438 appear to be important in the aa-tRNA recognition of EF-Tu2.
We found that all EF-Tu2 mutants bound to C.elegans EF-Ts (data not shown). This suggests that the mutants have a tertiary structure similar to the wild-type EF-Tu protein apart from the mutated or deleted region(s).
DISCUSSION
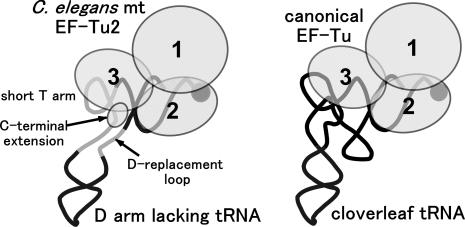
Based on our observations, the mechanism by which C.elegans mt EF-Tu2 recognizes Ser-tRNA can be summarized as follows (Figure 5). (i) EF-Tu2 interacts not only with the side of the acceptor and T stems that are also recognized by the canonical EF-Tu molecules, but it also interacts with the opposite side of the T stem (the 3′-phosphate of positions 49 and 50), which canonical EF-Tu molecules do not contact, (ii) while ‘the lack of a D arm’ on the tRNA is not necessary for EF-Tu2 binding, if the tRNA has a D arm, the T arm (or its 5′-side nucleoside at position 48) and the D arm should have only a weak tertiary interaction, (iii) at least seven amino acid residues of the C-terminal extension of EF-Tu2 are essential for its aa-tRNA recognition ability. In particular, the C-terminal extension residues K433 and K438 are likely to be involved in this recognition event.
Figure 5.
Model of the ternary complex of EF-Tu2 and D-armless Ser-tRNA. The bacterial ternary complex (3,4) (right); our model of the nematode mt ternary complex containing EF-Tu2 and D-armless Ser-tRNASer that bears a short T arm (left). GTP is omitted. EF-Tu is shown as a set of circles, each of which corresponds to the domain numbered in order. The tRNAs are portrayed as simplified backbones with the aminoacyl moiety at the 3′-terminus depicted by filled circles. The location of the C-terminal extension in EF-Tu2 is predicted from the results shown in Figure 1 and 2.
Why EF-Tu2 binds weakly only to wild-type E.coli Ser-tRNASer while strongly recognizing its G15 deletion, C48 deletion and G15A substitution mutants (Figure 2) can be explained as follows. With the wild-type E.coli Ser-tRNASer molecule, EF-Tu2 has trouble reaching the 3′-phosphate of positions 49 and 50 because of the D arm–T arm interaction. In particular, the interaction between positions 15 and 48 in the tRNA seems to prevent EF-Tu2 from binding. When the D arm–T arm interaction is weakened by mutation, this permits the unique binding sites to reach positions 49 and 50. This also explains the potent binding of EF-Tu2 to the D-armless Ser-tRNAs such as bovine mt and A.suum mt Ser-tRNAsSer, as the T stem of these tRNAs is free from such impeding tertiary interactions (26,27).
We initially speculated that EF-Tu2 may either recognize the D arm-replacement loop or a certain region that is normally masked by the D arm. The present study supports the latter case since we have observed that Ser-tRNAs bearing a D arm (but whose interaction with the T arm is weak) can actually bind to EF-Tu2 (Figure 2C–E).
The binding of EF-Tu2 to the tRNA acceptor stem was not apparent from the modification interference assay (Figure 1) because it is difficult to analyze the effects of modifying the 5′- and 3′-regions of the tRNA by this experiment, as has been described in (22,24). However, EF-Tu2 seems to bind to the acceptor stem in a similar manner to that of the canonical EF-Tu, as determined by co-crystallographic analyses of canonical EF-Tu/GTP/aa-tRNA (3,4), since the residues of EF-Tu2 and canonical EF-Tu that interact with the acceptor stem are highly conserved (14). Bacterial EF-Tu and EF-Tu2 are ∼70% identical with regard to the residues that interact with the acceptor stem, and the basic residues (K52, R59, K90 and R300 in T.aquaticus EF-Tu) that are known to interact directly with the tRNA backbone (3,4) are almost identical.
Most of the T stem-interacting residues on canonical EF-Tu molecules are not conserved in nematode mt EF-Tu2 (14) as domain 3 of T.thermophilus EF-Tu, which is known to contain the residues that interact with the T arm (3), and is only ∼30% identical to the equivalent domain in nematode mt EF-Tu2. This low homology may be responsible for the different ENU interference patterns around the T stem exhibited by EF-Tu2 and the canonical EF-Tu (Figure 1).
It seems that there is no mechanism that allows EF-Tu2 to recognize tRNA in a nucleotide specific manner. Among the sequences of tRNAsSer that could bind to EF-Tu2, there are no conserved residues except for U33 and A37 (Supplementary Figure 3). It is evident that EF-Tu2 cannot interact with the anticodon loop because the anticodon loop is too far from the acceptor and T stems with which canonical EF-Tu interacts. Thus, it appears that EF-Tu2 recognizes tRNA only in a structure specific manner, not in a nucleotide specific manner.
The unique aa-tRNA recognition mechanism of EF-Tu2 is probably due to its 17 amino acid C-terminal extension, which is one of the characteristic features of nematode mt EF-Tu2. It is unlikely that domains 1–2 of EF-Tu2, which are similar to the canonical EF-Tu, interact with positions 49 and 50 of the tRNA. Rather, the C-terminal extension is probably responsible for interacting with those positions. Supporting this is that the crystal structure of bovine mt EF-Tu has shown its C-terminal 11 amino acid extension has a helical structure, and it has been suggested that this extension may interact with the tRNA (at position 47 on the tRNA) (18). Although we found ENU-modification of this position and its vicinity did not interfere with the binding of bovine mt EF-Tu (Figure 1C), it is still possible that the extension interacts with position 47 or its vicinity in such a manner that is unaffected by ethylation of the phosphate group. Further supporting the notion that the C-terminal extension of nematode EF-Tu2 is probably responsible for interacting with positions 49 and 50 of the tRNA is that it is longer than the bovine mt EF-Tu extension, which suggests that it is more capable of reaching the tRNA. Moreover, secondary structure prediction programs (28,29) indicate that the C-terminal extension of EF-Tu2 is also an α-helix, which suggests that the EF-Tu2 C-terminus may point in the same direction towards the tRNA as the C-terminus of bovine mt EF-Tu observed in its crystal structure (18). Finally, our results using the deletion mutants of EF-Tu2 (Figure 4) strongly indicates that its C-terminal seven residues are essential for its recognition of aa-tRNA. Thus, it is highly probable that the C-terminus of EF-Tu2 binds to a unique region that cannot be bound by normal EF-Tu molecules. Notably, since many EF-1α have a C-terminal extension (relative to bacterial EF-Tu sequences), including several lysine residues, the C-terminal residues of eEF-1α may also interact with tRNA, as has been described previously by Andersen et al. (18).
Since modification of the 3′-phosphates of positions 49 and 65 in tRNA interfered with EF-Tu2 binding (Figure 1D), it appears that EF-Tu2 holds the root of the T stem, probably by using domain 3 and its C-terminal extension. This unique binding mechanism may be the result of co-evolution between nematode mt EF-Tu2 and tRNASer that compensates for the short T arm and lack of D arm of this tRNA. It appears that the binding of nematode mt EF-Tu1 to T-armless aa-tRNA follow a similar mechanism as EF-Tu2, as we have found EF-Tu1 needs its C-terminal 57 amino acid extension for recognizing T-armless tRNA (13) (Sakurai et al., in preparation). These findings suggest mt RNA-binding proteins encoded by the nuclear genome may become enlarged to compensate for the truncation of their mt genome-encoded cognate RNA species; this ultimately aids the function of mt RNAs in the mitochondrial biosynthesis system. Further studies of truncated tRNAs may be valuable in supporting the RNA world hypothesis, which needs evidence showing that the structure and function of RNAs can be substituted during evolution by proteins.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr Y. Watanabe (Graduate School of Medicine, University of Tokyo), Dr T. Suzuki (Graduate School of Engineering, University of Tokyo), and Prof. M. Sisido (Dept. of Bioscience and Biotechnology, Okayama University) for valuable discussions. We are also grateful to Prof. M. Sisido for facilities. This work was supported by Grants-in-Aid from the Ministry of Culture, Sports, Science and Technology, Japan to K. W. and T. O., and by the Kurata Memorial Hitachi Science and Technology Foundation to T.O. Funding to pay the Open Access publication charges for this article was provided by MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaziro Y. The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta. 1978;505:95–127. doi: 10.1016/0304-4173(78)90009-5. [DOI] [PubMed] [Google Scholar]
- 2.Sprinzl M. Elongation factor Tu; a regulatory GTPase with an integrated effector. Trends. Biochem. Sci. 1994;19:245–250. doi: 10.1016/0968-0004(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 3.Nissen P., Kjeldgaard M., Thirup S., Polekhina G., Reshetnikova L., Clark B.F., Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 4.Nissen P., Thirup S., Kjeldgaard M., Nyborg J. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure. 1999;7:143–156. doi: 10.1016/s0969-2126(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 5.Clark B.F.C., Kjeldgaard M., Barciszewski J., Sprinzl M. Recognition of aminoacyl-tRNAs by protein elongation factors. In: Soll D., RajBahndary U.L., editors. tRNA: Structure, Biosynthesis and Function. Washington, D.C.: American Society for Microbiology; 1995. pp. 423–442. [Google Scholar]
- 6.Forchhammer K., Leinfelder W., Bock A. Identification of a novel translation factor necessary for the incorporation of selenocystein into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 7.Kromayer M., Wilting R., Tormay P., Bock A. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J. Mol. Biol. 1996;262:413–420. doi: 10.1006/jmbi.1996.0525. [DOI] [PubMed] [Google Scholar]
- 8.Leibundgut M., Frick C., Thanbichler M., Bock A., Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sprinzl M., Vassilenko K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okimoto R., Macfarlane J.L., Clary D.O., Wolstenholme D.R. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics. 1992;130:471–498. doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe Y., Tsurui H., Ueda T., Furushima R., Takamiya S., Kita K., Nishikawa K., Watanabe K. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D Stem. J. Biol. Chem. 1994;269:22902–22906. [PubMed] [Google Scholar]
- 12.Keddie E.M., Higazi T., Unnasch T.R. The mitochondrial genome of Onchocerca volvulus: sequence, structure and phylogenetic analysis. Mol. Biochem. Parasitol. 1998;95:111–127. doi: 10.1016/s0166-6851(98)00102-9. [DOI] [PubMed] [Google Scholar]
- 13.Ohtsuki T., Watanabe Y., Takemoto C., Kawai G., Ueda T., Kita K., Kojima S., Kaziro Y., Nyborg J., Watanabe K. An ‘elongated’ translation elongation factor Tu for truncated tRNAs in nematode mitochondria. J. Biol. Chem. 2001;276:21571–21577. doi: 10.1074/jbc.M011118200. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsuki T., Sato A., Watanabe Y., Watanabe K. A unique serine-specific elongation factor Tu found in nematode mitochondria. Nature Struct. Biol. 2002;9:669–673. doi: 10.1038/nsb826. [DOI] [PubMed] [Google Scholar]
- 15.Yokogawa T., Watanabe Y., Kumazawa Y., Ueda T., Hirao I., Miura K., Watanabe K. A novel cloverleaf structure found in mammalian mitochondrial tRNASer (UCN) Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe Y., Kawai G., Yokogawa T., Hayashi N., Kumazawa Y., Ueda T., Nishikawa K., Hirao I., Miura K., Watanabe K. Higher-order structure of bovine mitochondrial tRNA(SerUGA): chemical modification and computer modeling. Nucleic Acids Res. 1994;22:5378–5384. doi: 10.1093/nar/22.24.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woriax V.L., Burkhart W., Spremulli L.L. Cloning, sequence, analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim. Biophys. Acta. 1995;1264:347–356. doi: 10.1016/0167-4781(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 18.Andersen G.R., Thirup S., Spremulli L.L., Nyborg J. High resolution crystal structure of bovine mitochondrial Ef-Tu in complex with GDP. J. Mol. Biol. 2000;297:421–436. doi: 10.1006/jmbi.2000.3564. [DOI] [PubMed] [Google Scholar]
- 19.Shimada N., Suzuki T., Watanabe K. Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial Seryl-tRNA synthetase. J. Biol. Chem. 2001;276:46770–46778. doi: 10.1074/jbc.M105150200. [DOI] [PubMed] [Google Scholar]
- 20.Pingoud A., Urbanke C., Krauss G., Peters F., Maass G. Ternary complex formation between elongation factor Tu, GTP and aminoacyl-tRNA: an equilibrium study. Eur. J. Biochem. 1977;78:403–409. doi: 10.1111/j.1432-1033.1977.tb11752.x. [DOI] [PubMed] [Google Scholar]
- 21.Silberklang M., Gillum A.M., RajBhandary U.L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977;4:4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vlassov V.V., Giege R., Ebel J.P. Tertiary structure of tRNAs in solution monitored by phosphodiester modification with ethylnitrosourea. Eur. J. Biochem. 1981;119:51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- 23.Riehl N., Giege R., Ebel J.P., Ehresmann B. Effect of elongation factor Tu on the conformation of phenylalanyl-tRNAPhe. FEBS Lett. 1983;154:42–46. doi: 10.1016/0014-5793(83)80871-0. [DOI] [PubMed] [Google Scholar]
- 24.Shi P., Maizel N., Weiner A.M. CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J. 1998;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke P. RNA footprinting and modification interference analysis. In: Haynes S., editor. RNA-Protein Interaction Protocols. Vol. 118. Totowa,N.J: Humana Press Inc.; 1999. pp. 73–91. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi I., Yokogawa T., Kawai G., Ueda T., Nishikawa K., Watanabe K. Assignment of imino proton signals of G-C base pairs and magnesium ion binding: an NMR study of bovine mitochondrial lacking the entire D arm. J. Biochem. 1997;121:1115–1122. doi: 10.1093/oxfordjournals.jbchem.a021703. [DOI] [PubMed] [Google Scholar]
- 27.Ohtsuki T., Kawai G., Watanabe K. The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNASerUCU having a short T arm and lacking the entire D arm. FEBS Lett. 2002;514:37–43. doi: 10.1016/s0014-5793(02)02328-1. [DOI] [PubMed] [Google Scholar]
- 28.Chou P.Y., Fasman G.D. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 29.Garnier J., Osguthorpe D.J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 30.Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17:1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.