Abstract
Drosophila telomeres have been maintained by retrotransposition for at least 60 MY, which predates the separation of extant species of this genus. Studies of D. melanogaster, D. yakuba, and D. virilis show that, in Drosophila, telomeres are composed of two non-LTR retrotransposons, HeT-A and TART. Far from being static, HeT-A and TART evolve faster than Drosophila euchromatic genes. In spite of their high rate of sequence change, HeT-A and TART maintain their basic structures and unusual individual features. The maintenance of their separate identities suggests that HeT-A and TART cooperate either in the process of retrotransposition onto the chromosome end, or in the formation of telomere chromatin by transposed DNA copies. The telomeric retrotransposons and the Drosophila genome constitute an example of a robust symbiotic relationship between mobile elements and the genome.
Drosophila telomeres are maintained by two non-LTR retrotransposons
Early studies by Muller on Drosophila (Muller and Herskowitz, 1954) and McClintock on maize (McClintock, 1942) concluded that telomeres are special structures that cap chromosome ends, protecting them from end-to-end fusions and distinguishing true chromosome ends from chromosome breaks. More recently, studies on many organisms have implicated telomeres in several cellular processes, such as the regulation of cell proliferation and oncogenesis. Although the mechanisms are not understood, it is clear that regulation of telomere length has multiple consequences for the cell and that cells invest significant resources in telomere maintenance (McEachern et al., 2000).
In most eukaryotes, the maintenance of the telomeres is done by a multi-subunit ribonucleoprotein, the enzyme telomerase. Telomerase is able to extend the sequence of the telomere by reverse transcribing a segment of its internal RNA template onto the end of the chromosome. The sequence of the telomerase template varies with species but is generally five to ten base pairs long and A+C-rich. Repeated additions of the sequence result in long arrays of telomeric repeats (Fig. 1A). Paradoxically, Drosophila, the organism in which telomeres were defined, does not completely follow this picture. Telomeres in all Drosophila species studied, and probably the entire genus, are composed of tandem arrays of repeated sequences formed by successive transpositions of two non-LTR retrotransposons, HeT-A and TART (Pardue and DeBaryshe, 2003). Thus, the Drosophila repeats are six to fourteen kilobases long, rather than five to ten base pairs long (Fig. 1B). Like telomerase repeats, the retrotransposons are reverse transcribed onto the end of the chromosome, thus forming a polar array with the 3′ ends of all elements oriented toward the centromere. In D. melanogaster and D. yakuba HeT-A and TART appear to be randomly intermixed in telomeric arrays (Pardue and DeBaryshe, 2002; Casacuberta and Pardue, 2002). Preliminary studies of D. virilis suggest that this species may have some bias in the chromosomal distribution of the two elements (Casacuberta and Pardue, 2003b).
Figure 1.
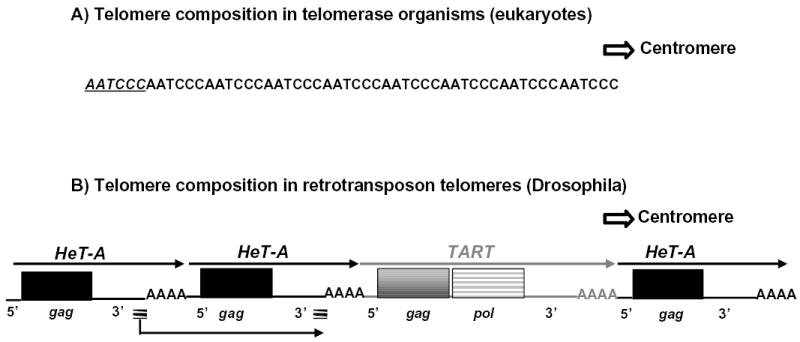
Schematic representation of telomere structure in A) Organisms with telomerase. The strand with the RNA template sequence, AATCCC, is shown. A single repetitive unit of the mammalian telomere sequence is shown in italics and underlined. B) Drosophila. The coding strand (equivalent to template strand) is shown. Centromere lies to the right in both cases.
Thin arrows above retrotransposon diagrams indicate 5′? 3″ direction of each element. Black arrow underneath the HeT-A element indicates that its transcription unit includes sequence from two adjacent elements (see discussion of HeT-A’s unusual promoter),. Striped short lines underneath HeT-A elements indicate sequences in the transcription unit that are duplicated because they derive from the 3′ end of two elements. 5′, and 3′ indicate untranslated regions, AAA, indicates oligo (A) adenine characteristic of non-LTR retrotransposons, gag, gag gene, pol, pol gene.
Although Drosophila telomeres might seem drastically different from telomerase telomeres, the two mechanisms are basically very similar. In both cases, an enzyme reverse transcribes its RNA template directly onto chromosomal DNA and elongates the telomere by addition of DNA repeats. Although the size of the repeats in the Drosophila telomere arrays is much greater than that of telomerase-generated repeats, the total amount of DNA on each Drosophila telomere is about the same as that of multicellular eukaryotes with telomerase (Pardue and DeBaryshe, 2002).
HeT-A and TART are clear examples of transposable elements completely dedicated to a cellular role. The symbiosis established between these retrotransposons and the Drosophila genome recalls McClintock’s idea that transposable elements have regulatory roles in the organism. It is interesting that McClintock first detected mobile DNA when she induced chromosome breaks to study the importance of telomeres. She suggested that the mobilization of the sequences was a cellular response to stress.
HeT-A AND TART: THE TWO TELOMERIC RETROTRANSPOSONS THAT HAVE CONSTITUTED DROSOPHILA TELOMERES FOR MORE THAN 60 MY.
HeT-A and TART share unusual features but also have significant differences
Although they are clearly non-LTR retrotransposons, HeT-A and TART share some features not seen in other elements of this class; they are the only Drosophila non-LTR retrotransposons specifically targeted to telomeres, they contain unusually long 3′UTR sequences, and they have closely related Gag proteins (see next section). These unusual characteristics are probably related to their role in telomere maintenance. However, in spite of their shared role at telomeres, HeT-A and TART differ in ways that suggest that they have different ancestries (Pardue and DeBaryshe, 2003).
TART has the structure of a conventional non-LTR element. It encodes both a Gag protein and a Pol protein with an endonuclease (E) domain and a reverse transcriptase (RT) domain (Fig 2). Apparently, the TART element contains all the features required for being an autonomous retroelement. The sequences of the TART proteins group into the Jockey clade of insect non-LTR elements (Malik et al., 1999). Surprisingly, HeT-A encodes only a Gag protein and therefore cannot supply its own enzymatic activities for transposition. In spite of this, HeT-A transposes efficiently and therefore must obtain the necessary activities from some other source. This lack of a pol gene is one of the defining features of HeT-A, seen in all species studied. The HeT-A Gag protein sequence is closely related to that of TART and also belongs to the Jockey clade (Casacuberta and Pardue, 2003b).
Figure 2.
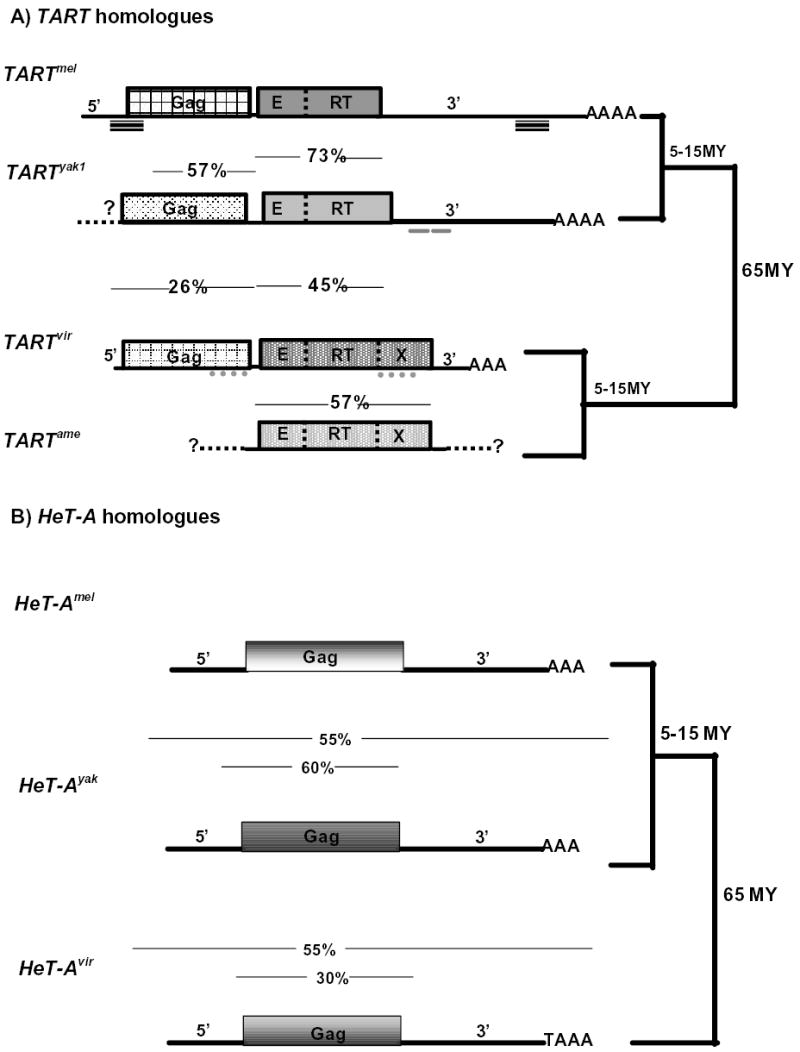
Diagrams of A) TART and B) HeT-A homologues in D.melanogaster, D. yakuba, D. virilis and D. americana. Drawn approximately to scale. Solid bars on the side indicate the phylogenic relationships. MY, million years. %, percentage of nucleotide identity of sequence between arrowheads: in A) 57% (Gag), 73% (Pol) mel to yak; 26% (Gag), 45% (Pol) vir to either mel or yak, 57% (Pol) vir to ame, in B) 55% (total, 60% (Gag) mel to yak; 55% (total), 30% (Gag) vir to either mel or yak. Striped lines under TARTmel indicate position of Perfect Non Terminal Repeats (PNTR). Small lines under TARTyak1 3′UTR indicate direct repeats. Dotted lines under TARTvir genes indicate concentration of poly-glutamine repeats. E, endo, RT, reverse transcriptase, X, extradomain of pol coding region. (Multiple subfamilies of TART have been found in D. melanogaster and D. yakuba. TARTmel represents a consensus of D. melanogaster subfamilies. TARTyak1 represents one subfamily of TARTyak elements.)
Another unusual aspect of HeT-A is the location of its promoter (Fig 1B), which is found at the extreme end of the 3′UTR and oriented so that each element drives transcription of the element immediately downstream (Danilevskaya et al., 1997). This mechanism is effective since HeT-A elements are found in tandem head-to-tail arrays and thus the promoter in one element drives transcription of its neighboring element. Because the 3′ promoter is identical to the 3′UTR of the element transcribed, the HeT-A transcription unit is analogous to the transcription unit of an LTR retrotransposon. Thus, the unusual HeT-A promoter resembles a possible evolutionary intermediate between a non-LTR and a LTR retrotransposon (Fig. 1B). TART has a very different promoter structure and transcription patterns. We believe that TART, like many other non-LTR elements, has a promoter in the 5′UTR that drives transcription of sense strand RNA. TART has a second promoter located in the 3′UTR but, in contrast to the HeT-A promoter, the TART promoter is oriented so that it drives antisense transcription of the element that contains it. The TART antisense promoter is very active. Antisense TART RNA has been detected in all the Drosophila species studied. In D. melanogaster, D. virilis, and D. americana these antisense transcripts are much more abundant than sense strand RNA (Danilevsakaya et al.,1999; Casacuberta and Pardue, 2003a), while in D.yakuba one subfamily, TARTyak1 yields approximately equal amounts of sense and antisense RNA (Casacuberta and Pardue, 2002). The function of the antisense RNA is not clear but its evolutionary conservation suggests it is important for the element, perhaps for transposition or some aspect of its regulation. In contrast, no antisense transcripts of HeT-A elements have been detected in any species.
The sequence at the 3′UTR in HeT-A, has a distinctive repeat pattern of A-rich regions. The exact sequence of the A-rich regions is not conserved but the spacing is and this produces a pattern of off-diagonal repeats in dot matrix comparisons of all HeT-A homologues. Although TART elements do not show this pattern of repeats in the 3′UTR, in all Drosophila species both HeT-A and TART have a strong A+C bias in their coding strand (Danilevskaya et al., 1998). A similar bias is seen in the telomerase RNA, which is equivalent to the sense strand of these retrotransposons.
Both HeT-A and TART are maintained in all Drosophila species
Both HeT-A and TART have been found in all Drosophila species, stocks, and cell lines studied (Pardue and DeBaryshe, 2003). This persistent association for at least 60 MY suggests that each element has individual characteristics that are needed for telomere maintenance and/or function. It would seem possible that a simple recombination between the two elements could produce a compound element competent for all functions and thus capable of replacing both HeT-A and TART. This has not been observed. Nevertheless, it has been shown that recombination between telomere arrays has a role in the maintenance of Drosophila telomeres (Melnikova and Georgiev, 2002; Siriaco et al., 2002)) and thus a mechanism for generating such recombinants exists. However, the only recombinant element we have found in any telomere array, Uvir, does not appear to be a significant component of telomeres (Casacuberta and Pardue, 2003b).
The recombinant element, Uvir, was isolated in a cloned segment of D. virilis telomeric DNA and named because it is previously unidentified, apparently unique, and highly unexpected element. Uvir appears to be a non-LTR element with a single coding sequence and a long 3′UTR. The sequence of the 5′UTR and the last approximately 1 kb of the 3′UTR are highly similar to HeT-Avir but the rest of the 3′UTR is novel sequence. The HeT-A gag gene has been replaced by an open and apparently complete pol gene followed by a segment of novel 3′UTR. The high level of identity with the HeT-A 5′ and 3′ UTR sequences suggests that this element is a chimera between HeT-Avir and an unknown pol coding sequence, from either another retrotransposon or a euchromatic gene. This element appears to be single copy and therefore does not transpose readily in the D.virilis genome. Although Uvir does not appear to be a significant component of D.virilis telomeres, it demonstrates that recombinant elements can be formed with telomere sequences.
IN SPITE OF HIGHLY DIVERGENT SEQUENCES, HeT-A AND TART HOMOLOGUES EACH SHOW STRONG CONSERVATION OF UNUSUAL FEATURES.
Conservation of the basic sequence organization has some exceptions
As just discussed, HeT-A elements from all species are characterized by a single coding region and a long 3′UTR with a pattern of A-rich regions, while TART homologues have both gag and pol coding regions (Fig 2). The sequences of these regions show many changes but are easily recognized because of the many conserved structural features of these elements. There are, however, some differences between TART in D. virilis and D. americana and their homologues in other species that should be noted.
The Pol protein of TART elements from D. virilis and D. americana contains an extra domain, X, located after the RT, causing these Pol proteins to extend ~ 400 amino acids beyond the C-terminus of the proteins from D. melanogaster and D. yakuba (Casacuberta and Pardue, 2003a). The X domain shows considerable amino acid conservation between TARTvir and TARTame suggesting that this extra domain is under functional constraints. Other retrotransposons in the Jockey clade contain only E and RT domains. Although the I factor retrotransposon has an RNase H domain at the C-terminus of its pol gene (Malik et al., 1999), we have seen no similarity between the X domain and the I factor, nor have we found hints of an alternative function for the X domain.
A second structural difference between TARTvir and its homologues in D. melanogaster and D. yakuba is in the 3′UTR. In those species, TART, like HeT-A, is characterized by a very long 3′UTR. Surprisingly this feature is not conserved in the TARTvir homologues. The unexpectedly short 3′UTRs of TARTvir imply that another distinct feature of TARTmel is not conserved in the virilis group, the presence of Perfect Non Terminal Repeats (PNTR). PNTR is a sequence from the 5′ UTR of TARTmel that is precisely repeated near, but not at, the 3′ end of the element(Pardue and DeBaryshe, 2002). Comparison of PNTR from different TART subfamilies within the D. melanogaster genome shows concerted evolution within subfamilies but significant sequence change between them. This concerted evolution may reflect a highly recombinogenic sequence. These conclusions apply only to TARTmel because all TARTyak elements cloned to date are all 5′-truncated into the gag coding region and it is not possible to determine if they contain PNTR sequences.
PNTR are also found in two other non-LTR retrotransposons, Tre5-A of Dictyostelium (Schumann et al., 1994) and TOC1 of Chlamydomonas (Day et al.,1988). These two elements have another characteristic in common with TARTmel elements, the antisense transcripts, suggesting that these elements identify a subclass of non-LTR retrotransposons. The relationship between PNTR and antisense transcripts is unclear. It has been proposed for Tre5-A, that both strands of RNA are necessary for the replication of the element. Another possibility is that PNTR are involved in self-regulation of the element through dsRNA that might be a target for the RNAi machinery.
The junctions between TART elements in the cloned D.virilis arrays are nearly identical, suggesting that the elements are complete and have not undergone the variable 5′ truncation frequently seen with non-LTR retrotransposons, yet we see no evidence of PNTR sequences (Casacuberta and Pardue, 2003a). This seems to indicate that whatever the function of the PNTR for the TARTmel elements, the TARTvir elements have evolved a different solution. One possible alternative solution for elements in tandem arrays, as TART sometimes is, would be for the 3′ end of one TART element to act as the PNTR for the adjacent downstream element. Such cooperation between neighbors would be similar to the HeT-A promoter.
The study of different TART homologues from these evolutionarily distant species shows that some traits can be acquired or lost in different genomes. Nevertheless, their basic structure and their specific and unusual genomic location unmistakably identify them as TART homologues.
HeT-A and TART have a high rate of sequence change but conserved phylogeny
Since HeT-A and TART have cellular roles, they must interact with, and probably be regulated by, cellular components. This might imply that their rate of evolution should be more similar to euchromatic genes than to faster changing retrotransposon or repetitive sequences. In order to address this question we compared the genes from HeT-A and TART homologues, with several euchromaticgenes representing equivalent genetic distances. For comparison we also included the genes from another non-LTR retrotransposon, the R1 element. Almost complete gag and RT sequences from R1 homologues in D. melanogaster, D. mercatorum and D. hydei, were available. R1 is a non-LTR retrotransposon that inserts in ribosomal RNA genes of organisms throughout insects (Burke et al., 1993). R1 does not belong to the Jockey clade, the phylogenic clade of HeT-A and TART; nevertheless, it is the best comparison available. The analyses (Table 1) show that, for both the gag and pol genes of HeT-A and TART, identity percentages are lower, and Ks and Ka values are higher, than for any of the euchromatic genes. These two results suggest that TART and HeT-A are evolving faster than euchromatic genes. In the case of R1, the gag gene also shows a high rate of change but the pol gene falls into the same range as the histone H1 and the rough gene, in agreement with the results of Eickbush et al. (1995).
Table 1. Comparisons of sequence identity and synonymous and nonsynonymous substitutions.
Nt, % of nucleotide identity; AA, % of amino acid identity. Values in parenthesis were calculated omitting the residues that fall in gaps. Values in brackets correspond to the MHR and zinc knuckle region only. Ks, synonymous substitutions, Ka, replacement substitutions (± SE). TARTvir gag and pol Ac: AY219708, AY219709, TARTame pol Ac: AY219710, R1 hyd Ac: U23196 R1mel Ac: P16424. R1mer Ac: AAB94026, D.virilis histone H1 Ac: L76558, D.melanogaster histone H1 Ac: X04073, D.hydei histone H3 Ac: X52576, D.melanogaster histone H3 Ac: X14215. D.melanogaster rough Ac: AAA56800, D.virilis rough Ac: M35372, D.melanogaster Adh Ac: X98338, D.virilis Adh Ac: AB033640. HeT-Amel Gag Ac: AAC17188, HeT-Ayak gag Ac: AAC01742. virilis, mercatorum, and hydei are all approximately equidistant from melanogaster. Table modified from Casacuberta and Pardue (2003 a and b).
| Nt (%) | AA (%) | Ks | Ka | |
|---|---|---|---|---|
| TART gag | ||||
| mel/vir | 26 (37) [47] | 13 (18) [34.8] | 1.79 ± 0.36 | 1.09 ± 0.2 |
| TART pol | ||||
| mel-vir | 45 (52) | 36 (45) | 1.8 ± 0.64 | 0.57 ± 0.17 |
| mel-ame | 44 (50) | 34 (38) | 1.65 ± 0.37 | 0.61 ± 0.03 |
| ame/vir | 57 | 54 | 1.5 ± 0.19 | 0.37 ± 0.02 |
| HeT-A gag | ||||
| mel-yak | 58 (62) [68] | 54 (57) [75.3] | 1.06 ± 0.07 | 0.31 ± 0.016 |
| mel-vir | 30 (30.3) [45] | 16 (18) [35.8] | 1.96 ± 0.49 | 1.27 ± 0.301 |
| yak-vir | 29 (29.5) [44] | 14.2 (17) [36] | 1.96 ± 0.71 | 1.27 ± 0.390 |
| R1 gag | ||||
| mel-mer | 39 (42) | 29 (31) | 1.68 ± 0.26 | 0.89 ± 0.056 |
| R1 pol | ||||
| mel/hyd | 60 | 59 | 1.3 ± 0.16 | 0.32 ± 0.03 |
| mel/mer | 58 | 56.7 | 1.4 ± 0.17 | 0.34 ± 0.02 |
| hyd/mer | 85 | 82.5 | 0.33 ± 0.04 | 0.09 ± 0.01 |
| Rough | ||||
| mel-vir | 55 (65.6) | 55.4 (66) | 1.20 ± 0.18 | 0.27 ± 0.024 |
| histone H1 | ||||
| mel-vir | 61.4 (65.4) | 62 (66) | 1.41 ± 0.21 | 0.27 ± 0.026 |
| histone H3 | ||||
| mel-hyd | 76.7 | 97 | ------ | ------ |
| Adh | ||||
| mel/vir | 75 | 79 | 1.04 ± 0.25 | 0.12 ± 0.03 |
As expected from retroviral sequences, the gag genes show the highest rate of change. This is especially well demonstrated by the elevated number of replacement substitutions, Ka. It has been shown that the average Ks/Ka is between 4–20 for Drosophila nuclear genes (Akashi, 1994). For the gag genes in the table, all Ks/Ka are under 2. Note that in spite of accepting a high number of amino acid replacements, the number of conservative substitutions, Ks, is still higher than Ka, and therefore no positive selection seems to be acting if the protein is considered as a whole. In analyzing the gag gene as a single unit, we are assuming that the whole gene is changing uniformly; however, there are indications that this is not true, at least for the telomeric retrotransposons. The identity values obtained for only the MHR and zinc knuckle regions of the proteins (brackets) show a higher rate of conservation. This domain of the Gag protein is important for the interactions between the telomeric Gag proteins (Rashkova et al., 2003), and thus it might be expected to change more slowly. It would be necessary to analyze smaller regions of the gag gene in order to determine which regions are evolving faster and whether there is positive selection within smaller domains.
The nucleotide sequence of the telomeric retrotransposons constitutes the actual telomeric repeats; this fact influences their composition and is reflected by the adenine-cytosine bias seen in the coding strand of all HeT-A and TART homologues. This particular feature of telomeric sequences might impose constraints on the evolution of HeT-A and TART genes. Nucleotide identities are almost twice as high as amino acid identities for the telomeric gag genes. The TART pol gene shows a similar trend although the differences are not as great. These differences could be related to the sequence bias of the telomere region. Finally, the two D.virilis homologues, HeT-Avir and TARTvir share a surprising characteristic not found in their homologues from D.melanogaster or D.yakuba. The genes of TARTvir and HeT-Avir contain a high content of CAX repeats in their sequences (Casacuberta and Pardue, 2003a,b). These repeats encode long stretches of glutamines with some interspersed histidines. The glutamine repeats tend to accumulate at the carboxy-termini of the proteins and do not interrupt any of the known functional domains. Long stretches of glutamine residues have a tendency to interact with each other and it has been suggested that they might constitute interactive domains (Perutz et al., 1994). It could be that the glutamine repeats in D.virilis HeT-A and TART constitute the interactive domains that in other species are made of different interacting residues.
Besides analyzing the rate of sequence change, we explored the phylogenetic relationships among TART and HeT-A homologues together with other non-LTR elements of the Jockey clade (Fig. 3). In order to analyze HeT-A and TART elements we used the only sequence common to both, the Gag protein. The resulting tree clearly separates the telomeric retrotransposons from the rest of the Jockey clade. The bootstrap value for this node is high (82), suggesting that the telomeric transposons constitute a subclade inside the Jockey clade. Moreover, the tree (Fig. 3A) reflects the high rate of sequence change of the gag gene. This low level of sequence conservation in the Gag protein is responsible for ungrouping the D.virilis telomeric elements from their respective homologues in the other species. Although both HeT-A and TART Gags diverge rapidly from their homologues in other species, within a species, the two proteins maintain a close relationship, possibly because of constraints related to their telomeric role.
Figure 3.
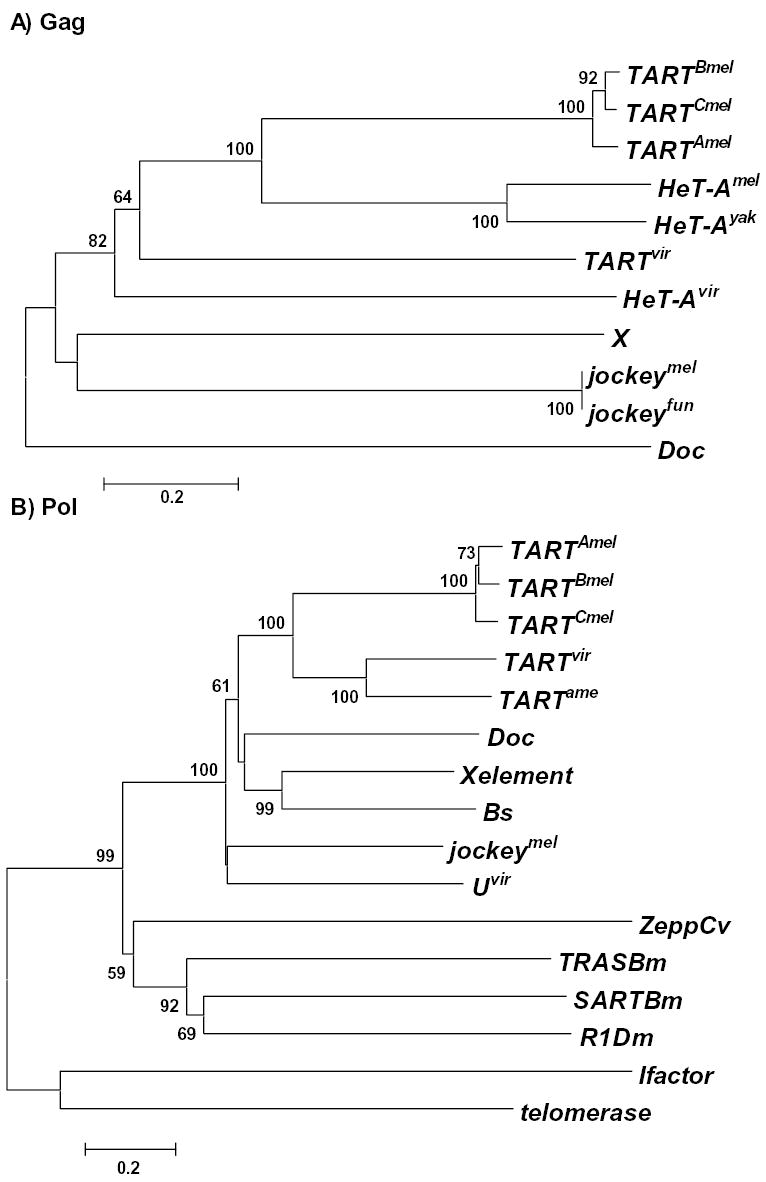
Phylogenetic relationships of Gag and Pol sequences. Neighbor-joining trees are shown. (UPGMA trees gave the same result). Bootstrap values (at corresponding nodes) calculated with 500 replications and a cutoff value of 50%. Scale bar indicates number of differences per residue. A) Gag proteins. B) Pol proteins. jockeymel Ac; M22874, jockeyfun Ac: PIR B38418, Doc Ac: CAA35587, X Ac: AF 237761, TARTBmel, Ac: U14101, TARTAmel: F. Sheen and R. Levis, TARTCmel : L. Tolar, J. Stolk, and R. Levis, TARTvir Ac: AAO67564, TARTame Ac: AAO67565, jockeymel Ac: AAA28675, SARTB.m Ac: T18196, TRASB.m Ac: T18199, ZeppC.v Ac: T00078, I factor Ac: AAA70222, R1D.m Ac: P16425, C.elegans telomerase Ac: NP_492374, Bs Ac: S55543, X element Ac: AAF81411.
THE RELATIONSHIP BETWEEN THE TELOMERIC RETROTRANSPOSONS AND THE GENOME
Do HeT-A and TART collaborate at telomeres?
With the recent evidence that both HeT-A and TART are maintained in all Drosophila stocks over 60 MY, it is puzzling that neither of the elements, nor a recombinant between the two elements, has taken over the job at the telomeres. One possible explanation is that both elements are individually needed for telomere maintenance and/or structure. Support for this suggestion comes from studies on D. melanogaster showing that Gag proteins from HeT-A and TART have the potential for cooperating in transposing to telomeres (Pardue and DeBaryshe, 2003).
The D. melanogaster studies have shown HeT-A Gag protein is very efficiently targeted to telomeres in interphase nuclei of mitotic cells. TART Gag protein does not associate with telomeres unless co-expressed with HeT-A Gag (Rashkova et al., 2002b). Thus, HeT-A Gag has the potential to deliver both HeT-A and TART transposition intermediates to their targets on the chromosome. TART, on the other hand, encodes a transposition enzymatic activity, which HeT-A lacks. If HeT-A provides telomere targeting and TART provides RT activity, transposition may only be possible if the two elements cooperate.
The evidence that HeT-Avir and TARTvir Gag proteins are as close to each other as to the Gag proteins of any of their homologues in the other species suggests that the two elements are evolving under similar constraints (Casacuberta and Pardue, 2003b), as would be expected if the elements are cooperating. Among those constraints must be the necessity to interact with the same components of the host cell and possibly with each other. Studies of D. melanogaster have shown that Gag proteins from HeT-A and TART are treated very differently in Drosophila cells than are Gags from other members of the Jockey clade (Rashkova et al., 2002a). Proteins from the telomeric elements are efficiently transported into the nucleus while most, if not all, of the proteins from the other elements remain in the cytoplasm. This shows that proteins from telomere elements have different interactions with cellular components than do proteins from presumably parasitic elements. Studies with deletion derivatives of the Gag proteins have identified specific regions of these proteins involved in different aspects of the intracellular localization and for HeT-A-TART interactions (Rashkova et al., 2003).
The identification of HeT-A and TART in D. virilis and D. yakuba will now allow us to determine what changes in their Gag proteins have been necessary to function in cells of their endogenous species and what parts of the protein maintain function in other species, in spite of sequence change.
HeT-A and TART appear to have converged on their telomeric roles from different origins
The close relationship of their Gag proteins and their telomeric targeting might suggest that HeT-A and TART have diverged from a common ancestor. However, the conserved differences in these elements, such as promoter placement, pattern of transcription (George and Pardue, 2003), and structure of the 3′UTR, lead us to believe that the elements have different origins but have converged on their roles at the telomere by acquiring related Gag proteins. This convergence must have occurred sometime before the separation of the extant Drosophila species because the two elements appear to be well-established at telomeres in all Drosophila.
Some insects maintain their telomeres with telomerase and thus Drosophila must have shared ancestors with them (Sahara et al., 1999). How did Drosophila come to have retrotransposon telomeres? Did the telomeric retrotransposons derive from components of the telomerase machinery or were they typical retrotransposons already present in the genome that somehow acquired telomere specificity? We speculate that HeT-A derived from telomerase while TART was present in the genome before it became targeted specifically to telomeres. TART has the two coding regions that typify many retrotransposons. It can also have the PNTR and dual promoters found in some other non-LTR retrotransposons. There are five non-LTR retrotransposons, SART and TRAS elements in Bombyx mori (Okazaki et al., 1995; Takahashi et al., 1997), GilT and GilM of Girardia lambia (Arkhipova and Morrison, 2001) and Zepp from Chlorella vulgaris (Higashiyama et al., 1997) that have been shown to have special affinity for telomeric repeats in species that have telomerase. For one of these non-LTR elements, TRAS, the telomeric sequence specificity has been shown to be due to the endonuclease domain, (Kubo et al., 2001). Therefore, we have analyzed the phylogenetic relationships of the Pol proteins from this particular group of non-LTR retrotransposons. The resulting tree (Fig. 3B) clearly demonstrates that the above mentioned non-LTR elements with specificity for telomerase repeats group together and apart from the TART elements. All the sequences from elements of the Jockey clade, including TART, group together with the maximum bootstrap value (100). The lack of relationship suggests that telomeric targeting of TART is not closely related to the targeting of the elements from species with telomerase. As explained above, we believe that the telomeric targeting for TART is due to the acquisition of a Gag protein related to HeT-A Gag protein.
HeT-A is more different from other retrotransposons than is TART. We have suggested that this element could have been derived from two of the transcription units that encode components of the telomerase complex (Pardue and DeBaryshe 2003). The best studied telomerase, that from Saccharomyces cerevisiae, has at least four components, the catalytic subunit, the RNA template, and two proteins needed for in vivo, but not in vitro, activity (Lendvay et al., 1996; Lingner et al., 1997). It seems reasonable to assume that one of the proteins necessary for in vivo activity acts in targeting the RNA template to the telomere (a “proto-Gag”). If the sequence coding for this protein were to become linked to the gene for the RNA template, perhaps by a chromosomal rearrangement, a transcript of this chimeric gene would, like HeT-A, have a Gag coding region joined to a long 3′ UTR, derived from the telomerase RNA template, which is never translated to protein. Translation of the proto-Gag coding region of this joint transcript would yield a protein capable of taking the RNA to telomeres where the 3′ UTR would fulfill its customary role in initiating reverse transcription.
The scenario proposed for the origin of HeT-A would leave HeT-A dependent on a catalytic subunit encoded elsewhere in the genome. This source might have been taken over by TART, which offers a multicopy source of RT. Still, the fact that HeT-A and TART are maintained as separate elements across more than 60 MY indicates that there is more to the story than this. Are HeT-A and TART Gag proteins different and both required for transposition? Does one or both play a role in telomere function? Does the presence of the RT coding sequence in TART prevent it from making perfect telomere chromatin? Are both 3′ UTR sequences required in telomere chromatin? Many important questions remain.
PERSPECTIVES
Our studies of different Drosophila species have shown both conserved and variable aspects of the telomeric retrotransposons. It is now necessary to understand the basis of both the conservation and the variability. It is known that telomeres and centromeres are rapidly evolving chromosome regions with highly species-specific sequences. The understanding of these sequences will clarify the insect history as well as retrotransposon and telomere evolution.
This work shows that the retrotransposon mechanism of telomere maintenance probably predates the separation of the Drosophila genus. We think it likely that other insects that lack telomerase may use a retrotransposon-based mechanism to maintain their chromosome ends. It will be very interesting to extend the study of non-telomerase telomere maintenance beyond the Drosophila genus.
Acknowledgments
We thank Ky Lowenhaupt, and the members of the Pardue laboratory for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM50315.
References
- Akashi, H Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova IR, Morrison HG. Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc Nat Acad Sci, USA. 2001;98:14497–502. doi: 10.1073/pnas.231494798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke WD, Eickbush DG, Xiong Y, Jakubczak J, Eickbush TH. Sequence relationship of retrotransposable elements R1 and R2 within and between divergent insect species. Mol Biol Evol. 1993;10:163–85. doi: 10.1093/oxfordjournals.molbev.a039990. [DOI] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L. Coevolution of the telomeric retrotransposons across Drsosophila species. Genetics. 2002;161:1113–1124. doi: 10.1093/genetics/161.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L. Transpososn telomeres are widely distributed in the Drosophila genus: TART elements in the virilis group. Proc Nat Acd Sci, USA. 2003;100:3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, Pardue M-L. HeT-A elements in D.virilis: Retrotransposon telomeres are conserved across the Drosophila genus. Proc Nat Acd Sci, USA (In press). [DOI] [PMC free article] [PubMed]
- Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue M-L. Promoting in tandem: the promoter of telomere transposon HeT-A and the consequences for the evolution of retroviral LTRs. Cell. 1997;86:647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- Danilevskaya ON, Lowenhaupt K, Pardue ML. Conserved subfamilies of the Drosophila HeT-A telomere-specific retrotransposon. Genetics. 1998;148:233–42. doi: 10.1093/genetics/148.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Traverse KL, Hogan NC, DeBaryshe PG, Pardue ML. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol Cell Biol. 1999;19:873–81. doi: 10.1128/mcb.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Schirmer-Rahire M, Kuchka MR, Mayfield SP, Rochaix JD. A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J. 1988;7:1917–27. doi: 10.1002/j.1460-2075.1988.tb03029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush, DG, Lathe IIIWC, Francino MP, Eickbush TH. R1 and R2 retrotransposons elements of Drosophila evolve at rates similar to those of nuclear genes. Genetics. 1995;139:685–695. doi: 10.1093/genetics/139.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JA, Pardue ML. The promoter of the heterochromatic Drosophila telomeric retrotransposon, HeT-A, is active when moved into euchromatic locations. Genetics. 2003;163:625–635. doi: 10.1093/genetics/163.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T, Noutoshi Y, Fujie M, Yamada T. Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J. 1997;16:3715–23. doi: 10.1093/emboj/16.12.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Okazaki S, Anzai T, Fujiwara H. Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in Lepidopteran insects. Mol Biol Evol. 2001;18:848–57. doi: 10.1093/oxfordjournals.molbev.a003866. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;27:6561–7. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- McClintock B. The fusion of ends of chromosome following nuclear fusion. Proc Nat Acd Sci, USA. 1942;28:458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure MA, Johnson MS, Feng D-F, Doolittle RF. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Nat Acd Sci, USA. 1998;85:2469–2463. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Ann Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- Melnikova L, Georgiev P. Enhancer of terminal gene conversion, a new mutation in Drosophila melanogaster that induces telomere elongation by gene conversion. Genetics. 2002;162:1301–1312. doi: 10.1093/genetics/162.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ, Herskowitz IH. Concerning the healing of chromosome ends produced by breakage in Drosophila melanogaster. Genetics. 1954;88:177–208. [Google Scholar]
- Okazaki S, Ishikawa H, Fujiwara H. Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori. Mol Cell Biol. 1995;15:4545–52. doi: 10.1128/mcb.15.8.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M-L, DeBaryshe PG. Telomeres and transposable elements. In: Mobile DNA II. (Craig N, Craigie R, Gellert M, Lambowitz A, eds.) American Society for Microbiology, Washington D.C. pp. 870–887. (2002).
- Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionary robust non-telomerase mechanism to maintain telomeres. Ann Rev Gen in press (2003). [DOI] [PubMed]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc Nat Acd Sci, USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Marec F, Traut W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999;7:449–460. doi: 10.1023/a:1009297729547. [DOI] [PubMed] [Google Scholar]
- Schumann G, Zundorf I, Hofmann J, Marschalek R, Dingermann T. Internally located and oppositely oriented polymerase II promoters direct convergent transcription of a LINE-like retroelement, the Dictyostelium repetitive element, from Dictyostelium discoideum. Mol Cell Biol. 1994;14:3074–84. doi: 10.1128/mcb.14.5.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriaco GM, Cenci G, Haoudi A, Champion LE, Zhou C, Gatti M, Mason JM. Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics. 2002;160:235–45. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Okazaki S, Fujiwara H. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 1997;25:1578–84. doi: 10.1093/nar/25.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]


